Abstract
A substantial body of evidence accumulated over the past 20 years supports the concept that gC1qR is a major pathogen-associated pattern recognition receptor (PRR). This conclusion is based on the fact that, a wide range of bacterial and viral ligands are able to exploit gC1qR to either suppress the host’s immune response and thus enhance their survival, or to gain access into cells to initiate disease. Of the extensive array of viral ligands that have affinity for gC1qR, the HIV-1 envelope glycoprotein gp41, and the core protein of hepatitis C virus (HCV) are of major interest as they are known to contribute to the high morbidity and mortality caused by these pathogens. While the HCV core protein binds gC1qR and suppresses T cell proliferation resulting in a significantly diminished immune response, the gp41 employs gC1qR to induce the surface expression of the NK cell ligand, NKp44L, on uninfected CD4+ T cells, thereby rendering them susceptible to autologous destruction by NKp44 receptor expressing NK cells. Because of the potential for the design of peptide-based or antibody-based therapeutic options, the present studies were undertaken to define the gC1qR interaction sites for these pathogen-associated molecular ligands. Employing a solid phase microplate-binding assay, we examined the binding of each viral ligand to wild type gC1qR and 11 gC1qR deletion mutants. The results obtained from these studies have identified two major HCV core protein sites on a domain of gC1qR comprising of residues 144–148 and 196–202. Domain 196–202 in turn, is located in the last half of the larger gC1qR segment encoded by exons IV–VI (residues 159–282), which was proposed previously to contain the site for HCV core protein. The major gC1qR site for gp41 on the other hand, was found to be in a highly conserved region encoded by exon IV and comprises of residues 174–180. Interestingly, gC1qR residues 174–180 also constitute the cell surface-binding site for soluble gC1qR (sgC1qR), which can bind to the cell surface in an autocrine/paracrine manner via surface expressed fibrinogen or other membrane molecules. The identification of the sites for these viral ligands should therefore provide additional targets for the design of peptide-based or antigen-based therapeutic strategies.
Keywords: gC1qR, PRR, Hepatitis C virus, HIV-1 gp41
1. Introduction
The receptor for the globular heads of C1q, gC1qR/p33 (also know as p32, HABP-1), is a ubiquitously distributed highly acidic (pI 4.15), multicompartmental and multifunctional cellular protein, which modulates a plethora of immunological functions including infection, inflammation, autoimmunity, and cancer (Ghebrehiwet et al., 2011). In addition to its known plasma ligands [C1q, high molecular weight kininogen (HK), fibrinogen, thrombin and multimeric vitronectin], gC1qR is also able to bind a diverse array of bacterial- (Braun et al., 2000; Nguyen et al., 2000; Ghebrehiwet et al., 2007) and viral-associated molecular ligands (Wang et al., 1997; Yu et al., 1995; Luo et al., 1994; Szabo et al., 2001; Fausther-Bonvendo et al., 2010; Bruni and Roizman, 1996; Mathews and Russell, 1998; Kittlesen et al., 2000; Mohan et al., 2002; Choi et al., 2009; Watthanasurorot et al., 2012) and as such is considered to be a significant pathogen recognition receptor (PRR). The core protein of the hepatitis C virus (HCV) and the envelope glycoprotein of the human immunodeficiency virus (HIV-1) gp41 are two examples of viral associated molecular ligands that employ gC1qR to induce immunosuppression by targeting CD4+ T cells and possibly monocytes. The HCV core protein binds gC1qR on T cells and inhibits their proliferation (Kittlesen et al., 2000) in a manner that is similar to the C1q-induced antiproliferative response reported by us earlier (Chen et al., 1994). Suppression of the host immune response in turn contributes to an extremely high rate of viral persistence and severe disease progression (Kittlesen et al., 2000; Yao et al., 2004). The HIV-1 transmembrane envelope protein gp41 has also been shown to bind to the globular heads of C1q (gC1q) via an external segment of the gp41 molecule with residues 601–613 (GIWGCS-GKLICTT) of gp160 (inclusive of gp41) having been identified as the potential C1q interaction site (Thielens et al., 1993; Quinkal et al., 1999). This interaction in turn has been shown to activate the classical pathway of complement (Thielens et al., 1993; Quinkal et al., 1999). More recent experiments have also demonstrated that gp41 is able to modulate T cell destruction thereby contributing to the persistently low level of T cells seen during the late phase of HIV-1 infection (Fausther-Bonvendo et al., 2010; Caffrey, 2001). An insight into the mechanism of this cell death was recently gained when it was shown that the interaction of gp41 and gC1qR plays a central role in this process (Fausther-Bonvendo et al., 2010). The 3S motif of HIV-1 gp41 comprising of residues: PWNASWSNKSLDDIWN – with the highlighted residues being critical – has been shown to bind to gC1qR on uninfected CD4+ T cells and to induce surface expression of NKp44L—a cellular ligand for the NK receptor, NKp44—thereby rendering them susceptible to autologous NK killing (Fausther-Bonvendo et al., 2010). Because molecules or peptides that inhibit the interaction between pathogen associated molecular ligands and gC1qR can potentially be used as templates for the development of therapeutic interventions, the present studies were undertaken to map out and refine the precise gC1qR interaction sites for these important molecules.
2. Materials and methods
2.1. Chemicals and general reagents
The following reagents and chemicals were purchased or obtained from the sources indicated: Dulbecco’s PBS (D-PBS) with and without calcium and magnesium (Mediatech Inc, Manassas, VA); Dulbecco’s Modified Eagles Medium (DMEM); RPMI 1640, 100× Penicillin/Streptomycin, (GIBCO-Invitrogen, Grand Island, NY); heat inactivated fetal bovine serum (FBS) (Hyclone, Logan, UT); human serum albumin (HSA) (Immuno-US, Rochester, MI); p-nitrophenyl phosphate (pNPP); alkaline phosphatase (AP)–conjugated rabbit anti-goat IgG (Pierce, Rockford, IL); C1q (CompTech); and D-phenylalanyl-L-prolyl-L-arginine chloromethyl ketone (PPACK)—a specific thrombin inhibitor (Sigma-Adlrich).
2.2. Expression of various versions of recombinant gC1qR proteins
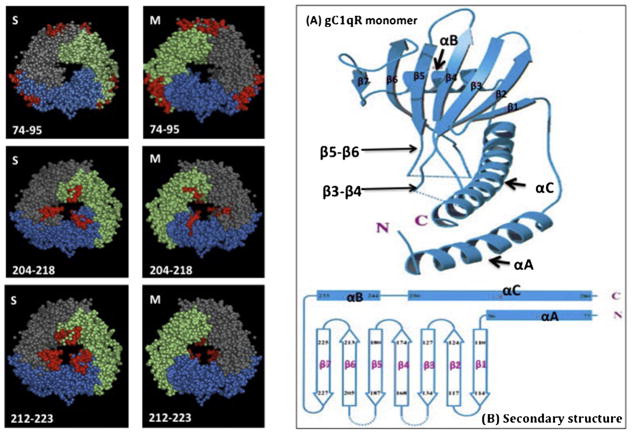
The strategy for the construction of plasmids containing the glutathione-S-transferase (GST)-gC1qR wild type (WT), as well as several important deletion (Δ) mutants lacking highly charged domains selected on the basis of their prominent position in the crystal structure (Jiang et al., 1999) have been described in detail in our earlier publications (Ghebrehiwet et al., 2011; Ghebrehiwet et al., 2002). In all cases, the final constructs were sequenced to verify sequence integrity. The fusion products were purified on glutathione-Sepharose 4B column, then cleaved by thrombin (3.2 μg/ml), and the GST-free gC1qR proteins purified on fast protein liquid chromatography (FPLC, Pharmacia) using a Mono-Q ion exchange column. After verification by ELISA and visualization by SDS-PAGE (Laemmli, 1970), the single peak containing the gC1qR protein was pooled, concentrated to 1–2 mg/ml, and stored at −80 °C in the presence of 50 nM PPACK (D-phenylalanyl-L-prolyl-L-arginine chloromethyl ketone)—a specific thrombin inhibitor. To estimate the organization and integrity of each recombinant protein, gel filtration of purified gC1qR (wild Type, WT) as well the deletion mutants was carried out by analytical gel filtration on Superose column (Ghebrehiwet et al., 2011). With the exception of those lacking residues 74–95, 204–218, and 212–223, all of the deletions mutants displayed a properly folded trimer as described in detail earlier (Ghebrehiwet et al., 2011).
2.3. Viral antigens, proteins and antibodies
Recombinant gp41 as well as mAb to gp-41 was obtained from the NIH AIDS repository Program (Germantown, MD). HCV core protein was purchased from Abcam (Cambridge, MA). Monoclonal and polyclonal antibodies to recombinant human gC1qR as well as to gC1qR peptides have been described in previous publications and represent part of the anti-gC1qR antibody databank in our laboratory (Ghebrehiwet et al., 1996). A 17-residue long synthetic peptide corresponding to the 3S motif of gp41, henceforth called gp41-3S (PWNASWSNKSLDDIWNC) with 1Cys at the C-terminus, as well as affinity purified rabbit anti-gp41-3S were made commercially (GenScript, Piscataway, NJ). Monoclonal antibody to Escherichia coli MBP (maltose binding protein) was purchased from Sigma.
2.4. Expression and purification of the wild type ghA module and its substitution mutants
The recombinant globular head protein, ghA, and its respective substitution mutants were expressed as a fusion with MBP in E. coli BL21 strain as described earlier (Kishore et al., 2003; Kojouharova et al., 2004). Briefly, bacterial cells were grown in 200 ml LB medium containing ampicillin (100 μg/ml) at 37 °C. Once grown to an OD of 0.6, cells were induced with 0.4 mM IPTG (isopropyl thiogalactoside) for 3 h and centrifuged (4500 rpm for 15 min). The cell pellet was suspended in 25 ml of lysis buffer (20 mM Tris pH 8.0, 0.5 M NaCl, 1 mM EDTA, 0.2% v/v Tween 20, 5% glycerol, 0.1 mM PMSF and 0.1 g lysozyme) and incubated at 4 °C for 1 h. The cells were then sonicated for 30 s with 2 min gaps for 10 cycles. After centrifugation (13,000 rpm, 15 min) the supernatant was diluted 5-fold in buffer I (20 mM Tris pH 8.0, 100 mM NaCl, 0.2% Tween 20, 1 mM EDTA and 5% glycerol) and passed through an amylose resin column that had been washed first with 3 bed volumes of buffer I followed by buffer II (250 ml of buffer I without Tween 20). The protein was then eluted with 10 mM maltose in 100 ml of buffer II. The ghA substitution mutants were generated as described earlier (Kishore et al., 2003; Kojouharova et al., 2004).
2.5. Cultured cells
The cell lines, MOLT-4 and U937 – representing CD4+ T cell and monocytic cell – were grown in suspension in RPMI 1640 containing 10% heat inactivated fetal bovine serum and 100 units/ml penicillin and 100 μg/ml streptomycin (GIBCO-Invitrogen, Grand Island NY) and maintained in a humidified air consisting of 5% CO2 and 95% air as described (Ghebrehiwet et al., 2011). Prior to each experiment, the viability of cells was verified by Trypan blue exclusion and only cultures with ≥95% viability were used for experiments.
2.6. Solid-phase microplate binding assay
The ability of the various gC1qR proteins to bind to HCV core protein or HIV-1 gp41 was assessed step-wise by solid-phase microplate binding assay. The overall strategy taken was to first screen all of the 10 deletion mutants and 1 substitution mutant (W233G) for their ability to bind to the target antigen, and once mutants that consistently showed diminished binding when compared to the WT gC1qR were identified, they were assessed more vigorously in a separate set of experiments. Briefly, microtiter plate wells were coated in duplicate (90 min, room temp or overnight, 4 °C) with 100 μl of either, 2 μg/ml HCV core protein, gp41, or BSA, in carbonate buffer, pH 9.6 (15 mM Na2CO3 and 35 mM NaHCO3). The unbound protein was removed; the wells washed 2× with TBST (20 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.05% Tween-20), and the unreacted sites blocked by incubation (30 min, room temp) with 300 μl of 5% non-fat dry milk or blotto (10 mg/ml casein in TBS, pH 8.0). After washing (2× with TBST), the microtiter plate bound HCV or gp41 was incubated (1 h, room temp.) with concentrations of biotinylated wild type (WT) gC1qR ranging from 0 to 5 μg/ml. This was followed by sequential reaction (1 h each, room temp) with alkaline-phosphatase conjugated streptavidin and pNPP. After firmly establishing dose-dependent binding (n = 3), experiments were repeated as above to compare the binding of either 5 μg/ml biotinylated WT gC1qR or deletion (Δ) mutants followed by sequential reaction with alkaline-phosphatase conjugated streptavidin and pNPP as described.
2.7. Solid-phase binding of gp-41 or gp41-3S to microplate-fixed cells
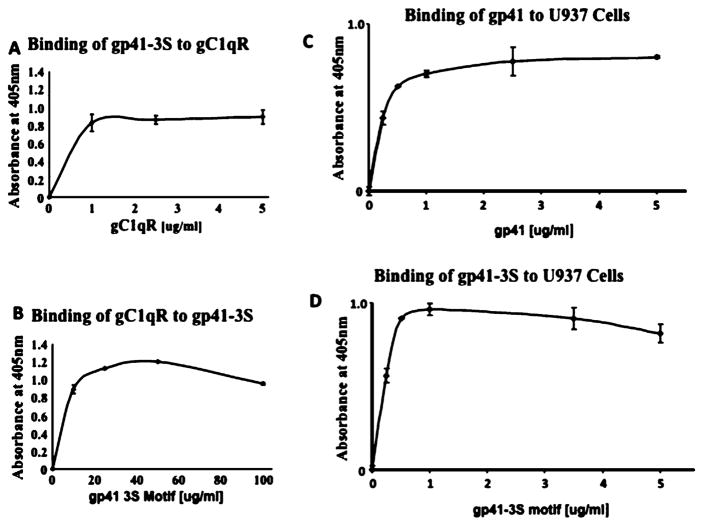
The ability of gp41 or gp41-3S peptide to bind to gC1qR expressing MOLT-4 or U937 cells was tested using microtiter plate fixed cells as described in our earlier publications (Ghebrehiwet et al., 2011; Ghebrehiwet et al., 1996). Briefly, intact cells (2 × 105 cells/well) were first attached (30 min, 22 °C) onto poly-Llysine (10 μg/ml in PBS, pH 7.4) coated duplicate ELISA wells. Subsequently, the cells were fixed (30 min, 22 °C) by addition of an equal volume of glutaraldehyde (0.5% solution in PBS), and the unreacted sites were quenched with glycine-BSA (100 mM Glycine, 0.1% BSA in PBS, pH 7.4). Next, gp41 or gp41-3S ranging from 0 to 5 μg/ml were added and incubated (60 min, 22 °C). Then, the bound proteins were detected by sequential interaction with either mAb to gp41 or pAb to gp4-3S followed by alkaline-phosphatase conjugated goat anti-mouse or goat ant-rabbit antibody (60 min, 22 °C). The reaction was further probed by incubation with pNPP and the color developed read spectrophotometrically at 405 nm.
2.8. Statistical analysis
Student t-tests were performed using statistical software (Excel; Microsoft, Redmond, WA, USA). A value of p = 0.05 was considered to be a significant difference. (n = separate experiments performed in duplicates)
3. Results
3.1. Identification of the gC1qR site for HCV core protein using gC1qR deletion mutants
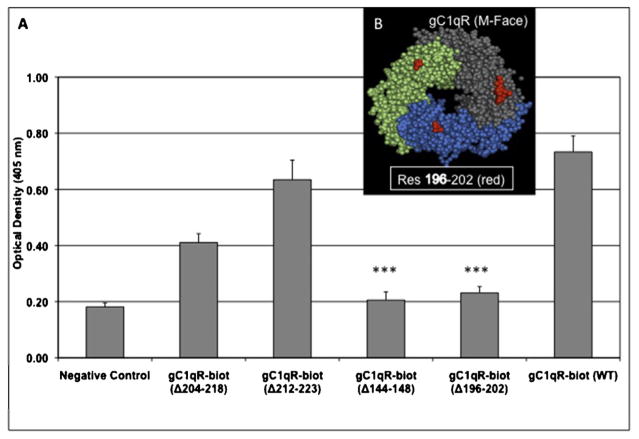
As shown in Fig. 1, the binding of HCV core to gC1qR is fast, reproducible, specific, and dose-dependent. Previous studies (Kittlesen et al., 2000) had suggested that the binding site for HCV core protein resides on a long stretch of gC1qR that is encoded by exons IV–VI (residues 159–282). Since this comprises of a large segment of the gC1qR molecule, we opted to verify and accurately define the HCV core protein site on gC1qR. To achieve this we used a solid-phase microtiter plate-binding assay and tested the interaction of several highly purified gC1qR deletion mutants including several deletions from the region encoded by exons IV–VI. The characteristics of the 10 deletion mutants and the point mutation used in these studies have been described in detail in a previous publication (Ghebrehiwet et al., 2011). When the binding of these gC1qR mutants was assessed using HCV coated wells, two mutants lacking residues 144–148 and 196–202 respectively, showed significantly (p ≤ 0.001) reduced interaction with HCV core when compared to the WT-gC1qR (Fig. 2). Deletion 204–218 also had a significantly diminished binding (p ≤ 0.05). However, we think this lack of binding might be due to the fact that deletion of this region is known to disrupt the trimerization of the molecule, which is important for the integrity and binding to ligands.
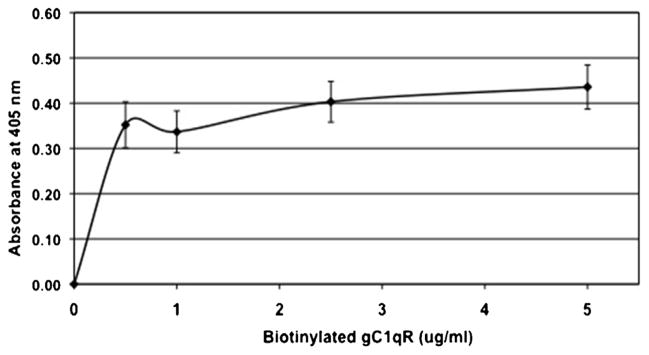
Fig. 1.
Dose–dependent binding of gC1qR to HCV core protein.
Microtiter platewells were first coated with 2 μg/ml HCV core protein (1 h, 37 °C). After washing and blocking with 5% non-fat dry milk, the bound HCV core protein was reacted (overnight, 4C) with concentrations of biotinylated WT gC1qR ranging from 0 to 5 μg/ml followed by sequential reaction with alkaline-phosphatase conjugated streptavidin and pNPP following the standard procedure for ELISA. Each data point is a mean ± SD of duplicate experiments and the figure is a representative of three such experiments.
Fig. 2.
Comparative binding of WT gC1qR and gC1qR deletion mutants with HCV core protein.
Preliminary microplate binding studies were first performed as above to compare the binding of biotinylated WT gC1qR and the 10 deletion and 1 substitution mutants to microtiter plate coated HCV core protein as described in the legend to Fig. 1. After three such tests, the deletion mutants that showed consistently diminished binding to HCV core were selected and further analyzed. Data shown in (A) include data of only the deletion mutants (out of 11 mutants) that reacted weakly with HCV core protein. The residue numbers indicate the segment of the deleted gC1qR region (Δ) and the location in the gC1qR 3D structure of one such deleted region is shown in red (B). The significance of data is indicated as: *p ≤ 0.05 and ***p ≤ 0.001.
3.2. Identification of the gC1qR site for the HIV-1 gp41
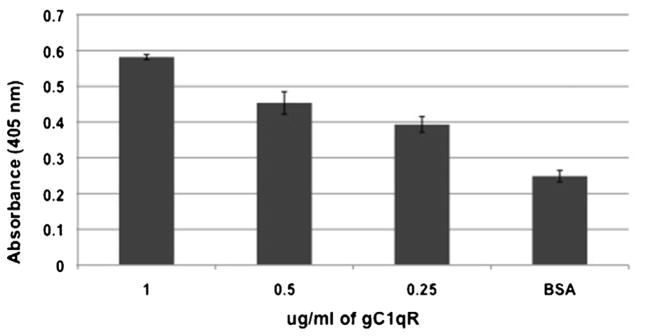
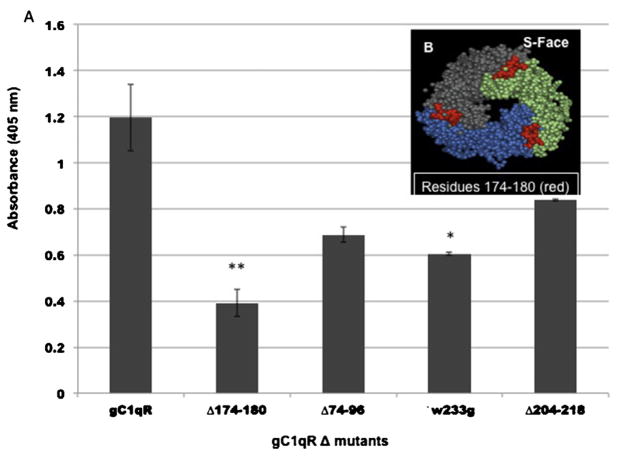
Similarly, the interaction between gC1qR and gp41 is also dose-dependent (Fig. 3). Subsequent binding assays comparing WT-gC1qR and its deletion mutants showed (Fig. 4) that only gC1qRΔ174–180 (p ≤ 0.01) and to a lesser extent W233G (p ≤ 0.05) had a significantly reduced binding to gp41. However, although deletion 74–96 and 204–218 also showed reduced binding, this is probably due to the disruption of the trimeric integrity, caused by deletion of these segments.
Fig. 3.
Binding of gC1qR to the HIV-1 envelope glycoprotein, gp41.
ELISA plate wells were first coated with 100 μl per well of 2 μg/ml of gp41 (1 h, 37 °C). Following blocking with 5% non-fat dry milk for 1 h, wells were washed (3× with TBS-T) and incubated with concentrations of gC1qR (1 μg, 0.5 μg and 0.25 μg/ml) in TBS, (1 h, 37 °C). Bound gC1qR was then detected using mAb 74.5.2 anti-gC1qR (0.5 μg/ml) followed by sequential reaction with alkaline phosphatase goat anti-mouse IgG and pNPP. The absorbance of the color developed after 30 min was measured spectrophotometrically at 405 nm. Each data bar is a mean ± SD of each experiment run in duplicates (n = 3).
Fig. 4.
Identification of gp41 site on gC1qR using gC1qR deletion mutants.
The same strategy as in Fig. 2 was applied in that the deletion mutants that showed consistently diminished binding to gp41-coated wells were selected and compared. Microtiter plate wells were first coated with 2 μg/ml HIV-gp41 (1 h, 37 °C) as described in the legend to Fig. 3 and after washing and blocking, the bound gp41 was reacted (O/N, 4C) with 2.5 μg/ml of biotinylated WT gC1qR or the selected biotinylated gC1qR deletion (Δ) mutants. The single point mutation is indicated as G substitution for W at position 233 (W233G). The position of residues 174–180, which showed significantly reduced binding to gp41 (A), is shown in the gC1qR 3D structure (B). The significance of the data is indicated as: * p ≤ 0.05 and **p ≤ 0.01.
3.3. Interaction of gp41 and gp41-3S with isolated and membrane expressed gC1qR
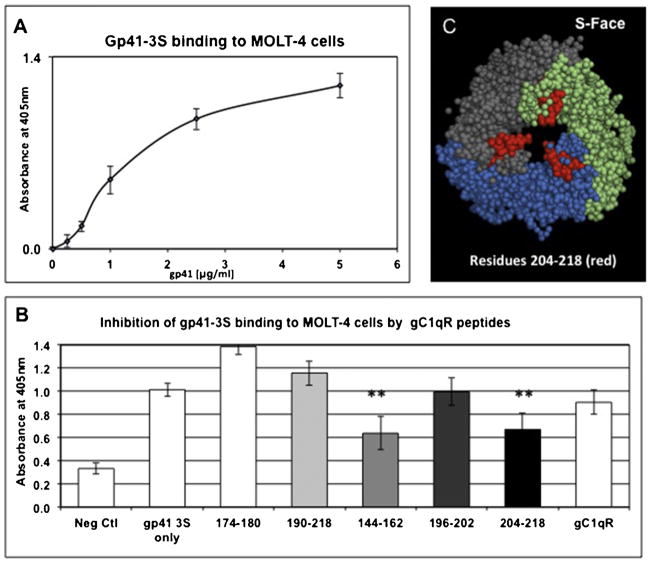
The conserved region of gp41 residues 604–609 (NH2-SWSNKS-COOH) has been reported to contain the binding site for gC1qR (Fausther-Bonvendo et al., 2010). Therefore we generated (GenScript, USA Inc, Piscataway, NJ) a 17-residue synthetic peptide, designated gp41-3S (PWNASWSNKSLDDIWNC) with several residues flanking the putative gC1qR binding site including an introduction of Cys at the C-terminus to allow conjugation to other molecules (e.g. KLH, FITC or biotin). When the ability of this peptide to bind to gC1qR was analyzed, a strong and dose-dependent interaction between gC1qR and the peptide over a wide-range of concentrations was observed (Fig. 5A) and did not matter which of the reactants was coated onto the microtiter plate (Fig. 5A vs B). More importantly, both gp41 (Fig. 5C) as well the gp-41-3S peptide (Fig. 5D) were able to bind to U937 cell surface-expressed gC1qR in a reproducible (n = 3) and dose-dependent manner. In addition, gp41-3S peptide was able to bind to MOLT-4 cells (n = 3), but interestingly this binding was inhibited by two peptides corresponding to gC1qR residues 144–162 and 204–218 and not, as expected by peptide 174–180 (Fig. 6). Residues 144–162 in turn comprise a longer stretch of gC1qR residues within which is contained the 144–148 peptide that, showed inhibitory activity for HCV core protein (Fig. 2).
Fig. 5.
Interaction between HIV-1 gp41-3S and gp41 and recombinant gC1qR and cell surface expressed gC1qR.
(A) Duplicate wells of a microtiter plate were coated with 100 μl of 2 μg/ml gC1qR and incubated (60 min, 37C). After blocking, the bound gC1qR was reacted (1 h, 37 °C) with concentrations of synthetic gp41 3S peptide ranging from 1 to 5 μg/ml (A) and the bound peptide was detected using an affinity purified rabbit anti-gp41 peptide IgG. In (B) the reaction sequence was reversed in that the wells were first coated with 100 μl of 100 μg/ml gp41-3S peptide, then was reacted with increasing concentrations of biotinylated gC1qR (0–100 μg/ml) and the bound gC1qR was detected using alkaline phosphatase conjugated streptavidin. The experiments in (C) and (D) are similar except that microtiter plate-fixed cells were used and reacted with either various concentrations of purified gp41 (C) or the gp41-3S peptide (D) and the bound protein was detected using mAb to gp41 or immunoaffinity purified anti-gp41-3S peptide antibody. Each data point is a mean ± SD run in duplicates (n = 3).
Fig. 6.
Inhibition of gp41-3S peptide binding to MOLT-4 cells by gC1qR peptides.
First the binding of gp41-3S peptide to MOLT-4 cells was verified using microtiter plate fixed MOLT-4 cells (A) as described in detail in Material and Methods. The bound gp41-3S peptide was detected using rabbit anti-gp41-3S antibody. Then the ability of selected gC1qR peptides (corresponding to the deleted regions) to inhibit the binding of gp41-3S peptide to MOLT-4 cells was carried out by preincubation of 100 μl of 5 μg/ml of each peptide with MOLT-4 cells (O/N, 4 °C). Then the unbound gC1qR peptides were removed and the wells further incubated with gp41-3S peptide (1 h, room temp). After incubation the bound gp41-3S peptide was detected with affinity purified anti-gp41-3S peptide, and the reaction developed following the protocol for a standard ELISA (B). For visual clarity, the location of one of the inhibitory peptides (aa204-218) within the gC1qR 3D structure is shown in (C). The significance of the data is indicated as: **p ≤ 0.01; n = 3.
3.4. The ghA domain of C1q contains the binding site(s) for both gC1qR and gp41
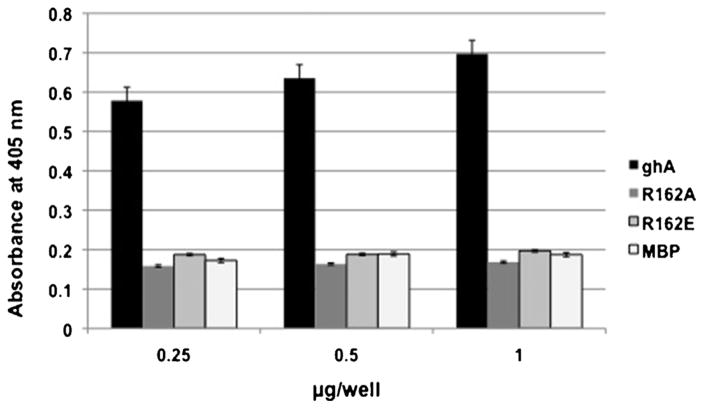
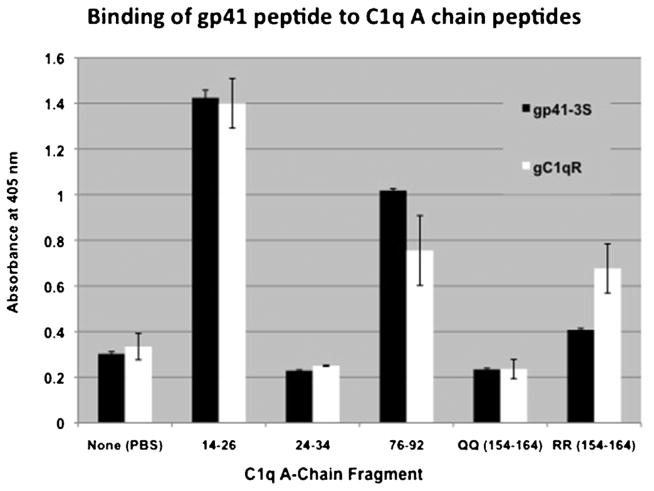
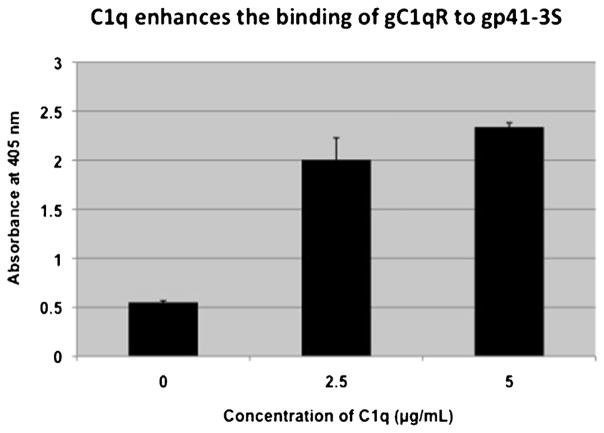
Previous studies using both ligand blot and peptide mapping analyses have shown that gC1qR binds to gC1q predominantly via the A chain (ghA residues 155–164) and moderately to ghB and ghC chains. Furthermore it was shown that the two adjacent Arg residues at positions 162 and 163 in the ghA chain are critical for this binding (Ghebrehiwet et al., 2001). Indeed ghA mutants in which, the Arg at position 162 was substituted with either Ala (R162A) or Glu (R162E) failed to bind gC1qR (Fig. 7) thus confirming earlier results (Ghebrehiwet et al., 2001). Since the HIV-1 gp41 also binds to the A-chain of C1q (Thielens et al., 1993; Quinkal et al., 1999; Kojouharova et al., 2004), we examined if the two proteins bind to the same or overlapping regions of C1q. Employing several synthetic peptides derived from the A-chain of C1q spanning the collagen region (residues 14–26), the globular head region (residues 155–164) as well as a region at the interface between the globular and collagen regions (residues 76–92), we compared the binding of gp-41 peptide and gC1qR. The surprising finding as shown in Fig. 8 is that, while both gC1qR and gp41 were found to bind strongly (p ≤ 0.05; n = 3) to the A-chain residues 14–26, and 76–92 only gC1qR was found to bind to the globular head region residues 154–164 of C1q as expected. Furthermore, the binding of gC1qR to gp41 peptide coated plates was found to be enhanced rather than inhibited when the plates were first pre-incubated with increasing concentrations of C1q (Fig. 9), suggesting that either the gC1qR is binding to the gp41 bound C1q itself or to both the C1q and gp41 suggesting that the binding sites for gp41 and gC1qR on gC1q may not overlap.
Fig. 7.
Interaction of ghA mutants with gC1qR.
Microtiter wells were coated with 1.0, 0.5 or 0.25 μg/well of gC1qR in carbonate buffer and the plate incubated at 4 °C overnight. The next morning, contents were discarded and wells were blocked for 2 h with 2% BSA at 37 °C. After washing with PBS + 0.05% Tween, 2.5 μg/well of ghA wild type, R162A, R162E and MBP was added and the plate was incubated 1.5 h, 37 °C and 1.5 h at 4 °C. Wells then were washed and anti-MBP (1 g/ml) was added, incubated for 1 h 37 °C and the bound protein was detected using horseradish peroxidase conjugated IgG. After addition of OPD buffer, the color developed was read at 450 nm. Each data bar is a mean ± SD run in duplicates (n = 3).
Fig. 8.
Localization of the gp-41 binding site on the A-chain of C1q.
Various peptides from selected regions of the A-chain of C1q were synthesized and 100 μl of 1 mg/ml of each was coated onto ELISA wells in duplicates following the standard ELISA described above. After blocking, 100 μl of either 5 μg/ml of gC1qR or 200 μg/ml of gp41-3S were added and incubated overnight at 4 °C. After removal of the excess unbound reactants and washing, the bound gC1qR and gp41-3S were detected using mAb 74.5.2 or anti-gp41-3S antibody respectively, following standard ELISA procedure. The designation QQ (residues 155–164) indicates that two Q residues at positions 162 and 163 substituted the two R residues. The significance of binding of gp41-3S to C1q peptide 14–16 was (p ≤ 0.05; n = 3) whereas to 76–92 was (p ≤ 0.001).
Fig. 9.
Effect of preincubation of gp41-3S with C1q on the binding of gC1qR to gp41-3S.
Standard ELISA was performed in which microtiter bound gp41-3S peptide (2 μg/ml) was first incubated with increasing concentrations of C1q before addition of a concentration of gC1qR that gives optimal binding under similar experimental conditions. The bound gC1qR in turn was detected with mAb 74.5.2 and further developed following standard ELISA protocol. Each data point is a mean ± SD run in duplicates (n = 3).
4. Discussion
There is a growing list of pathogenic microorganisms (Ghebrehiwet et al., 2011; Ghebrehiwet et al., 1996) that target the gC1qR molecule for their pathogenic design. Among these are two important pathogens: hepatitis C virus (HCV) and HIV-1, both of which suppress or destroy T cells via gC1qR (Fausther-Bonvendo et al., 2010; Chen et al., 1994; Yao et al., 2004). Hepatitis C virus is a major cause of chronic hepatitis worldwide, and is the leading cause of hepatocellular carcinoma and to date there is no effective treatment (Cummings et al., 2009). Similarly, the mortality and morbidity caused by HIV infection is staggering. According to the WHO in 2014 alone close to 37 million people have been diagnosed with HIV-1 infection and approximately 1.5 million have died of AIDS or AIDS-related diseases of which almost 70% are in sub-Saharan Africa. Therefore, understanding at the molecular level, the virus-host cell interaction and, in particular, identification of novel molecular targets that contribute to the pathogenesis of HIV-1 AIDS, will not only give us insight into how they manipulate and escape the onslaught of the innate immune defense mechanism, but may provide us with alternative approaches for designing better or adjuvant therapeutic options.
Previous studies have shown that the 22 KDa, unglycosylated non-enveloped HCV core protein binds gC1qR on CD4+ T cells and induces an anti-proliferative or pro-apoptotic response. Suppression of T cells in turn, is postulated to contribute to an extremely high rate of viral persistence and severe disease progression (Kittlesen et al., 2000; Yao et al., 2004; Cummings et al., 2009). Furthermore, gC1qR has been shown to play a role in the pathogenesis of HCV-related cryoglobulin vascular damage (Sansonno et al., 2009). Mixed cryoglobulinemia (MC) is a lymphoproliferative disorder observed in 10–15% of patients with HCV and the non-enveloped HCV core protein is detected in cryoprecipitable immune complexes and interacts with gC1qR on cells. More importantly, the levels of circulating gC1qR in chronically infected patients with MC is significantly higher than in infected patients without MC, and the circulating gC1qR is found predominantly in complex with C1q and HCV core protein. Together these data suggest that gC1qR contributes to vascular cryoglobulin-induced damage via activation of the classical pathway of complement (Sansonno et al., 2009). Although the binding of HCV core protein has been previously suggested to be in a domain within a long stretch of gC1qR comprising of residues 188–259 (Yao et al., 2004), the precise binding site has not been identified. In this study, we not only confirmed that the interaction site is indeed within the previously suggested region of gC1qR, but also mapped more precisely the HCV core protein site(s) to two gC1qR domains comprising of residues 144–148 and 196–202. These residues make up the loops in each gC1qR monomer that connect β3 to β4 and β5 to β6 (Fig. 10), respectively in the structure of the protein (Jiang et al., 1999; Ghebrehiwet et al., 2002). These prominent loops (which cover up much of the doughnut hole of the gC1qR homotrimer), are highly charged and probably form a binding “cluster in space” in the native molecule. Interestingly, the gC1qR doughnut hole of which these residues are part of also overlaps with the binding site (residues 204–218) for high molecular weight kininogen (Ghebrehiwet et al., 2002), which is recognized by mAb 74.5.2. Similarly, the envelope glycoprotein gp41 of human immunodeficiency virus type 1 (HIV-1) interacts with gC1qR on CD4+ T cells. This interaction is considered to enhance the destruction of uninfected T cells by activated NK cells thereby contributing to the persistently low level of T cell count seen in the late phase of the disease (Fausther-Bonvendo et al., 2010). While the gC1qR interaction site on gp41 has been mapped to the 3S motif comprising of gp41 (Fausther-Bonvendo et al., 2010), the complementary binding site for gp41 on gC1qR has not been identified. In the present study, we have identified gC1qR residues 174–180 as a major binding site for HIV-1 gp41. Domain 174–180, which has recently been shown to contain the cell surface-binding site for soluble gC1qR, is a highly conserved loop, which connects β4 to β5 in the 3D structure of gC1qR (Ghebrehiwet et al., 2011; Jiang et al., 1999; Ghebrehiwet et al., 2002). Previous studies by others have also shown that C1q, the natural ligand for gC1qR, binds to gp41 and the binding site was mapped to a domain, which is adjacent to the 3S motif of gp41 (Thielens et al., 1993; Quinkal et al., 1999). Although the biologic significance still remains to be assessed, it is intriguing to find that the binding sites for gC1qR and C1q be localized in the same stretch of the gp41-3S domain. Furthermore, both gp41-3S and gC1qR appear to compete for the same binding site(s) on the ghA chain of C1q (Fig. 8). The biological implication of a simultaneous binding of gp41 and C1q to gC1qR expressing CD4+ T cells or the binding of gp41 to monocytes such as the monocytic cell line U937, which express membrane-associated C1q in addition to gC1qR should be intriguing and will be vigorously pursued in future studies
Fig. 10.
Location of the gC1qR residues that disrupt trimer formation.
All the deletions retained the trimeric integrity of the gC1qR molecule with the exception of residues 74–95, 204–218 and 212–223, which did not. The location of these residues (shown in red) within the 3D structure as seen from both the S- and M-faces is shown on the left panel. Figures (A and B) on the right panel show the secondary structure of a single monomer of gC1qR showing that the loops connecting β3 to β4 and β5 to β6 strands occupy a large part of the doughnut hole in the homotrimer (A). In (B) the residues comprising the entire monomer are shown for reference. The structures in A and B were adapted from Jiang et al. (1999).
Viral envelope proteins such as the outer envelope protein gp120 of HIV-1 have been recognized to share some structural and functional similarities with C1q (Szabo et al., 2001). These structural similarities had in fact allowed us to predict that proteins like gC1qR that are able to interact with C1q may also interact with isolated gp120 or even with the whole HIV-1 virus. Previous experiments that were performed to test this hypothesis in fact led to the observation that recombinant gC1qR could effectively and dose-dependently inhibit the production of one T-lymphotropic (X4) and one macrophage-tropic (R5) strain in human T cell lines (MT-4 and H9) and human monocyte-derived macrophage cultures, respectively (Szabo et al., 2001). Interestingly, the extent of this inhibition was enhanced when gC1qR was first incubated with and then removed from the target cell cultures before virus infection, compared to that when the cells were infected with gC1qR-HIV mixtures (Szabo et al., 2001). Furthermore, these experiments showed that although gC1qR did not bind to a solid-phase recombinant gp120, it showed a strong and dose-dependent binding of gC1qR to solid-phase CD4 (Szabo et al., 2001). Since gp120 within the gp120/gp41 complex determines viral tropism and the role of gp41 is to mediate fusion between viral and cellular membranes (Caffrey, 2001), gC1qR may prevent the latter process by binding to gp41 within the complex in the early phase of viral infection. While these results imply a beneficial role of gC1qR for the host, a darker side to this interaction is when gp41, potentially released during infection, interacts with cell surface gC1qR to favor viral survival through destruction of CD4+ cells (Stocker et al., 1999; Vieillard et al., 2005). Since the great majority of dying cells during the latter phase of HIV-1 infection are the uninfected CD4+ T cells, the interaction of gp41 and gC1qR can play a significant role in perpetuating the disease process by enhancing the autologous destruction of not only these cells (Fausther-Bonvendo et al., 2010; Sansonno et al., 2009; Stocker et al., 1999), but also of other cells including malignant ones. The destruction of uninfected T cells is therefore postulated to be in part responsible for the persistently low T cell count during viral infection (Fausther-Bonvendo et al., 2010; Stocker et al., 1999; Vieillard et al., 2005). The identification of gC1qR domain 174–180 as a major binding site for gp41, therefore presents an alternative template for the design of either peptide-based or antibody-based therapy that salvages CD4+ T cells from gp41 mediated destruction. However, it should also be pointed out that although peptide 174–180 showed significantly reduced binding to microtiter plate bound gp41 (Fig. 4), it did not block the binding of gp-41S to MOLT-4 cells. Instead peptides 144–162 and 204–218 appear to block this binding on intact cells (Fig. 6) indicating that the interaction of gp41 with cell surface-expressed gC1qR may be different than the binding to isolated gC1qR. Since gC1qR is highly expressed on malignant cells, and it has been shown to induce NKp44L expression, it also has the potential to serve as a molecular target to induce on tumor cells NKp44L expression resulting in autologous NK-cell-mediated tumor cell destruction (Fausther-Bonvendo et al., 2010).
The present studies also identify three potential gp41 binding sites on the A chain of C1q which more or less paralleled that of gC1qR, although gC1qR was not known to bind to the collagen tail of C1q. Interestingly the strongest interaction was seen with the amino terminal peptide 14–26 followed by peptide 76–92, of C1q A chain. Peptide 14–26, contains two adjacent arginine residues (AGRPGRRGRPGLK) and has been shown to be a promiscuous binding site for several important ligands including CRP (Jiang et al., 1992a) DNA (Jiang et al., 1992c), S. Minnesota-derived LPS (Jiang et al., 1992b), and human serum amyloid protein (SAP) (Ying et al., 1993). The second peptide 76–92, which is located at the interface between the collagen and globular head region of C1q (GIKGTKGSPGNNIKDQPR), is also a promiscuous binding site for several non-immunoglobulin molecules including the same proteins that bind to peptide 14–26 mentioned above albeit, with much lower affinity (Trinder et al., 1993). Therefore in addition to the globular head region of its A-chain, C1q possesses at least two sites in its collagen-like region, which support classical pathway complement activation, and is therefore likely to be exploited by pathogen associated molecular ligands and thereby modulate the host immune response.
In summary, the present studies have identified three important gC1qR sequences (Fig. 10) that may have relevance in HCV core binding (residues 144–148 and 196–202) and gp41 binding (residues 174–180). As described in detail in our earlier publication (Ghebrehiwet et al., 2002), all three sequences are part of a highly acidic region of gC1qR that are localized in the S-face (Solution Face) of the 3D structure. In fact residues 144–148 and 196–202 form part of the highly charged residues [141–146 (PTFDGE) and 191–196 (DEVGQE)] that have high solvent mobility and are therefore unlocated in all three chains of the 3D structure (Ghebrehiwet et al., 2002). Residues 174–180 (IKNDDGKKA) on the other hand are part of a highly conserved stretch found in many organisms with only one or two amino acid substitutions (bold and underlined). These include Trichomonas vaginalis G3 (IKNDDLKKA) Borrelia valaisiana (IKNDDIKKA), Borrelia burgdorferi B31 (IKNEDGKKV) and translation initiation factor (IF-2) in zebra fish (IKNDDGRKA). The fact that this sequence is highly conserved across species suggests that it may fulfill highly relevant and possibly multiple biological functions. Furthermore, although the sequences identified indicate that they play a role in the respective bindings of HCV core and gp41, the exact binding sites may involve other residues, which together could form a “cluster-in-space” that defines the perimeter of the binding pocket. However, verification of this postulate could only be accomplished by solving the 3D structure of gC1qR in complex with the HCV core protein and gp41, which we intend to do in the near future. Regardless, the present information should provide an initial platform for the design of more refined peptide-based or antibody-based inhibitors that block interaction of gC1qR with its viral ligands: HCV core and HIV-1 gp-41.
Acknowledgments
This work was supported in part by grants from the National Institutes of Allergy and Infectious Diseases (R01 AI 060866 and R01 AI-084178 to BG), and NIH/NCI Cancer Center support grantP30 CA008748 to MSKCC). This publication represents work that was performed by L.P. while a visiting Graduate Student at Stony Brook University, and was presented in part at the XXIV International Complement Workshop (Chania, Crete, Greece; Oct 10–15, 2012) and published in an abstract form (Mol. Immunol).
Abbreviations
- gC1qR
receptor for the globular “heads” of C1q
- ghA
globular head A chain
- PRR
pathogen recognition receptor
- MBP
maltose binding protein
Footnotes
Conflict of interest
The authors have no financial or personal interests to disclose.
References
- Braun L, Ghebrehiwet B, Cossart P. gC1q-R/p32: a C1q-binding protein is a novel receptor for Listeria monocytogenes. EMBO J. 2000;19:1458–1466. doi: 10.1093/emboj/19.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni R, Roizman B. Open reading frame P-a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M. Model for the structure of the HIV gp41 ectodomain: insight into the intermolecular interactions of the gp41 loop. Bioch Biophys Acta. 2001;1536:116–122. doi: 10.1016/s0925-4439(01)00042-4. [DOI] [PubMed] [Google Scholar]
- Chen A, Gaddipati S, Volkman DJ, Peerschke EIB, Ghebrehiwet B. Human T cells possess specific receptors for C1q: role in activation and proliferation. J Immunol. 1994;153:1430–1440. [PubMed] [Google Scholar]
- Choi Y, Kwon YC, Kim SI, Park JM, Lee KH, Ahn BY. A hantavirus causing hemorrhagic fever with renal syndrome requires gC1qR/p32 for efficient cell binding and infection. Virology. 2009;381:178–183. doi: 10.1016/j.virol.2008.08.035. [DOI] [PubMed] [Google Scholar]
- Cummings KL, Rosen HR, Hahn YS. Frequency of gC1qR+CD4+ T cells increases during acute Hepatitis C virus infection and remains elevated in patients with chronic infection. Clin Immunol. 2009;132:401–411. doi: 10.1016/j.clim.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausther-Bonvendo H, Vieillard V, Sagan S, Bismuth G, Debre P. HIV gp41 engages gC1qR on CD4+ T cells to induce the expression of an NK ligand through the PIP3/H2O2 pathway. PLoS Pathog. 2010:e1000975. doi: 10.1371/journal.ppat.1000975. http://dx.doi.org/10.1371/journal.ppat.1000975. [DOI] [PMC free article] [PubMed]
- Ghebrehiwet B, Lu PD, Zhang W, Lim BL, Eggleton P, Leigh LEA, Reid KBM, Peerschke EIB. Identification of functional domains on gC1q-R, a cell surface protein, which binds to the globular heads of C1q, using monoclonal antibodies and synthetic peptides. Hybridoma. 1996;15:333–342. doi: 10.1089/hyb.1996.15.333. [DOI] [PubMed] [Google Scholar]
- Ghebrehiwet B, Lim BL, Kumar R, Feng X, Peerschke EIB. gC1q-R/p33: a member of a new class of multifunctional and multicompartmental cellular proteins (MMCPs) is involved in inflammation and infection. Immunol Rev. 2001;180:65–77. doi: 10.1034/j.1600-065x.2001.1800106.x. [DOI] [PubMed] [Google Scholar]
- Ghebrehiwet B, Jesty J, Peerschke EIB. gC1q-R/p33: structure-function predictions from the crystal structure. Immunobiology. 2002;205:421–432. doi: 10.1078/0171-2985-00143. [DOI] [PubMed] [Google Scholar]
- Ghebrehiwet B, Tantral L, Titmus MA, Panessa-Warren B, Tortora GT, Wong SS, Warren J. The exosporium of B.cereus contains a binding site for gC1qR/p33: Implications in spore attachment and/or entry. Curr Topics Innate Immun. 2007;598:180–197. doi: 10.1007/978-0-387-71767-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebrehiwet B, Jesty J, Xu S, Vinayagasundaram R, Vinayagasundaram U, Ji Y, Valentino A, Hosszu KK, Mathew S, Joseph K, Kaplan AP, Peerschke EIB. Structure-function studies using deletion mutants identify domains of gC1qR/p33 as potential therapeutic targets for vascular permeability and inflammation. Front Inn Mol Immunol. 2011;2:1–9. doi: 10.3389/fimmu.2011.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Robey FA, Gewurz H. Localization of the sites through which C-reactive protein binds and activates complement to residues 14–26 and 76–92 of the human C1q A chain. J Exp Med. 1992a;175:1373. doi: 10.1084/jem.175.5.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Ying S-C, Kim SB, Gewurz H. Endotoxin activates the classical complement pathway via residues 14–26 of the C1q A chain and peptide 14–26 inhibits this activation. In: Loos M III, editor. International Workshop on C1-The First Component of Complement. Behring Institute Mitteilungen; Mainz Germany: 1992b. p. p47. [Google Scholar]
- Jiang H, Cooper B, Robey FA, Gewurz H. DNA binds and activates complement via residues 14–26 of the human C1q A chain. J Biol Chem. 1992c;267:25597. [PubMed] [Google Scholar]
- Jiang J, Zhang Y, Krainer A, Xu RM. Crystal structure of p32: a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA. 1999;96:3572–3577. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore U, Gupta SK, Perdikoulis MV, Kojouharova MS, Urban BC, Reid KBM. Modular organization of the carboxyl-terminal, globular head region of human C1q A, B, and C chains. J Immunol. 2003;171:812–820. doi: 10.4049/jimmunol.171.2.812. [DOI] [PubMed] [Google Scholar]
- Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, Hahn YS. Interaction between complement receptor gC1q-R and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest. 2000;106:1239–1249. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojouharova MS, Gajeva MG, Tsacheva IG, Zlatarova A, Roumenina LT, Tchorbadjieva MI, Atansov BP, Walters P, Urban BC, Sim RB, Reid KBM, Kishore U. Mutational analyses of the recombinant globular regions of human C1q A, B, and C chains suggest an essential role for arginine and histidine residues in the C1q-IgG Interaction. J Immunol. 2004;172:4351–4358. doi: 10.4049/jimmunol.172.7.4351. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luo Y, Yu H, Peterlin BM. Cellular protein modulates effects of human immunodeficiency virus type 1 TRev. J Virol. 1994;68:3850–3856. doi: 10.1128/jvi.68.6.3850-3856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DA, Russell WC. Adenovirus core protein V interacts with p32-a protein, which is associated with both the mitochondria and the nucleus. J Gen Virol. 1998;79:1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- Mohan KVK, Som I, Ghebrehiwet B, Atreya CD. Cellular gC1q-R/p33 interacts with rubella virus capsid protein and enhances viral infectivity. Virus Res. 2002;85:151–161. doi: 10.1016/s0168-1702(02)00030-8. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Ghebrehiwet B, Peerschke EIB. Staphylococcus aureus protein A recognizes platelet gC1q-R/p33: A novel mechanism for staphylococcal interactions with platelets. Infect Immun. 2000;68:2061–2068. doi: 10.1128/iai.68.4.2061-2068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinkal I, Hernandez JF, Chevallier S, Arlaud GJ, Vernet T. Mapping of the interaction between the immunodominant loop of the ectodomain of HIV-1 gp41 and human complement protein C1q. Eur J Biochem. 1999;265:656–663. doi: 10.1046/j.1432-1327.1999.00757.x. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Tucci FA, Ghebrehiwet B, Lauletta G, Peerschke EIB, Conteduca V, Russi S, Gatti P, Sansonno L, Dammaco F. Role of the receptor for the globular domain of C1q protein in the pathogenesis of hepatitis C virus-related cryoglobulin vascular damage. J Immunol. 2009;183:6013–6020. doi: 10.4049/jimmunol.0902038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Scheller C, Jassoy C. Destruction of primary CD4+ T cells by cell–cell interaction in human immunodeficiency virus type 1 infection in vitro. J Gen Virol. 1999;8:1907–1911. doi: 10.1099/0022-1317-81-8-1907. [DOI] [PubMed] [Google Scholar]
- Szabo J, Cervenak L, Toth FD, Prohaszka Z, Horvath L, Kerekes K, Beck Z, Bacsi A, Erdei A, Peerschke EI, Füst G, Ghebrehiwet B. Soluble gC1q-R/p33, a cell protein that binds to the globular heads of C1q, effectively inhibits the growth of HIV-1 strains in cell cultures. Clin Immunol. 2001;99:222–231. doi: 10.1006/clim.2001.5013. [DOI] [PubMed] [Google Scholar]
- Thielens NM, Bally IM, Ebenbichler CF, Dierich MP, Arlaud GJ. Further characterization of the interaction between C1q subcomponent of human C1 and the transmembrane envelope glycoprotein gp41 of HIV-1. J Immunol. 1993;151:6583–6592. [PubMed] [Google Scholar]
- Trinder PKE, Maeurer MJ, Kaul M, Petry F, Loos M. Functional domains of the human C1q A-chain. Behring Inst Mitt. 1993;93:180–188. [PubMed] [Google Scholar]
- Vieillard V, Strominger JL, Debré P. NK cytotoxicity against CD4+ T cells during HIV-1 infection: a gp41 peptide induces the expression of an NKp44 ligand. Proc Natl Acad Sci USA. 2005;102:10981–10986. doi: 10.1073/pnas.0504315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Finnan JE, Middeldorp JM, Hayward SD. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- Watthanasurorot A, Jiravanichpaisal P, Soderhall I, Soderhall K. A gC1qR prevents white spot syndrome virus replication in the freshwater crayfish Pacifastacus leniusculus. J Virol. 2012;20:10844–10851. doi: 10.1128/JVI.01045-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yao ZQ, Eisen-Vandervelde A, Waggoner SN, Cale EM, Hahn YS. Direct binding of Hepatitis C virus core to gC1qR on CD4+ and CD8+ T cells leads to impaired activation of Lck and Akt. J Virol. 2004;78:6409–6419. doi: 10.1128/JVI.78.12.6409-6419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SC, Gewurz AT, Jiang HX, Gewurz H. Human serum amyloid P component oligomers bind and activate the classical complement pathway via residues 14–26 and 76–92 of the A chain of the collagen-like region of C1q. J Immunol. 1993;150:169. [PubMed] [Google Scholar]
- Yu L, Lowenstein PM, Zhang Z, Green M. In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J Virol. 1995;69:3017–3023. doi: 10.1128/jvi.69.5.3017-3023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]