Summary
Group A Streptococcus (GAS, Streptococcus pyogenes) is a human-restricted pathogen with a capacity to both colonize asymptomatically and cause illnesses ranging from pharyngitis to necrotizing fasciitis. An understanding of how and when GAS switches between genetic programs governing these different lifestyles has remained an enduring mystery and likely requires carefully tuned environmental sensors to activate and silence genetic schemes when appropriate. Herein, we describe the relationship between the Control of Virulence (CovRS, CsrRS) two-component system and the Rgg2/3 quorum-sensing pathway. We demonstrate that responses of CovRS to the stress signals Mg2+ and a fragment of the antimicrobial peptide LL-37 result in modulated activity of pheromone signaling of the Rgg2/3 pathway through a means of proteolysis of SHP peptide pheromones. This degradation is mediated by the cytoplasmic endopeptidase PepO, which is the first identified enzymatic silencer of an RRNPP-type quorum-sensing pathway. These results suggest that under conditions in which the virulence potential of GAS is elevated (i.e. enhanced virulence gene expression), cellular responses mediated by the Rgg2/3 pathway are abrogated and allow individuals to escape from group behavior. These results also indicate that Rgg2/3 signaling is instead functional during non-virulent GAS lifestyles.
Keywords: quorum quenching, protease, carriage, stress response, LL-37
Introduction
Group A Streptococcus (GAS, Streptococcus pyogenes) is an obligate human pathogen that presents clinically as the cause of a variety of diseases, ranging from uncomplicated superficial manifestations (pharyngitis) to serious, invasive infections of soft tissues and blood (necrotizing fasciitis and sepsis) (Cunningham, 2000; Cunningham, 2008). Post-infection sequelae, such as acute rheumatic fever or glomerulonephritis, may follow superficial infections (Marijon et al., 2012; DeMuri and Wald, 2014). Relatively high levels of asymptomatic GAS carriage among the general population, primarily in children, have led to the postulate that the pathogen alternates between states of carriage and disease (recently reviewed in Wollein Waldetoft and Råberg, 2014). The switch is thought to be the consequence of changes in microbial gene expression in response to as-of-yet poorly characterized environmental conditions, host immune responses, and likely other factors contributing to stress and resource limitation.
Gene regulatory networks that coordinate behaviors across bacterial populations provide benefits to a microbial community by sharing responsibilities to conserve or compete for limited resources, or to defend against host anti-microbial assaults (Kreikemeyer et al., 2003; Jimenez and Federle, 2014). One mechanism by which GAS coordinates gene expression at the population level is through intercellular chemical signaling, commonly referred to as quorum sensing (QS). To date, four general types of QS regulatory circuits have been described in GAS: Sil, lantibiotic-related, LuxS/AI-2, and the Rgg paralogs (reviewed in Jimenez and Federle, 2014). Among these systems, the Rgg pathways are conserved among all sequenced isolates and recent studies indicate roles for a pathway in which Rgg2 and Rgg3 (Rgg2/3) contribute to the regulation of biofilm development and lysozyme resistance (Chang et al., 2011, Chang et al., 2015). Both Rgg2 and Rgg3 are transcription factors whose activities are controlled through the direct binding of peptide pheromones (SHP2 and SHP3) that are produced by GAS and other streptococcal species (Cook et al., 2013; Lasarre et al., 2013; LaSarre et al., 2013). SHP pheromones are ribosomally produced, linear peptides that are exported and processed by at least one proteolytic enzyme (Eep) prior to becoming mature signals capable of diffusion and interaction with nearby cells. The predominant form of mature SHP peptides consist of the eight C-terminal amino acids (SHP-C8) of shp coding sequences, but longer variants including nine and ten residues are also produced (Aggarwal et al., 2014). The mature pheromones must be imported to the cytoplasm via the oligopeptide permease (Opp) in order to engage the Rgg receptors (Chang et al., 2011, Aggarwal et al., 2014). As pheromones accumulate and reach a sufficient concentration, they bind to Rgg receptors, presumably promoting conformational changes in them. Our studies indicate that SHP2-C8 (DILIIVGG) and SHP3-C8 (DIIIIVGG) are each capable of engaging either Rgg2 or Rgg3 with nearly equivalent binding affinities and activation potentials (Aggarwal et al., 2014). Given their seemingly redundant activities, it remains unknown what advantage retaining both shp2 and shp3 genes in all GAS genomes provides. However, production of each peptide in culture does not appear to be equivalent, and their disparity is likely controlled both at transcriptional and post-transcriptional levels (Lasarre et al., 2013, Chang et. al, 2015). It is also reasonable to assume that peptide signals are subject to degradation and thus their net concentration must account for production and turnover.
The effect by which SHP pheromones exert their regulatory role in GAS is realized in two ways. At pheromone concentrations below a level favoring an interaction with Rgg receptors, Rgg3 predominates as a negative transcriptional regulator, suppressing transcription from the shp promoters (Pshp2 and Pshp3) (Chang et al., 2011). When pheromones accumulate to levels that result in receptor binding, they cause Rgg3 to release DNA and as a consequence de-repress transcription of target promoters. Concurrently, SHP-bound Rgg2 serves as a transcriptional activator and causes a strong increase in Pshp expression. Thus, the net effect of SHP binding to Rgg2 and Rgg3 is robust induction of transcription of the operons encoding each shp, and this induction is sustained by a resulting positive-feedback regulatory loop made possible through further pheromone production.
GAS also contains several regulatory circuits that sense a variety of stress conditions present in the host and responds by regulating the expression of factors that enhance survival. Among the best-characterized regulatory systems is the CovRS (CsrRS) two-component signal-transduction pathway. In this circuit, the membrane-localized kinase, CovS, acts as an environmental sensor. In response to changing external stimuli, such as high salt, low pH, or exposure to the antimicrobial peptide LL-37, CovS is able alter CovR phosphorylation (Levin and Wessels, 1998; Federle et al., 1999; Gryllos et al., 2003; Gryllos et al., 2007; Gryllos et al., 2008; Horstmann et al., 2014; Velarde et al., 2014, Dalton and Scott, 2004). Following phosphorylation, DNA binding of the transcriptional regulator CovR is enhanced, affecting expression of 15–20% of GAS genes, including direct repression of the has hyaluronate-capsule synthesis operon, the IL-8 protease spyCEP (prtS, scpA), the DNAse sda1, and many other genes considered virulence factors. Importantly, many invasive strains of GAS are found to have mutations in covR, leading to unrestrained expression of the CovRS regulon. These isolates have been the subject of extensive research as they appear to be ‘locked’ in a virulent phenotype and are implicated in the vast majority of highly invasive infections (Engleberg et al., 2001; Graham et al., 2002; Sumby et al., 2006; Cole et al., 2011).
Here we demonstrate that signaling by the Rgg2/3 QS pathway is deficient under conditions that promote CovRS-dependent virulence induction. Using a forward genetic screen, we identify PepO to be a CovRS-regulated aminopeptidase and demonstrate its ability to degrade mature SHP pheromones and limit quorum-sensing responses. We also find that PepO preferentially degrades SHP2 over SHP3, providing further evidence that production and stability of the SHP peptides differ in GAS, despite having nearly equivalent activation potentials and binding affinities for their receptors (LaSarre et al., 2013; Aggarwal et al., 2014). Together, these findings suggest that QS is inhibited during physiological states that correlate with acute infections and that the Rgg2/3 pathway may instead be relevant during asymptomatic colonization of the host.
Experimental Procedures
Bacterial strains and culture media
Bacterial strains used in these studies are described in Table 1. S. pyogenes are regularly grown in Todd Hewitt medium (BD Bioscience) supplemented with 0.2% (w/v) yeast extract (Amresco). Plates were made with the addition of 1.4% agar. All luminescence assays were conducted in chemically defined medium (CDM) with 1% (w/v) glucose or 1% mannose and 0.05% glucose (Rijn and Kessler, 1980). CDM plates were made by mixing filter-sterilized 2X CDM with an autoclaved mixture of 2.8% agar + 0.01% yeast extract. 5-bromo-4-chloro-3-indolyl β-D-glucuronide (X-gluc) was added to a final concentration of 300 μg/ml. Antibiotics were used at the following concentrations: chloramphenicol, 2 μg/ml; erythromycin, 0.5 μg/ml; kanamycin, 100 μg/ml; spectinomycin, 100 μg/ml. Escherichia coli strain BH10C and 5-α(New England Bioscience) were used for cloning, and the E. coli strain C41 (DE3) was used for protein expression. Strains were grown in Luria-Bertani broth (BD Bioscience) supplemented with antibiotics as indicated: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml; erythromycin, 500 μg/ml; kanamycin, 100 μg/ml; spectinomycin, 100 μg/ml.
Table 1.
Strains and plasmids used in this study
| Strain/plasmid | Description | Reference |
|---|---|---|
| S. pyogenes | ||
| NZ131 | Wild-type M49 S. pyogenes isolate | (Simon and Ferretti, 1991; McShan et al., 2008) |
| Δrgg3 shp3-luxAB | NZ131 Δrgg3::cat with luxAB inserted downstream of shp3; CmR | This study |
| Δrgg3 shp3-gus | NZ131 Δrgg3::cat with gus inserted downstream of shp3; CmR | This study |
| Δrgg3 shp3-gus ΔcovR | NZ131 Δrgg3::cat ΔcovR with gus inserted downstream of shp3; CmR | This study |
| ΔcovR | NZ131 with unmarked deletion of covR | This study |
| Δrgg3ΔcovR | NZ131 Δrgg3::cat ΔcovR; CmR | This study |
| Δrgg3ΔcovRΔspyCEP | NZ131, Δrgg3::cat ΔspyCEP::aphA3 ΔcovR; CmR KanR | This study |
| Δrgg3ΔcovRΔpepO | NZ131 Δrgg3::cat ΔpepO::aphA3 ΔcovR; CmR KanR | This study |
| ΔpepO | NZ131 ΔpepO::aphA3; KanR | This study |
| shp2GGGshp3GGG | NZ131 Start codons of each shp gene mutated to GGG | (Lasarre et al., 2013) |
| ΔpepOshp2GGGshp3GGG | NZ131 ΔpepO::aphA3. Start codons of each shp gene mutated to GGG; KanR | This study |
| ΔoppDshp2GGGshp3GGG | NZ131 with unmarked deletion of oppD. Start codons of each shp gene mutated to GGG; Unmarked | This study |
| ΔoppDΔpepOshp2GGGshp3GGG | NZ131 with unmarked deletion of oppD. Start codons of each shp gene mutated to GGG, ΔpepO::aphA3; KanR | This study |
| MGAS5005 ΔpepO | MGAS5005 ΔpepO::aphA3; CmR | This study |
| HSC5 ΔpepO | HSC5 ΔpepO::aphA3; KanR | This study |
| E. coli | ||
| BH10c | Cloning strain | (Howell-Adams and Seifert, 2000) |
| C41 (DE3) | Protein overexpression strain | (Miroux and Walker, 1996) |
| Plasmids | ||
| pCovRKO | Fragments upstream and downstream of covR cloned into pFED760; ErmR | This study |
| pET21a | Cloning/expression vector with IPTG-inducible T7 promoter and N-terminal HIS-6 | Novagen |
| pFED760 | Shuttle vector pGH9-ISS1 with ISS1 element deleted; temperature-sensitive; ErmR | (Mashburn-Warren et al., 2010) |
| pGH9-ISS1 | Shuttle vector bearing ISS1 insertion sequence; temperature-sensitive; ErmR | (Maguin et al., 1996) |
| pJC191 | Fragment encompassing oppD fused by PCR to make unmakared oppD deletion | (Chang et al., 2011) |
| pJC227 | Fragments upstream and downstream of shp3, with Vibrio fischeri luxAB genes inserted directly downstream of shp3; cloned into pFED760; ErmR | This study |
| pJC258 | Fragment encompassing spyCEP, with spyCEP replaced by aphA3; cloned into pFED760; ErmR KanR | This study |
| pJC273 | Fragment encompassing Δrgg3::cat and shp3, with gus inserted directly downstream of shp3; cloned into pFED760; ErmR CmR | This study |
| pKS310 | Source of luxABCDE fragment | Lynn Hancock Lab, unpublished |
| pLZ12-Sp | Shuttle vector; SpecR | (Husmann et al., 1995) |
| pOsKaR | Source of aphA3 fragment | (Le Breton and McIver, 2013) |
| pRVW14 | shp3 promoter fused to luxABCDE, cloned into p7INT; ErmR | This study |
| pRVW19 | Fragment encompassing pepO, with pepO replaced by aphA3; cloned into pFED760; ErmR KanR | This study |
| pRVW21 | recA promoter region fused to pepO, cloned into pLZ12-Spec; SpecR | This study |
| pRVW23 | pepO cloned into pET21a expression plasmid; AmpR | This study |
| pRVW24 | pepO promoter region fused to luxABCDE, cloned into p7INT; ErmR | This study |
| pRVW25 | Fragment encompassing pepO, with pepO replaced by cat; cloned into pFED760; ErmR, CmR | This study |
| pRVW27 | spyCEP promoter fused to luxABCDE, cloned into p7INT; ErmR | This study |
Transposon screen for mutants defective in Rgg-SHP signaling
Transposon mutagenesis was performed in NZ131 Δrgg3 reporter strains containing either luciferase (luxAB) or β-glucuronidase (gus) genes inserted in the genome directly downstream of shp3; this genetic background was chosen because the deletion of the Rgg3 repressor results in constitutive Rgg-SHP signaling. Reporter strains were constructed using the method described below and pJC227 (luxAB) or pJC273 (gus). Mutagenesis was conducted using the protocol for highly transformable GAS (Le Breton and McIver, 2013). Briefly, the transposon-donating plasmid, pOsKar, was electroporated into reporter strains, and cells were allowed to grow at the permissive temperature (30° C) for four hours before being plated on selective THY agar at 37° C to select for transposition events. Individual colonies were subsequently patched on CDM agar + 0.2% mannose + 0.5% yeast extract + kan. To detect luxAB expression, colonies were exposed to the luxAB substrate, decyl aldehyde, luminescence was monitored via a cooled CCD camera (Syngene). To detect gus activity, X-gluc was added to the CDM agar and white or light blue colonies were selected for further validation. The site of transposon insertion in each clone was identified using an arbitrary PCR method as described (Le Breton and McIver, 2013). Briefly, genomic DNA prepared from each transposon mutant, subjected to a PCR with a transposon-specific primer and a degenerate primer, a subsequent PCR with nested primers to amplify target DNA then sequenced with a nested transposon-specific primer. Sequence reads were compared to the published NZ131 genome.
Construction of mutant strains
Mutants described in these studies were derived from NZ131, a serotype M49 strain (Simon and Ferretti, 1991; McShan et al., 2008). To delete pepO, 1 kbp upstream of pepO were amplified with RW36 and RW37 and downstream 1 kbp were amplified with RW38 and RW49 (Table 2). The aphA3 cassette was amplified with JC397 and JC304. The three fragments were assembled using the restriction sites indicated in the primers then amplified using the outside primers RW36 and RW49. The resulting fragment was cloned into the EcoRI and SalI sites of pFED760 to create pRVW19. pRVW19 was electroporated into NZ131 and a two-step temperature-dependent process was used to replace pepO with aphA3 (Degnan et al., 2000). In brief, NZ131 cells containing the deletion vector and grown at the permissive temperature (30°C) were shifted to the non-permissive temperature (37°C) for 2–3 hours then plated on selective media at 37°C. After confirming of the presence of the aphA3 cassette in clones that grew at the restrictive temperature, bacteria were passaged a minimum of 6 times at permissive temperatures in the absence of antibiotics to allow the plasmid to recombine and excise from the genome. Loss of vector antibiotic resistance was used to identify mutants. Mutants were confirmed by PCR using primers RW62 and RW63. Deletion of pepO in MGAS5005 followed a similar strategy using plasmid pRVW25. Deletion of covR was accomplished in a similar fashion. For the former, primers LW2-1, LW2-2, LW2-3, and LW2-4 were used to amplify upstream and downstream flanking regions, which were then ligated and cloned into pFED760 to make pCovRKO; following the temperature-dependent process described above, colonies were screened for a mucoid phenotype, and the deletion was confirmed by PCR. Deletion of spyCEP also used this temperature dependent process and knockout plasmid pJC258, in which an aphA3 cassette was inserted between upstream and flanking downstream sequences.
Table 2.
Primers used in this study
| Plasmid | Primer | Sequence | Description |
|---|---|---|---|
| pCovRKO | LW 2-1 | GCGTGGCGGCCGCGAGAATATCTAGCTGATGCTA | Sense primer, covR upstream flanking region; NotI |
| LW 2-2 | GCGTGGGATCCTTATACCAACCCTTATCCTC | Antisense primer, covR upstream flanking region; BamHI | |
| LW 2-3 | GCGTGGGATCCTAAGTCATATGGAAAATCAG | Sense primer, covR downstream flanking region; BamHI | |
| LW 2-4 | GCGTGGGTACCCTTCTTGTTTGCCAGTCACTGAAAG | Antisense primer, covR downstream flanking region; KpnI | |
| pJC227 | JC195 | CATGGCGGCCGCAGAAGCTCAAGAGATGACC | Sense primer, shp3 upstream flanking region; NotI |
| JC284 | CATGAGATCTTTACCCACCAACAATAATG | Antisense primer, shp3 upstream flanking region; BglII | |
| JC273 | CATGACGCGTTACTTAGGTGTATACCTAAG | Sense primer, shp3 downstream flanking region; MluI | |
| Spy0533_S2 | GCGTGGCGGCCGCGAAAAAAAATATT | Antisense primer, shp3 downstream flanking region; NotI | |
| JC285 | CATGAGATCTATTAATCACCAAAAAG | Sense primer for luxA; BglII | |
| JC275 | CATGACGCGTTTATGGTAAATTCATTTCGAT | Antisense primer for luxB; MluI | |
| pJC258 | JC256 | CATGGCGGCCGCGGTTATATTGCTAGTGGTGC | Sense primer; spyCEP upstream flanking region; NotI |
| JC259 | CATGGCGGCCGCAGGCATCAAAGCATACTCG | Antisense primer; spyCEP downstream flanking region; NotI | |
| JC344 | CATGTTAATTAACCTGACGAGTACCAGTGC | Antisense primer; spyCEP upstream flanking region; PacI | |
| JC345 | CATGTTAATTAAGCAGACTGGCTTGTCACT | Sense primer; spyCEP downstream flanking region; PacI | |
| JC320 | CATGTTAATTAACGATACTATGTTATACGC | Sense primer for aphA3 cassette; PacI | |
| JC321 | CATGTTAATTAAAGCGAACTTTTAGAAAAG | Antisense primer for aphA3 cassette; PacI | |
| pJC273 | JC369 | CATGGCGGCCGCTGCTGGTATAGAGATTTCTG | Sense primer, shp3 upstream flanking region; NotI |
| JC284 | See above | ||
| JC342 | CATGTTAATTAAGTTACTTAGGTGTATACCTAAG | Sense primer, shp3 downstream flanking region; PacI | |
| Spy0533_S2 | See above | ||
| JC340 | CATGAGATCTAGAAGGAGGAAAAATATGTTA | Sense primer for gus; BglII | |
| JC343 | CATGTTAATTAATCATTGTTTGCCTCCCTG | Antisense primer for gus; PacI | |
| pRVW14 | JC147 | CATGGAATTCGTGTCGGAAAGTAAACATGC | Sense primer for shp3 promoter; EcoRI |
| RW13 | GTATGTAAGCAAAAAGTTTCCAAATTTCATAAGTGGACTTCCTTTCAG | Antisense primer for shp3 promoter; luxA overlap underlined | |
| JC106 | ATGAAATTTGGAAACTTTTTGCTTACATAC | Sense primer for luxA | |
| RW14 | CATGGGATCCACTAGTGACTATCAAACGCTTCGG | Antisense primer for luxE; BamHI | |
| pRVW19 | RW36 | ATCGGAATTCTCAGAAATGGACTATTATGGCTTT | Sense primer, pepO upstream flanking region; EcoRI |
| RW37 | ATCGTTAATTAATCGTATCTCCTTATATCATTTTAAAT | Antisense primer, pepO upstream flanking region; PacI | |
| JC397 | GCATGTTAATTAAATGGCTAAAATGAGAATATCACC | Sense primer for aphA3; PacI | |
| JC304 | GCATGACGCGTCCCTCCTTGTATTTTCAATAAT | Antisense primer for aphA3; MluI | |
| RW38 | ATCGACGCGTTTAGGTAGTTGATAAACCTTCAAATGC | Sense primer, pepO downstream flanking region; MluI | |
| RW49 | CTAGGTCGACCGTTGCTGCTGTTGTTGA | Antisense primer, pepO downstream flanking region; SalI | |
| pRVW21 | RW44 | ATCGAGATCTAATATGGTCCGTAAAACTTTATTAAAACTTG | Sense primer for recA promoter; BglII |
| RW41 | TTGATAAGTTGTCATTACTTTCTCCTCTTTTTTAACAAACCATCG | Sense primer for pepO; recA promoter overlap underlined | |
| RW43 | AAAGAGGAGAAAGTAATGACAACTTATCAAGATGATTTTTACCAG | Antisense primer for recA promoter; pepO overlap underlined | |
| RW42 | ATCGGAATTCTTACCAAATAATCACACGGTCTTT | Antisense primer for pepO; PstI | |
| pRVW23 | RW58 | TTTAAGAAGGAGATATACAATGACAACTTATCAAGATGATTTTTAC | Sense primer for pepO. Upstream plasmid flanking region underlined |
| RW59 | CAGTGGTGGTGGTGGTGGTGCTCGAGCCAAATAATCACACGGTC | Antisense primer for pepO. Downstream plasmid flanking region underlined | |
| pRVW24 | RW55 | ATCGGAATTCCTAAAAGCCTGAGGTTAGGCGAATTC | Sense primer for pepO promoter region; EcoRI |
| RW56 | AAGTTTCCAAATTTCATTCGTATCTCCTTATATC | Antisense primer for pepO promoter region fusion to luxA | |
| RW57 | GATATAAGGAGATACGAATGAAATTTGGAAACTTTTTGCT | Sense primer for luxA. pepO promoter region overlap underlined | |
| RW14 | CATGGGATCCACTAGTGACTATCAAACGCTTCGG | Antisense primer to amplify luxE; BamHI | |
| pRVW25 | RW60 | CATGACGCGTGCGAAAAAGGAGAAGTCGGTTCAGAA | Sense primer to amplify cat; MluI |
| RW61 | CATGTTAATTAAGCTTGATGAAAATTTGTTTGATTT | Antisense primer to amplify cat; PacI | |
| pRVW27 | RW66 | CCTCTAGACTCGAGGTAAAACAGACAAAAACCTCTG | Sense primer for spyCEP promoter; flanking plasmid region underlined |
| RW67 | CAAATTTCATCTGATACCCTCCTAAATGTTTTAAC | Antisense primer for prtS promoter; flanking luxA region underlined. | |
| RW68 | GGTATCAGATGAAATTTGGAAACTTTTTGC | Sense primer for luxA. Flanking prtS promoter region in lowercase. | |
| RW69 | GTTGGGAGCTCTCCGACTATCAAACGCTTCGGTTAAG | AS primer for luxE. Flanking plasmid region underlined. | |
| Other Primers | |||
| RW62 | CAGGATGATTCGTTTATGAC | Sense primer, 1100 bp upstream pepO, for deletion verification | |
| RW63 | GATGGTGACATGGACAAAG | Antisense primer, 1100 bp downstream pepO, for deletion verification | |
| ISS1-FOUT1 | GCAAGAACCGAAGAAATGGAACG | Sense primer for ISS1 amplification, (Maguin et al., 1996) | |
| ISS1-ROUT1 | GATTGTAACGTAGATAATAACCAACAGC | Antisense primer for ISS1 amplification, (Maguin et al., 1996) | |
| ISS1-ROUT2 | AATAGTTCATTGATATATCCTCGCTGTCA | Sequencing primer for ISS1 junction site identification, (Maguin et al., 1996) |
Construction of complementation plasmid
For the complementation plasmid, pRVW21 (pLZ12-PrecA-pepO), pepO was amplified using the primers RW41 and RW42. The constitutive recA promoter was amplified from NZ131 genomic DNA using RW43 and RW44. Equimolar amounts of the PrecA fragment and the pepO fragment were mixed then amplified using the outside flanking primers to generate a PrecA-pepO fusion product, which was cloned into pLZ12-Spec using EcoRI and BamHI. The construct was verified by restriction digest and sequencing.
Construction of luxABCDE reporters
All luxABCDE (lux) reporters used in these experiments were constructed using the integrative plasmid p7INT, capable of stably integrating into the genome at the T12 phage site attB (McShan et al., 1998). The Photorhabdus luminescens lux operon–was amplified from pKS310 using JC106 and RW14. Pshp3 was amplified using JC147 and RW13. Promoter and reporter fragments were fused as described above and cloned into p7INT to make pRVW14. A similar strategy was employed to in the construction of pRVW24, the PpepO reporter. PpepO was amplified using RW55 and RW56. luxABCDE were amplified using RW57 and RW14. Resulting fusion fragments were cloned into p7INT. The constructs were verified by restriction digest and sequencing. The PspyCEP reporter, pRVW27, was constructed using Gibson Assembly (Gibson et al., 2009). PspyCEP was amplified using primers RW66 and RW67. luxABCDE were amplified with RW68 and RW69. p7INT was cut with EcoRI and BamHI, and all three fragments were assembled using Gibson Assembly master mix (Gibson et al., 2009). Constructs were verified by sequencing. Following verification of reporter plasmids, NZ131 wild type or noted mutants were transformed with indicated reporter plasmids. Integration was confirmed by selection on Erm and ability to generate luciferase.
Luminescence transcriptional reporter assay
To monitor Rgg2/3 induction as well as PpepO and PspyCEP expression via luminescent transcriptional reporter, strains were grown in THY overnight and diluted 1:100 into CDM in the morning. The resulting cultures were monitored for growth and luminescence on a Synergy 2 plate reader (Biotek). Cultures were incubated at 37° C. The optical density of visible light (OD600) and luminescence (counts per second, CPS) were measured every 10 minutes, with shaking for 30 seconds prior to each reading. Relative light units (RLU) were calculated by normalizing luminescence values to corresponding OD600 readings (CPS/OD600). Some strains did not grow well in microtiter conditions (e.g., those containing ΔcovR mutations); experiments using these strains and experiments in which mannose was provided as the primary carbon source were conducted with a larger volume (>5 ml) in glass tubes using a Spectronic 20D spectrometer (Thermo) and luminometer (Veritas). Readings were taken hourly. To collect induction kinetics data for WT, Δrgg3 and ΔpepO strains, overnight cultures were spun down, washed twice in PBS and diluted 1:100 into CDM in glass tubes. Readings were taken ever 30 minutes as described above.
Conditioned supernatant experiments
Strains were grown in CDM to an optical density (OD600) = 0.3–0.5. Culture supernatant was separated from cells by centrifugation and erythromycin was added (0.5 μg/ml final). To compare SHP activity across different GAS strains, conditioned supernatants were normalized to OD600 = 0.3. Conditioned supernatants were subsequently inoculated with indicated reporter strains with a 1:100 dilution of overnight culture and incubated at 37° C. Luminescence and growth was monitored as described above.
Synthetic peptides
Synthetic peptides were purchased from Neo-Peptide, Cambridge MA. Desiccated peptides were suspended in DMSO and stored at −20° C
Transposon screen for rescue of Rgg2/3 induction
A transposon screen using pGH9-ISS1 was conducted as previously described (Maguin et al., 1996). In brief, NZ131 Δrgg3ΔcovR with the β-glucuronidase (gus) gene inserted in the genome directly downstream of shp3 were electroporated with 5 μg of pGH9-ISS1 plasmid, and transformants were selected by growth on THY agar + erm at 30° C. A single colony was grown in THY + erm overnight, then diluted 1:100 into THY without antibiotics, incubated for 3.5 hours at 30° C and then shifted to 37° C for 2.5 hours. Dilutions were plated onto THY agar, THY agar + erm and CDM agar + erm + X-gluc and incubated overnight at 37° C. Colonies on THY plates were enumerated to calculate transposition efficiency. X-gluc plates were screened for positive transposants (indicated by blue coloration). Genomic DNA was prepared from positive clones (MasterPure Gram Positive Genomic DNA Kit, Epicentre). 5 μg of genomic DNA was digested with HindIII, circularized via ligation and transposon insertion sites were amplified via inverse PCR using primers ISS1-FOUT1 and ISS1-ROUT1 (Maguin et al., 1996). Sequencing with primer ISS1-ROUT2 identified transposon junction sites, which were mapped to the NZ131 genome.
Activity-based screening for PepO localization
Indicated strains were grown to mid-logarithmic phase (OD600 = 0.3–0.5). Cells were pelleted by centrifugation and conditioned supernatants were collected. Cells were suspended in PBS and aliquots used for whole-cell analysis. Equivalent numbers of cells were lysed by addition of phage lysin (PlyC) and DNAseI. 50 nM sSHP2-C8 was added to all samples, and samples were incubated at 30°C for 4 hours. PlyC was inactivated by incubation at 65°C for 10 minutes. sSHP activity was detected via the addition of a ΔpepO shp2GGGshp3GGG Pshp3-lux reporter strain as previously described.
Western blots
Wild-type NZ131 cultures were treated with indicated peptides and allowed to grow to mid-logarithmic phase (OD600 = 0.4–0.5). Samples were pelleted by centrifugation, washed once with PBS, and lysed by resuspension in 100 μl PBS containing Phage Lysin C (Nelson et al., 2006) and 0.5 units DNAseI followed by incubation at 37° C for 15 minutes. Total protein was determined by Bradford, and samples were normalized. 8 μg of lysate was run on 10% polyacrylamide gels then transferred to PVDF membranes. Following blocking, the membrane was incubated over night at 4° C with a 1:1000 dilution of rabbit anti-PepO raised against pneumococcal PepO (Agarwal et al., 2013). Anti-PepO was detected using goat anti-rabbit HRP secondary antibody and SuperSignal West Dura Extended Duration Substrate (Thermo). Blots were imaged on a FluorChem E imaging system (ProteinSimple), and band intensity was quantified using in-system software. PepO protein levels were normalized to a non-specific band appearing in all samples. To determine cellular localization of PepO, mid-logarithmic phase WT NZ131 and ΔpepO mutants were washed twice in lysis buffer (50 mM Tris, 5 mM MgSO4 pH=8). Pellets were resuspended in 250 μL of lysis buffer containing Phage Lysin C (PlyC) and DNAseI and incubated at 37° C for 15 minutes. Samples were spun at 21,000 x g for 1.5 hrs to separate soluble from insoluble cellular components. The resulting insoluble fraction was washed twice in lysis buffer before resolubilization in 1X SDS-PAGE sample buffer. Sample loading was normalized based on apparent protein concentration on coomassie staining, and proteins were blotted and probed as previously described.
Purification of recombinant PepO
The pepO (spy49_1735c) gene from NZ131 was amplified using the primers RW58 and RW59 and cloned into pET21a upstream of a 6-HIS tag using XhoI and NdeI and Gibson Assembly. The assembled construct was transformed into DH5α E. coli cells (NEB). The resulting plasmid, pRVW23 was verified by restriction digest and sequencing prior to transformation into E. coli C41(DE3) cells. Once cells had reached OD600 of ~0.7, expression of pepO was induced with 0.5 mM IPTG and cells were incubated at 30° C for an additional six hours. Cells were harvested and suspended in Buffer A (0.02 M phosphate buffer, 0.0054 M potassium chloride, 0.274 M sodium chloride, 20 mM imidazole, 0.07% β-mercaptoethanol), with the addition of 1X protease inhibitor cocktail (Pierce). Cells were lysed via sonication on ice and debris was removed via centrifugation at 40,000 × g for 25 minutes at 4° C. The resulting lysate was adsorbed onto a HisTrap-HP Nickel column (GE Bioscience) and washed with Buffer A. rPepO was eluted using 10% Buffer B (0.02 M phosphate buffer, 0.0054 M potassium chloride, 0.274 M sodium chloride, 500 mM imidazole, 0.07% β-mercaptoethanol). Purification was assessed by SDS-PAGE and fractions containing rPepO were pooled. A 50,000 MWCO column (Amicon) was used to concentrate rPepO, and elution buffer was exchanged for Buffer C (0.02 M phosphate buffer, 0.0054 M potassium chloride, 0.274 M sodium chloride). rPepO was concentrated to 1 mg/ml in 20% glycerol. Aliquots were flash frozen and stored at −80° C.
SHP degradation experiments
500 nM of synthetic SHP2-C8 (sSHP2-C8) or sSHP3-C8 were incubated with rPepO at indicated concentrations in reaction buffer (20 mM Tris HCL, pH 7.0) for 5 hours at 30° C, then added to an early-logarithmic phase ΔpepO shp2GGG shp3GGG (incapable of synthesizing endogenous SHPs) reporter strain at OD600 of approximately 0.1. Samples were observed for light production and growth as described above.
Results
Rgg2-Rgg3 quorum sensing activation is restricted in covR mutants
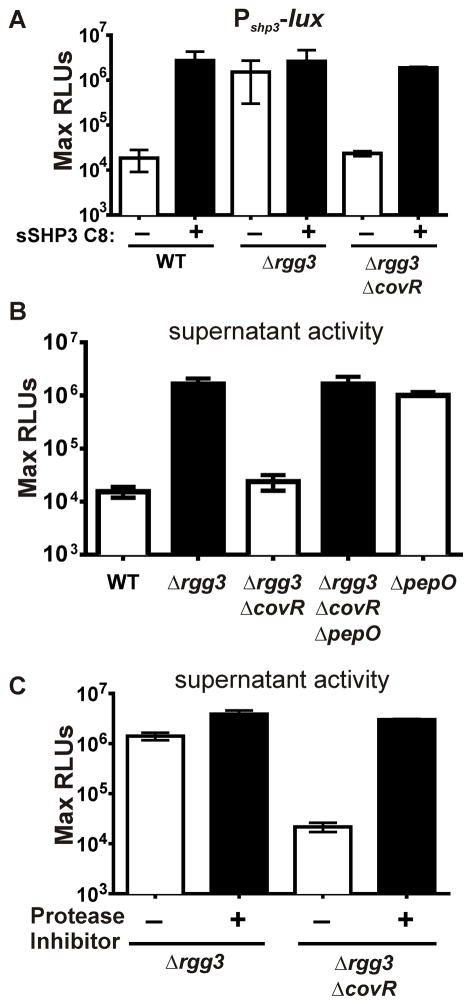
Using a transposon-mutagenesis strategy to identify genes contributing to the processing, export, or detection of the SHP2 and SHP3 pheromones, we found among ~10,000 independent mutants, twelve with insertions in the covRS locus that displayed defective activity of the Rgg2/3 circuit. To confirm this observation and to explore the relationship between the CovRS two-component system (TCS) and the Rgg2/3 quorum sensing (QS) circuit, we generated a covR deletion (ΔcovR) in the Δrgg3 background and assessed Rgg-SHP signaling using a Pshp3-lux reporter, which provides a highly sensitive readout. As described previously, Rgg3 serves to repress the QS circuit by blocking transcription from shp promoters. Deleting rgg3 leads to robust activation of the circuit that does not require exogenous addition of pheromone to induce the system but does require endogenous accumulation of pheromone within cultures and an intact positive feedback system to realize full transcriptional potential (Chang et al., 2011). However, consistent with the phenotype of covR transposon-insertion mutants, the Δrgg3ΔcovR strain failed to induce Pshp3-lux in the absence of exogenous peptide (Figure 1A). This suggested a possible regulatory link between genes under CovRS control and induction of the Rgg2/3 circuit.
Figure 1. Inactivation of covR limits SHP activity.
(A) The maximum luciferase activity, normalized to cell density and reported as relative light units (RLUs), of the Pshp3-lux transcriptional reporter integrated into the chromosome of WT (NZ131), Δrgg3, or Δrgg3ΔcovR strains, and supplemented with 125 nM sSHP3-C8 where indicated. (B) Clarified culture supernatants of wild type and mutant strains were inoculated with the WT NZ131 Pshp3-lux transcriptional reporter. (C) A protease-inhibitor cocktail was added to culture supernatants and inoculated with WT NZ131 Pshp3-lux. All experiments are representative of at least three independent experiments.
To elucidate the means by which CovRS affected the Rgg2/3 circuit, we first investigated whether the covR mutant was compromised in its ability to generate, sense, or respond to SHP pheromones. Transcription of Pshp3-lux in the Δrgg3ΔcovR strain in response to exogenous addition of 125 nM synthetic SHP3-C8 (sSHP3-C8) was similar to that seen in the Δrgg3 strain without exogenous pheromone (Figure 1A). This indicated the Δrgg3ΔcovR mutant’s lack of reporter activity was not due to a defect in pheromone uptake or transcriptional activation. Instead, the mutant appeared to be deficient in producing pheromones since cell-free culture supernatants from the Δrgg3ΔcovR mutant did not induce luciferase activity in a SHP-responsive reporter strain, unlike supernatants prepared from the Δrgg3 strain (Figure 1B). These results suggest that the signaling impairment of the Δrgg3ΔcovR mutant could be due to either a defect in pheromone synthesis or altered pheromone stability. As prior studies have found that covR mutants display increased expression of several proteases, we hypothesized that the deficiency of SHP pheromones in supernatants of Δrgg3ΔcovR might occur through a mechanism of enhanced SHP degradation by a CovRS-regulated protease (Graham et al., 2006; Cole et al., 2011). In support of this hypothesis, addition of a protease-inhibitor cocktail restored the Pshp3-lux-inducing activity of Δrgg3ΔcovR culture supernatants to the level of Δrgg3 supernatants (Figure 1C).
The aminopeptidase PepO limits SHP activity
The CovRS pathway differentially regulates the IL-8 protease spyCEP (spy49_0336, also known as prtS or scpC) (Graham et al., 2006). To test the possibility that SpyCEP might be responsible for SHP degradation or instability, we targeted spyCEP for mutagenesis. An in-frame deletion of spyCEP was generated in combination with covR and rgg3 (Δrgg3 ΔcovR ΔspyCEP) and culture supernatants from this mutant were assessed. Pshp3-lux expression from the reporter strain incubated in these supernatants remained as low as the Δrgg3ΔcovR mutant, indicating stability of SHP peptides was not affected by SpyCEP (Figure 2B). Given these negative results, we decided to employ a genetic screen to identify factors responsible for SHP instability in ΔcovR mutants.
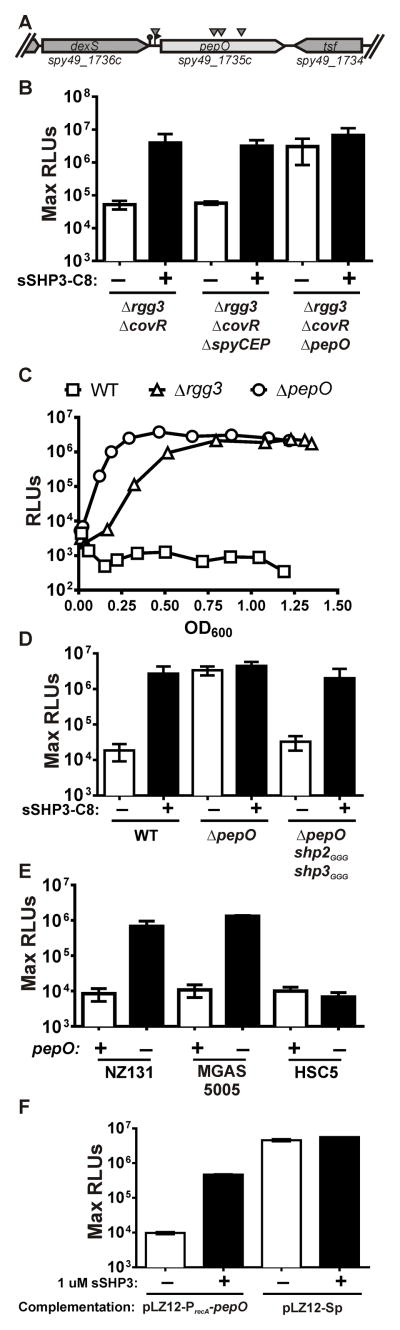
Figure 2. The peptidase pepO limits SHP stability.
(A) Arrowheads indicate ISS1 insertions in pepO and the upstream putative promoter region. (B) The maximum light activity, normalized to cell density (RLU), of Δrgg3 ΔcovR, Δrgg3ΔcovRΔspyCEP and Δrgg3ΔcovRΔpepO with the integrated Pshp3-lux transcriptional reporter. 125 nM sSHP3-C8 added where indicated. (C) Induction kinetics of WT, Δrgg3 and ΔpepO strains containing Pshp3-lux, indicating rapid accumulation of SHP in pepO-null strains. (D) The maximum RLU of WT, ΔpepO and ΔpepO shp2GGGshp3GGG strains containing integrated Pshp3-lux reporters. 125 nM sSHP3-C8 added as indicated. (E) CDM was conditioned by WT or ΔpepO mutant variants of indicated strains and assessed for SHP activity using the Pshp3-lux reporter described above. The maximum RLUs observed are reported. (F) Maximum RLU of ΔpepO mutants complemented with pepO in multi-copy (pLZ12-PrecA-pepO), or provided with the empty vector (pLZ12-Sp), and supplemented with 1 μM sSHP3-C8 as indicated. Panels (B), (D), (E) and (F) are results of at least three independent experiments [error bars represent standard deviation]; data in (C) is a representative of at least three independent experiments.
Since CovRS regulates 15–20% of the GAS genome, including multiple proteases (Graham et al., 2002; Sumby et al., 2006), we devised a screen that would identify mutations restoring SHP-dependent signaling in the Δrgg3ΔcovR mutant. To facilitate screening, we constructed a Pshp3-β-glucuronidase (Pshp3-gus) transcriptional reporter that would produce a blue colony phenotype if the shp3 promoter expression were elevated. The Pshp3-gus reporter placed in the Δrgg3ΔcovR mutant does not confer a blue colony phenotype unless excess sSHP-C8 is provided exogenously. We anticipated that if a CovR-repressed protease was responsible for the degradation of SHP pheromones, then a transposon insertion in the protease gene would result in endogenous SHP stabilization and accumulation to levels sufficient to trigger the positive-feedback regulatory loop, producing a blue phenotype. The mobile element pGH9-ISS1 was used to generate insertion mutations throughout the GAS genome (Maguin et al., 1996). Among ~11,000 colonies containing ISS1 insertions, four independent blue colonies were identified. The genomic regions flanking the insertion site of the isolated mutants were amplified and sequenced. Three of four insertions occurred within the pepO open reading frame (spy49_1735c), while the remaining one inserted 40 base pairs upstream of the pepO coding sequence, within a region likely to contain a promoter (Figure 2A).
An in-frame deletion of pepO (ΔpepO) was generated in the Δrgg3ΔcovR background and the resulting strain was tested for its ability to induce Pshp3-lux. Like the Δrgg3 single mutant, Δrgg3ΔcovRΔpepO exhibited high levels of reporter activity, confirming the results of the transposon screen (Figure 2B). Consistent with this result, the Pshp3-lux-inducing activity present in culture supernatants prepared from the triple mutant was also high (Figure 1B). Surprisingly, deletion of pepO in a wild-type background led to a rapid increase in Pshp3-lux reporter activity, demonstrating maximum induction at an OD600 of approximately 0.25 (Figure 2C). In comparison, an Δrgg3 mutant induces more gradually, with an increase beginning around OD600 0.25 but not reaching peak luminescence until an OD600 of approximately 0.75. Both the Δrgg3 and ΔpepO strains reached equivalent maximum levels of induction. Pshp3 -lux activity of the wild-type reporter strain remained low, suggesting that SHP peptides are produced at substantial levels but their production does not match rates of peptide turnover in pepO-expressing cells, at least not to a threshold level that leads to auto-induction. The auto-induction phenotype that was apparent in pepO mutants was confirmed to be dependent upon pheromone production since ΔpepOshp2GGGshp3GGG mutants (in which start codons of both shp genes have been disrupted and are unable to undergo translation) did not exhibit Pshp3-lux induction (Figure 2D). To determine if pepO plays a similar role in limiting QS induction in other strains of GAS, we generated in-frame deletions of pepO in two other GAS species – MGAS5005 and HSC5 and assessed SHP production and stability by conditioning CDM with WT and mutant strains, followed by detection with a Pshp3-lux ΔpepOshp2GGGshp3GGG reporter. MGAS5005 ΔpepO demonstrated the production of active SHPs, while the HSC5 ΔpepO mutant did not (Figure 2E). Finally, as compared to wild-type cultures, levels of Pshp3-lux-inducing activity were elevated in conditioned supernatants from the pepO strain (Figure 1B). The pepO deletion in the WT background was complemented in trans by cloning pepO on a multi-copy plasmid and expressing it under the constitutive recA promoter (pRVW21, pLZ12-PrecA-pepO). The resulting strain did not exhibit auto-induction of Pshp3-lux, yet remained partially responsive to the addition of 1 μM sSHP3-C8, demonstrating that the Rgg2/3 circuit remained intact but was severely limited by the over-expression of pepO (Figure 2F). When pLZ12 without PrecA-pepO was transferred to the ΔpepO mutant the auto-inductive phenotype persisted.
PepO acts intracellularly
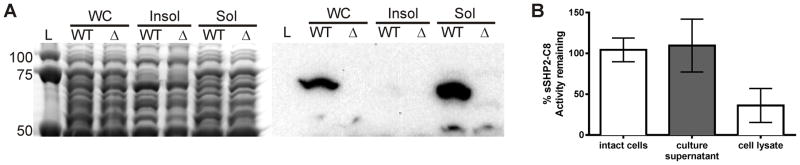
As the cellular location of PepO shapes its role in affecting QS, we sought to determine the cellular address of PepO activity in GAS. In silico analysis did not predict the presence of a secretion signal sequence, transmembrane helices, or non-canonical secretion pathway utilization, leading to the hypothesis that PepO is active in the cytoplasm of GAS (Secretome 2.0, PSORTb, TMHMM 2.0) (Krogh et al. 2001; Sonnhammer et al. 2001; Bendsten et al. 2005, Yu et al. 2010). This prediction was supported by localizing PepO to the cytoplasm using rabbit antisera raised against pneumococcal PepO (Figure 3A) (Agarwal et al., 2013). No PepO was found associated with membrane or cell-wall-associated fractions of cellular extracts or in cell-free culture supernatants. These results were corroborated by activity-based assays of cellular fractions using oppD mutants which are unable to transport extracellular SHP to the cytoplasm. In these assays synthetic SHP2-C8 was incubated with intact cells, lysed cells, or conditioned supernatants and assessed for which fraction led to a PepO-dependent loss of SHP activity, as determined by subsequent application of the reaction products to reporter cells capable of inducing Pshp3 -lux in response to intact SHP. PepO-dependent degradation of SHP was found only in the cytoplasmic fraction (Figure 3B). These results suggest strongly that SHP pheromones are degraded by PepO only within the cytoplasm.
Figure 3. PepO degrades SHP-C8 in the cytoplasm.
(A) Cellular compartments of WT or ΔpepO of washed-cell extracts were separated into membrane-associated (insoluble) and cytoplasmic (soluble) fractions and proteins were separated by SDS-PAGE (left). An equivalent gel was blotted and immunostained with anti-PepO antisera. (B) 50 nM sSHP2-C8 was incubated with intact cells, conditioned culture supernatant, or lysates of pepO+ and pepO− cultures with genetic backgrounds rendering cells incapable of producing or importing SHP peptides (ΔoppD shp2GGG shp3GGG). Following incubation, SHP activity remaining in each reaction was assessed using a Pshp3-lux shp2GGG shp3GGG ΔpepO reporter strain. SHP-C8 decay due to PepO is presented as a percent of maximum SHP activity retained in corresponding fractions of a pepO mutant.
PepO expression is altered by the CovRS two-component system
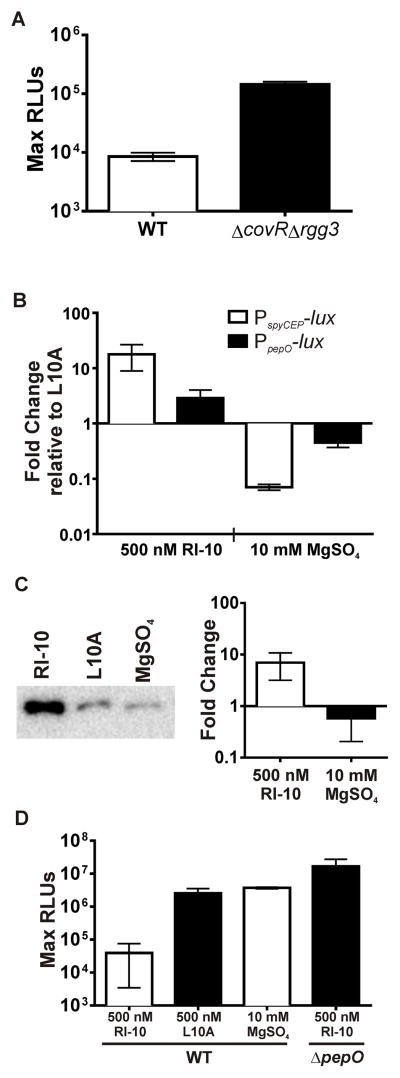
Since disruption of covR led to abrogation of SHP signaling, we suspected pepO would follow the paradigm of genes being directly repressed at the transcriptional level by CovR; however, inspection of the ~200 bp intergenic region between pepO and the upstream gene dexS (spy49_1736c) did not reveal any conserved CovR binding sequences (ATTARA) that would account for direct repression (Federle and Scott, 2002). The region upstream from pepO includes a putative promoter located ~85 bp from the start codon of pepO and a predicted transcriptional terminator that follows dexS. To confirm the relationship between CovR and pepO expression, we compared the transcriptional activity generated from this region by constructing a luciferase reporter (PpepO-lux) and comparing luminescence levels between WT and ΔcovR Δrgg3 backgrounds. Results show that expression of PpepO-lux was 10-fold higher in the covR mutant compared with WT grown in equivalent conditions (Figure 4A), confirming that CovR represses (directly or indirectly) transcription of this region. We therefore hypothesized that modulation of the CovRS signaling pathway, i.e. altering phosphorylation of CovR, would affect pepO transcription. Previously, it was demonstrated that Mg2+ is a potent environmental signal that leads to repression of many CovRS-regulated genes by means of enhanced phosphorylation of CovR (Gryllos et al., 2007). In contrast, the peptide RI-10 (a nontoxic fragment of the human antimicrobial peptide LL-37) signals through CovS to dephosphorylate CovR, leading to up-regulation of many genes, whereas an engineered variant of RI-10 (L10A) was shown to not affect CovRS regulation (Gryllos et al., 2008; Velarde et al., 2014). We tested the effects of these stimuli on GAS in CDM by constructing and monitoring the expression of a spyCEP luciferase reporter (PspyCEP-lux) and observed a >100-fold difference in expression between RI-10 and Mg2+ treatments, which is consistent with published results and indicates that phospho-CovR represses expression of spyCEP (Figure 4B). To evaluate whether pepO transcription was modulated by these stimuli, we generated a luciferase reporter to the 200 bp intergenic region containing the putative pepO promoter (PpepO) and placed this in single copy in the wild type (NZ131). Cultures were grown in CDM and treated with MgSO4, RI-10, or L10A. The observed differences in transcription of pepO between conditions amounted to an approximate six-fold change between Mg2+ and RI-10, suggesting that phosphorylated CovR repressed transcription of pepO (Figure 4B).
Figure 4. Rgg2/3 induction can be altered by CovRS modulation.
(A) Expression of PpepO-lux in NZ131 WT and ΔcovRΔrgg3. (B) Expression of PspyCEP-lux and PpepO-lux reporters integrated into WT NZ131 upon addition of 500 nM RI-10 or 10 mM MgSO4 and presented as fold changes relative to expression following addition of 500 nM L10A. (C) Left: representative Western blot indicating levels of PepO in cell lysates following the addition of 500 nM RI-10 or 10 mM MgSO4. Right: Relative amounts of PepO quantified by image densitometry of western blot and presented as fold-change compared to L10A treatment. (D) The maximum RLU for either WT or ΔpepO mutants grown in 1% mannose with 0.05% glucose, and treated as indicated. All strains contain chromosomally-integrated Pshp3-lux reporter. All data show results of at least three independent experiments.
To assess further PepO regulation by CovR, we analyzed relative protein levels of PepO in cell lysates of cultures treated with RI-10, L10A, or Mg2+ using anti-PepO rabbit antisera (Agarwal et al., 2013). Again, we found that levels of PepO in cells stimulated by RI-10 were approximately 6 to 10-fold increased over those treated with magnesium (Figure 4C). These combined experiments indicate that, despite the pepO promoter lacking a defined CovR binding site, pepO expression is responsive to signals known to modulate CovRS.
Rgg2/3 QS is modulated by CovRS
With the finding that CovRS regulates PepO levels, and that PepO diminishes SHP pheromones, we wondered if modulating CovRS activity with RI-10 or Mg2+ could affect QS through alteration of SHP pheromone concentration. To test this possibility, the wild-type Pshp3-lux reporter was grown in CDM containing 1% mannose, which we have recently reported serves as an environmental signal to induce Rgg-SHP signaling (Chang et. al, 2015). We predicted that treatment of cells with the RI-10 peptide would increase PepO production as a result of diminished repression by CovR, and therefore increase SHP degradation. The addition of RI-10 peptide to cultures of the reporter strain prevented Pshp3-lux induction, unlike cultures treated with the inactive L10A variant or Mg2+ (Figure 4D). The addition of RI-10 to the ΔpepO reporter strain had no effect, confirming that modulation of Rgg-SHP signaling through CovRS stimulation requires PepO.
PepO degrades sSHP2-C8 more efficiently than sSHP3-C8
To assess whether the PepO aminopeptidase was capable of directly degrading SHP peptides, a recombinant C-terminal 6xHIS-tagged PepO (rPepO) was expressed and purified. To assess activity of pheromones treated with rPepO, a ΔpepO reporter strain incapable of producing endogenous pheromones (ΔpepO shp2GGG shp3GGG) was employed; this strain responds to sSHP2-C8 and sSHP3-C8 similarly, as expected (Figure 5A). Titrations of rPepO incubated with 500 nM of SHP2-C8 or SHP3-C8 resulted in a dose-dependent loss of each pheromone’s activity; however, unexpectedly, a clear difference in deactivation was observed between the two SHP substrates (Figure 5B). Ten times more rPepO was required to degrade sSHP3-C8 than the equivalent amount of sSHP2-C8. This observed difference in catalysis of one peptide over the other was somewhat surprising given that these peptides differ in only one hydrophobic amino acid (isoleucine versus leucine).
Figure 5. rPepO degrades sSHP-C8.
(A) Luminescence activity of an integrated Pshp3-lux reporter in ΔpepO shp2GGGshp3GGG shows equivalent induction patterns when stimulated by either sSHP2-C8 or sSHP3-C8. (B) Synthetic C8 peptides were incubated with a range of concentrations of rPepO and added to a ΔpepO shp2GGGshp3GGG strain containing an integrated Pshp3-lux reporter. Maximum luminesce normalized to cell density (RLUs) were recorded. All data show results of at least three independent experiments.
Discussion
Herein we describe a mechanism by which the M49 GAS strain NZ131 and the M1T1 GAS strain 5005 govern their ability to respond to intercellular signals through a regulated degradation of peptide pheromones (Figure 6). Pheromone accumulation in cultures provides the opportunity to achieve adequate levels to engage Rgg receptors, induce transcription from shp promoters, and trigger positive feedback (auto-induction). We observe that PepO, an aminopeptidase, is capable of inactivating mature SHP pheromones, leading to a diminished ability to induce the Rgg2/3 quorum-sensing circuit. Conversely, the ΔpepO mutant or cells in which pepO expression is repressed by phosphorylated CovR show increased levels of SHP pheromones. Though PepO activity is clearly controlled by CovRS, our studies indicate that changes in pepO transcription and protein levels are limited to a more narrow dynamic range than that seen for another CovRS-regulated protease, SpyCEP. Regardless, the level to which PepO is produced is adequate to significantly alter quorum sensing responses, at least under conditions tested in the laboratory. Thus, PepO serves as a potent modulator of cell-cell communication involving peptide signals in Streptococcus.
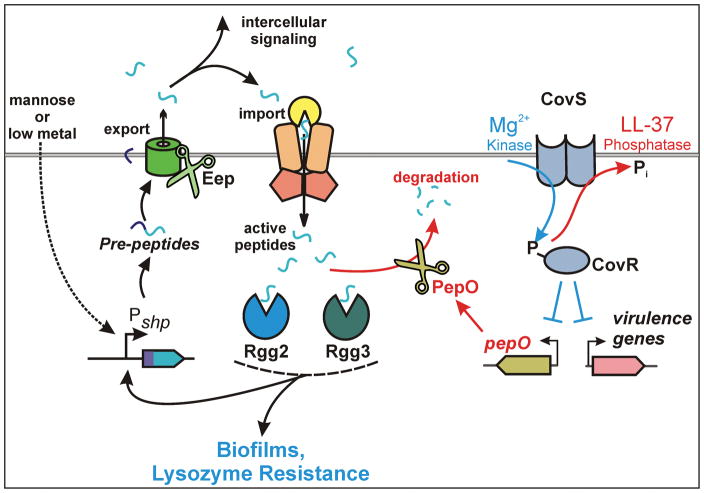
Figure 6. Model of the interaction between the Rgg2/3 quorum sensing pathway and the CovRS TCS via the aminopeptidase PepO.
Quorum sensing is activated under low metal conditions or when cells are provided with mannose as a primary carbon source. Activation of the Rgg2/3 pathway can be limited by exposure to CovRS-stimulants such as the LL-37 fragment RI-10, which in turn increases levels of the aminopeptidase PepO. PepO is able to directly degrade the active form of the SHP pheromones, blocking quorum-sensing activation.
Determinants that account for the wide varieties of disease due to GAS remain unclear. An appealing explanation for the seemingly capricious nature of this pathogen states that GAS exists in two primary adaptive states in the host: an avirulent carrier state and a virulent, superficial symptomatic state (Wollein Waldetoft and Råberg, 2014). Unique genetic programs govern these different lifestyles, and to maximize benefit to the bacteria, activation of each program must be tightly regulated. Accordingly, the avirulent carrier state supports a lower bacterial burden, with longer generation times, and is best suited to evading or limiting immune detection in the human host. Under other conditions, induction of a more virulent lifestyle is beneficial, as the ability to induce an inflammatory response allows GAS to recruit additional nutrients, support a greater bacterial burden and improve environmental spread. In order to take advantage of both lifestyles, GAS must maintain mechanisms to detect environmental stimuli, as well as activate the appropriate genetic programs to induce each lifestyle. The CovRS two-component system is a potent regulator of GAS virulence, known to be responsive to high pH, high temperature, and antimicrobial peptide exposure (Gryllos et al., 2003; Gryllos et al., 2007; Gryllos et al., 2008; Velarde et al., 2014). Our work on the Rgg2/3 QS circuit suggests that its regulatory program is consistent with a lifestyle of asymptomatic carriage, since activation leads to biofilm formation and increased resistance to lysozyme, and the pathway is induced by exposure to specific nutrient conditions GAS is likely to encounter in the nasopharynx (Chang et al., 2015). However, any interplay between CovRS and Rgg2/3 has not been previously revealed. The discovery that PepO is controlled by CovRS and is able to limit Rgg2/3 induction is a novel association between these two otherwise discrete systems, and provides an important mechanistic link in support of the hypothesis that these two systems contribute to unique bacterial lifestyles. By these early accounts, it appears pathway interaction is unidirectional, where CovRS suppresses quorum sensing through use of a protease. Whether quorum sensing influences CovRS signaling is not apparent at this time.
The use of proteases to modify quorum-sensing peptides has thus far been mainly documented for the maturation process of pheromones rather than for their elimination. Within GAS and Enterococcus, the protease Eep is required for pheromone development, likely during the process of export (Chandler and Dunny, 2007; Fleuchot et al., 2011; Chang et al., 2011). In Bacillus subtilis, the proteases subtilisin, Vpr and Epr are important in the maturation of the CSF peptide to its active pentapeptide form (Lanigan-Gerdes et al., 2007). Type I and II signal peptidases and other specialized endopeptidases (e.g., AgrB and SepM) are also important in development of signaling peptides in Staphylococcus aureus, B. subtilis, and streptococci (Thoendel and Horswill, 2009; Thoendel and Horswill, 2010; Hossain and Biswas, 2012). Surprisingly, however, identification of enzymes leading to degradation of peptide signals has only been implicated in S. pneumoniae where HtrA (DegP homolog) mediates CSP turnover during non-stress conditions (Cassone et al., 2012). Perhaps better characterized is the active degradation of homoserine lactone signals used by Gram-negative bacteria, suggested to function both in limiting endogenous QS responses as well as in disrupting communication of competing species. For these reasons their use in antivirulence strategies is appealing (Reviewed in LaSarre and Federle, 2013). It is somewhat surprising that HSC5 ΔpepO mutants fail to produce active SHPs in culture supernatants, as assessed using a reporter assay with sufficient sensitivity to observe pheromones produced by pepO mutants in NZ131 and MGAS5005, especially considering that HSC5 is a robust producer of pheromones when grown mannose or low metal conditions (Chang et al., 2015). However, it seems likely that other pathways contribute to pheromone induction and turnover dynamics and these systems vary from strain to strain. Nevertheless, PepO is the first example to our knowledge of a regulated protease that degrades mature QS peptides of the Rap/Rgg/Npr/PrgX/PlcR (RRNPP) family.
PepO is conserved among all Lactococci and Streptococci and has been most extensively studied in Lactococcus lactis, Streptococcus pneumoniae and Streptococcus parasanguis. The PepO protein is highly conserved amongst the species in which it has been characterized, with 72% identity to S. parasanguis, 44% identity to Lactococcus lactis MG1363 and 66% identity to S. pneumoniae D39, all compared to NZ131, and comparing the 80 residues surrounding the catalytic HEXXH domain, identity increases to 87%, 71% and 100%, respectively (Froeliger et al., 1999). Studies of PepO in Lactococcus have primarily focused on the role of the endopeptidase on protein catabolism. Lactococcus lactis uses a series of protease and peptidases to harvest energy from complex polypeptides. Though mutations in L. lactis pepO, or the aminopeptidase pepN, were not found to affect growth during culture in milk, the combination of pepO and pepN mutations led to a significant growth defect. This observation led to the conclusion that PepO may play a role in the derivation of essential amino acid nutrients from the complex polypeptide mixtures found in casein, a conclusion reinforced by the later observation that pepO in L. lactis is in an operon under the regulation of the branched-chain amino acid sensitive regulator CodY (Mierau et al., 2004). In S. pneumoniae, Agarwal et al found that PepO is capable of binding numerous human proteins, such as fibronectin and plasminogen, as well as blocking complement activation via interaction with C1q (Agarwal et al., 2013; Agarwal et al., 2014). We have not investigated whether these relationships hold true in GAS, but if so, would raise the prospect that PepO contributes to virulence. The studies of PepO in S. parasanguis have primarily focused on the characterization of the enzyme, and have found that PepO behaves most similarly to Endothelin Converting Enzyme 1, a mammalian enzyme that limits vasodilation (Froeliger et al., 1999; Oetjen et al., 2001; Oetjen et al., 2002). It is interesting to note that PepO has significant homology with the Neprilysin family of mammalian peptidases, which are responsible for the degradation of bioactive opioid peptides (Meyer et al., 2004). The homology between streptococcal PepO and mammalian analogs has been cited as evidence for horizontal gene transfer between prokaryotes and eukaryotes (Froeliger et al., 1999). Homologs of PepO are found in several species of bacterial pathogens, but are notably absent from taxa such as the archaea and fungi (Froeliger et al., 1999).
The presence of two SHP pheromones with approximately equivalent inductive capacity remains one of the unique and poorly understood features of the Rgg2/3 circuit. Previous work has shown that while SHP2-C8 and SHP3-C8 are capable of binding Rgg2 and Rgg3 with equivalent affinity, the deletion of shp3 limits system induction (Aggarwal et al., 2014, LaSarre et al., 2013). Further experiments demonstrated that the difference in the inductive capacity of the two shp genes lay in the N-terminal end of the pheromones, regions not involved in Rgg2/3 binding. It was hypothesized that the two pheromones may be differentially processed during maturation. Despite these discoveries, we still lack a biological explanation for the presence of two SHP pheromones. However, the finding that PepO preferentially degrades SHP2, even though the peptides differ at only one residue (DILIIVGG versus DIIIIVGG), provides a new understanding into the difference between these pheromones, and opens new avenues of investigation into an explanation as to why GAS is the only species of Streptococcus containing both SHP2 and SHP3. At a biochemical level, we speculate that SHP3-C8 is a poorer substrate for PepO simply due to steric hindrances caused by the asymmetric sidechain of isoleucine compared with leucine. Further enzymological studies would be required to determine whether this is the case. However, the biochemistry alone will not be sufficient to understand the physiological relevance of the two different pheromones. No other streptococcal species contains both shp2 and shp3, but several contain one pheromone type (e.g. S. agalactiae produces SHP2), and the preference of PepO to degrade SHP2 may indicate an ability to interfere with specific signals used by other organisms (Cook et al., 2013). Another possibility is that the SHP peptides, in addition to acting as QS signals, may act as environmentally-stimulated competitors of other substrates of PepO. We have noticed that in CDM, ΔcovRΔpepO mutants demonstrate a pronounced growth defect when compared to the ΔcovR parent strain. Surprisingly, this defect is not present in peptide-rich media (unpublished data) and suggests additional roles for PepO in peptide-free environments. As we have shown that the Rgg2/3 system is regulated by nutritional signals, it seems possible that the PepO-SHP relationship provides a yet-to-be described environmental sensory role. We intend to identify and characterize other substrates for PepO, which will allow us to refute or support this possibility.
The intracellular location of PepO presents an interesting aspect relating to its ability to degrade intercellular signals. Though conflicting reports addressing the cellular localization of PepO across other streptococcal species have been reported, our studies indicate PepO is cytosolic in GAS based on sub-cellular fractionation with subsequent immunoblotting and activity-based trials. Oetjen et al. reports that PepO is cytosolic in S. parasanguis, while Agarwal et al. argue that PepO is active as both a secreted protease and a membrane-bound protease (Oetjen et al., 2001; Agarwal et al., 2014). In Lactococcus, PepO has been determined to be cytoplasmic (Monnet, 1995; Gobbetti and Corsetti, 1996). Unlike membrane-diffusible signals such as homoserine lactones, peptide pheromones of the RRNPP family are actively transported to the cytosol before engaging their cognate receptors, and do so by a relatively non-specific oligopeptide transport systems (Chang et al., 2011). As PepO is located in the GAS cytoplasm, we deduce that it degrades imported SHP pheromones while en route to interaction with Rgg2 or Rgg3 or possibly targeting pre-peptides prior to their secretion. Thus, when present, PepO reduces the concentration of SHPs within the cell to a level below that needed to engage Rgg proteins; this results in Rgg3-mediated repression of target promoters, disruption of the SHP positive-feedback loop, and exit from the quorum-sensing response. Importantly, under these circumstances, the activity of destroying internal quorum-sensing signals is a decision made by, and potentially affecting, only the individuals in the population expressing PepO. This is in contrast to a scenario in which PepO were to be located outside the cell and capable of affecting the concentration of pheromones available to all members of the population, and thereby influencing decisions carried out by the group. Instead, it appears that individuals or a subset of the population that expresses PepO consider stress (detected by CovS) a condition under which participation in SHP-dependent coordination of gene expression is no longer a beneficial activity. This scenario is consistent with a hypothesis that the Rgg2/3 pathway assists in the establishment or maintenance of an asymptomatic relationship with the host through regulation of biofilm development, lysozyme resistance and an avirulent lifestyle (Chang et al., 2011; Chang et al., 2015; Marks et al., 2014). Although cooperative behavior among individuals may enhance fitness by sharing resources or unifying defense strategies, the perception of stress, possibly generated by host assaults, may present a circumstance where individual behavior and heterogeneous gene expression at the population level provides a better strategy for survival, dispersal, or transmission.
In conclusion, we have shown that PepO is an aminopeptidase capable of degrading active pheromones of the Rgg2/3 quorum-sensing system. These data provide a mechanistic link between the CovRS two-component system, which regulates PepO expression, and the Rgg2/3 QS circuit. The findings also provide new approaches for investigating the Rgg2/3 circuit, as pepO mutants are capable of autoinduction. Finally, the observation that PepO preferentially degrades SHP2 over SHP3 is consistent with the notion that each pheromone instills unique information content for GAS.
Acknowledgments
We are especially grateful to the Anna Blom Lab (Lund University, Lund, Sweden) for providing PepO anti-sera, the Kevin McIver and Daniel Nelson labs (University of Maryland) for phage lysin and pOsKar, and the Lynn Hancock Lab (University of Kansas) for pKS310. We are also grateful to Laura Cook for manuscript review, and Lauren Mashburn-Warren for generation of the ΔcovR strain.
Funding
This work was funded by NIH R01-AI091779 and the Burroughs-Welcome Fund Investigators in the Pathogenesis of Infectious Diseases Award. RW was partially supported by the American Heart Association Pre-doctoral Fellowship 15PRE22710027.
Bibliography
- Agarwal V, Kuchipudi A, Fulde M, Riesbeck K. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen-and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J Biol Chem. 2013;288:6849–63. doi: 10.1074/jbc.M112.405530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Sroka M, Fulde M, Bergmann S, Riesbeck K, Blom AM. Binding of Streptococcus pneumoniae endopeptidase O (PepO) to complement component C1q modulates the complement attack and promotes host cell adherence. J Biol Chem. 2014;289:15833–44. doi: 10.1074/jbc.M113.530212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal C, Jimenez J, Nanavati D, Federle MJ. Multiple length peptide-pheromone variants produced by Streptococcus pyogenes directly bind Rgg proteins to confer transcriptional regulation. J Biol Chem. 2014;289:22427–36. doi: 10.1074/jbc.M114.583989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrløv Bendtsen J, Kiemer L, Fausbøll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiology. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone M, Gagne AL, Spruce LA, Seeholzer S, Sebert MS. The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J Biol Chem. 2012;287:38449–59. doi: 10.1074/jbc.M112.391482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Dunny G. Characterization of the Sequence Specificity Determinants Required for Processing and Control of Sex Pheromone by the Intramembrane Protease Eep and the Plasmid-Encoded Protein PrgY. J Bacteriol. 2007;190:1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 2011;7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Jimenez JC, Federle MJ. Induction of a quorum sensing pathway by environmental signals enhances group A streptococcal resistance to lysozyme. MolMicrobiol. 2015 doi: 10.1111/mmi.13088. Published ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol. 2011;9:724–36. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- Cook L, LaSarre B, Federle M. Interspecies communication among commensal and pathogenic streptococci. MBio. 2013;4:00382–13. doi: 10.1128/mBio.00382-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections and their sequelae. Adv Exp Med Biol. 2008;609:29–42. doi: 10.1007/978-0-387-73960-1_3. [DOI] [PubMed] [Google Scholar]
- Dalton TL, Scott JR. CovS Inactivates CovR and Is Required for Growth under Conditions of General Stress in Streptococcus pyogenes. J bacteriol. 2004;186:3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, et al. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect Immun. 2000;68:2441–2448. doi: 10.1128/iai.68.5.2441-2448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMuri G, Wald E. The Group A Streptococcal Carrier State Reviewed: Still an Enigma. J Ped Infect Dis. 2014;3:336–342. doi: 10.1093/jpids/piu030. [DOI] [PubMed] [Google Scholar]
- Engleberg CN, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. Journal of Infectious Diseases. 2001;183:1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- Federle M, Scott J. Identification of binding sites for the group A streptococcal global regulator CovR. b Microbiol. 2002;43:1161–1172. doi: 10.1046/j.1365-2958.2002.02810.x. [DOI] [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–57. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, et al. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum sensing mechanism in streptococci. Mol microbiol. 2011;80:1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x. [DOI] [PubMed] [Google Scholar]
- Froeliger E, Oetjen J, Bond J, Fives-Taylor P. Streptococcus parasanguis pepO encodes an endopeptidase with structure and activity similar to those of enzymes that modulate peptide receptor signaling in eukaryotic cells. Infect immun. 1999;67:5206–14. doi: 10.1128/iai.67.10.5206-5214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–347. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gobbetti M, Smacchi E, Fox P, Stepaniak L, Corsetti A. The sourdough microflora. Cellular localization and charecterization of proteolytic enzymes in lactic acid bacteria. LWT- Food Science and Technology. 1996;29:561–569. [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, et al. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2002;99:13855–60. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Gardner DJ, Long RD, Welty DM, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol. 2006;169:927–42. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryllos I, Grifantini R, Colaprico A, Jiang S, DeForce E, Hakansson A, et al. Mg2+ signalling defines the group A streptococcal CsrRS (CovRS) regulon. Mol Microbiol. 2007;65:671–683. doi: 10.1111/j.1365-2958.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- Gryllos I, Levin JC, Wessels MR. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+ Proc Natl Acad Sci USA. 2003;100:4227–32. doi: 10.1073/pnas.0636231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryllos I, Tran-Winkler HJ, Cheng MFF, Chung H, Bolcome R, Lu W, et al. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci USA. 2008;105:16755–60. doi: 10.1073/pnas.0803815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann N, Saldaña M, Sahasrabhojane P, Yao H, Su X, Thompson E, et al. Dual-Site Phosphorylation of the Control of Virulence Regulator Impacts Group A Streptococcal Global Gene Expression and Pathogenesis. PLoS Pathog. 2014;10:e1004088. doi: 10.1371/journal.ppat.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Biswas I. An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J bacteriol. 2012;194:5886–5896. doi: 10.1128/JB.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell-Adams B, Seifert HS. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol microbiol. 2000;37:1146–1158. doi: 10.1046/j.1365-2958.2000.02067.x. [DOI] [PubMed] [Google Scholar]
- Husmann LK, Scott JR, Lindahl G. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Federle M. Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol. 2014;4 doi: 10.3389/fcimb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–32. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lanigan-Gerdes S, Dooley AN, Faull KF, Lazazzera BA. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis. Mol Microbiol. 2007;65:1321–33. doi: 10.1111/j.1365-2958.2007.05869.x. [DOI] [PubMed] [Google Scholar]
- Lasarre B, Aggarwal C, Federle MJ. Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes. MBio. 2013;3:e00333–12. doi: 10.1128/mBio.00333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre B, Chang JC, Federle MJ. Redundant group a streptococcus signaling peptides exhibit unique activation potentials. J Bacteriol. 2013;195:4310–8. doi: 10.1128/JB.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y, McIver KS. Genetic Manipulation of Streptococcus pyogenes (The Group A Streptococcus, GAS) Curr Protoc Microbiol. 2013;30:9D.3.1–9D.3.29. doi: 10.1002/9780471729259.mc09d03s30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–19. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Maguin E, Prevost H, Ehrlich SD. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marijon E, Mirabel M, Celermajer DS, Jouven X. Rheumatic heart disease. Lancet. 2012;379:953–964. doi: 10.1016/S0140-6736(11)61171-9. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan WM, Ferretti JJ, Karasawa T. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J Bacteriol. 2008;190:7773–7785. doi: 10.1128/JB.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan WM, McLaughlin RE, Nordstrand A. Vectors containing streptococcal bacteriophage integrases for site-specific gene insertion. Methods cell sci. 1998;20:51–57. [Google Scholar]
- Meyer DH, Oetjen J, Fives-Taylor PM. Neprilysin homolog of streptococci. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of proteolytic enzymes. 2. San Diego: Elsevier Academic Press; 2004. pp. 443–445. [Google Scholar]
- Mierau I, Kok J, Monnet V. Oligopeptidase O. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of proteolytic enzymes. 2. San Diego: Elsevier Academic Press; 2004. pp. 445–447. [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J mol bio. 1996;260:289–98. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Monnet V. Oligopeptidases from Lactococcus lactis. Methods in Enzymology. 1995;556:579–592. doi: 10.1016/0076-6879(95)48037-4. [DOI] [PubMed] [Google Scholar]
- Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci USA. 2006;103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen J, Fives-Taylor P, Froeliger EH. The divergently transcribed Streptococcus parasanguis virulence-associated fimA operon encoding an Mn2+-responsive metal transporter and pepO encoding a zinc metallopeptidase are not coordinately regulated. Infect immun. 2002;70:5706–5714. doi: 10.1128/IAI.70.10.5706-5714.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen J, Fives-Taylor P, Froeliger E. Characterization of a Streptococcal Endopeptidase with Homology to Human Endothelin-Converting Enzyme. Infect Immun. 2001;69:58–64. doi: 10.1128/IAI.69.1.58-64.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijn VI, Kessler RE. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun. 1980;27:444–448. doi: 10.1128/iai.27.2.444-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D, Ferretti JJ. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS microbiol lett. 1991;82:219–224. doi: 10.1016/0378-1097(91)90336-9. [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M, Horswill AR. Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J Biol Chem. 2009;284:21828–21838. doi: 10.1074/jbc.M109.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M, Horswill AR. Biosynthesis of peptide signals in gram-positive bacteria. Adv Appl Microbiol. 2010;71:91–112. doi: 10.1016/S0065-2164(10)71004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde J, Ashbaugh M, Wessels M. The Human Antimicrobial Peptide LL-37 Binds Directly to CsrS, a Sensor Histidine Kinase of Group A Streptococcus, to Activate Expression of Virulence Factors. J Biol Chem. 2014;289:36315–36324. doi: 10.1074/jbc.M114.605394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollein Waldetoft K, Råberg L. To harm or not to harm? On the evolution and expression of virulence in group A streptococci. Trends Microbiol. 2014;22:7–13. doi: 10.1016/j.tim.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]