Abstract
BTK kinase is a member of the TEC kinase family and is a key regulator of the B-cell Receptor (BCR)-mediated signaling pathway. It is important for B-cell maturation, proliferation, survival and metastasis. Pharmacological inhibition of BTK is clinically effective against a variety of B-cell malignances, such as MCL, CLL and AML. MNK kinase is one of the key downstream regulators in the RAF-MEK-ERK signaling pathway and controls protein synthesis via regulating the activity of eIF4E. Inhibition of MNK activity has shown moderate efficacy for AML cell lines proliferation. Through a structure-based drug design approach, we have discovered a selective and potent BTK/MNK dual kinase inhibitor (QL-X-138), which exhibits covalent binding to BTK and non-covalent binding to MNK. Compared to the BTK kinase inhibitor (PCI-32765) and the MNK kinase inhibitor (cercosporamide), QL-X-138 displays a stronger anti-proliferative effect against a variety of B-cell cancer cell lines, as well as AML and CLL primary patient cells. The agent can effectively arrest the growth of lymphoma and leukemia cells at the G0–G1 stage and can induce strong apoptotic cell death. These results demonstrated that simultaneous inhibition of BTK and MNK kinase activity might be a new therapeutic strategy for B-cell malignances.
Keywords: Lymphoma, Leukemia, B-cell malignances, BTK, MNK, dual inhibition, kinase inhibitors
Introduction
Bruton’s tyrosine kinase (BTK) is a non-receptor tyrosine kinase discovered in the early 1990s during investigation of inherited immunodeficiency disease X-linked agammaglobulinaemia (XLA).1, 2 It belongs to the TEC tyrosine kinase family and is predominantly expressed in B cells.3, 4 BTK kinase is a key element of B cell receptor (BCR) signaling and plays important roles in the regulation of B-cell survival, activation, proliferation and differentiation.5, 6, 7 Upon BCR activation, BTK kinase translocates to the membrane and gets phosphorylated by Src family kinases at Y551, followed by auto-phosphorylation at Y223, which in turn induces the activation of PLCγ and other signaling pathways, including PI3K/Akt/mTOR, Stat5 and NF-κB.4, 8 Defective BTK function has been linked to a variety of B-cell malignances, such as Chronic Lymphocytic Leukemia (CLL), Mantle Cell Lymphoma (MCL), Diffuse Large B-Cell Lymphoma (DLBCL), Waldenstrom macroglobulinemia (WM) and Multiple Myeloma (MM).8 Pharmacological targeting of BTK kinase has led to the discovery of the kinase inhibitor ibrutinib (PCI-32765), which has been clinically investigated for activity against CLL, MCL, WM, ABC-DLBCL, and MM and recently received regulatory approval for MCL and CLL.9, 10, 11, 12 Recently, PCI-32765 has also demonstrated beneficial effects in AML.13 Other BTK inhibitors, such as GDC-0834 and HM-7124, are being clinically tested for activity in Rheumatoid Arthritis, and CC-292 and NO-774 are being tested against CLL.12
Mitogen-activated protein kinase interacting kinase 1 and 2 (MNK1/2) are serine/threonine kinases ubiquitously expressed in mammalian cells.14 MNK kinases are substrates of ERK and p38 kinases and regulate translation through phosphorylation of serine 209 of eukaryotic initiation factor 4E (eIF4E). eIF4E is a key regulator of translational processes that can induce cell transformation by augmenting translation of proliferation- and survival- promoting proteins such as cyclins, c-Myc and Bcl-xL.15, 16 Elevated eIF4E levels/activity have been shown to promote tumor development and progression in vivo and are observed in about 30% cancers, including those of the colon, breast, lung, and also Hodgkin’s lymphomas.17, 18, 19 Studies show that MNK-mediated phosphorylation of S209 is essential for eIF4E’s role in oncogenic transformation, but not for normal physiological processes; therefore pharmacological inhibition of MNKs may be an attractive approach for cancer therapy.14 MNK kinase inhibitors, such as CGP57380 and cercosporamide, can block MNK-mediated eIF4E phosphorylation and induce dose-dependent inhibition of proliferation as well as increased apoptosis in HCT-116 and B16 cell lines.20 Recently, cercosporamide has been shown to exhibit anti-tumor activity in MV4-11 AML models. In addition, inhibition of MNK kinase has been shown to be effective against the blast crisis stage of chronic myeloid leukemia (CML).21 Collectively, these findings suggest that pharmacological blockage of MNK may be beneficial for some B-cell- mediated malignances.
Despite the significant clinical efficacy of BTK inhibitors and pre-clinical effects observed with MNK inhibitors in B-cell mediated malignances, it is surprising that these inhibitors were found to exhibit more modest activity against cell line models in vitro compared to other targeted inhibitors. Both BTK and MNK inhibitors have been combined with other agents to enhance overall efficacy.22, 23 Given the fact that BTK kinase-mediated BCR signaling is upstream of PI3K/Akt/mTOR signaling and MNK kinase-mediated eIF4E signaling is downstream of RAS/RAF/MEK/ERK and PI3K/Akt/MTOR signaling, we hypothesized that simultaneously inhibiting BTK and MNKs kinases would exert greater anti-proliferation effects than targeting these kinases individually. Here, we present the first potent and selective BTK/MNK dual kinase inhibitor, QL-X-138, through a rational drug design approach. We demonstrate that the dual inhibition leads to induction of greater anti-proliferation effects in lymphomas, leukemia cell lines and CLL/AML primary patient cells. Our findings introduce a novel multi-targeted treatment approach for B-cell malignancies.
Materials and Methods
Chemical reagents
QL-X-138 was synthesized in the lab with the procedure provided in the Supplemental Materials section.
Cell lines
The human AML lines, OCI-AML3, SKM-1, NOMO-1, and NB4 were obtained from Dr. Gary Gilliland. HEL cells were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The human AML-derived, FLT3-ITD-expressing line, MOLM14, was provided to us by Dr. Scott Armstrong, Dana Farber Cancer Institute (DFCI), Boston, MA. The human ALL cell lines, derived from the pleural effusion of a child with T-cell ALL, and NALM6 (pre-B) were generous gifts from Dr. Thomas Look and Dr. David Weinstock, respectively. HEL, MOLM14, NOMO-1, NB4, SKM-1, and NALM6 cells were cultured with 5% CO2 at 37°C, at a concentration of 2×105 to 5×105 in RPMI (Mediatech, Inc., Herndon, VA) with 10% fetal bovine serum (FBS) and supplemented with 2% L-glutamine and 1% penicillin/streptomycin. OCI-AML3 cells were cultured in alpha MEM media (Mediatech, Inc, Herndon, VA) with 10% FBS and supplemented with 2% L-glutamine and 1% pen/strep.
We have authenticated the following cell lines through cell line short tandem repeat (STR) profiling (DDC Medical, Fairfield, OH): MOLM14, NOMO-1, HEL, SKM-1, OCI-AML3, and NB4. All cell lines matched >80% with lines listed in the DSMZ Cell Line Bank STR Profile Information.
Primary cells
Mononuclear cells were isolated from AML patients. Mononuclear cells were isolated by density gradient centrifugation through Ficoll-Plaque Plus (Amersham Pharmacia Biotech AB, Uppsala, Sweden) at 2000 rpm for 30 minutes, followed by two washes in 1X PBS. Freeze-thawed cells were then cultured in liquid culture (DMEM, supplemented with 20% FBS). All blood and bone marrow samples from AML patients were obtained through written consent under approval of the Dana-Farber Cancer Institute Institutional Review Board. The ethics committees approved the consent procedure.
Peripheral blood mononuclear cells (PBMCs) from individuals with CLL were isolated by density centrifugation through Ficoll and frozen for each subject. Those subjects with low white counts whose CLL cell purity was expected to be < 85% underwent B cell isolation using RosetteSep. The protocol was approved by the Dana-Farber Harvard Cancer Center Institutional Review Board and all subjects signed written informed consent prior to participation.
Antibodies
P-BTK Y551 (#441355) antibody was obtained from Invitrogen (Carlsbad, CA). eIF4E antibody was obtained from BD Biosciences (San Jose, CA). P-BTK Y223 (#5082), BTK (#3533), peIF4E Ser209 (#9741), P-P70S6K Thr389 (#9234), P70-S6K (#2708), P-4EBP1 Thr37/46 (#2855), 4EBP1(#9644), P-SYK Thr352 (#2717), SYK(#2712), P-MNK Thr197/202(#2111), MNK (#2195), p-PLCγ2 Y1217 (#3871), PLCγ2(#3872), PARP(#9542), Caspase 3(#9662) antibodies were obtained from Cell Signaling Technology (Danvers, MA).
EGFR (T790M) crystallography complexes with QL-X-138
Construct preparation, protein expression and purification of EGFR 696-1022 T790M were done as previously reported.24 The EGFR 696-1022 T790M crystals were grown in the absence of QL-X-138 by hanging drop vapor diffusion. The reservoir solution for crystallization was 0.1M HEPES (pH 7.5), 21% PEG6000, 0.3 M NaCl, and 5 mM tris (2-carboxyethyl)-phosphine (TCEP). The compound was then incorporated by soaking the crystals in the soaking buffer (the reservoir solution supplemented with 1.0 mM QL-X-138) overnight. The complex crystals were flash-frozen in the cryo-protectant that was a 3:1 mixture of the soaking buffer and ethylene glycol. The diffraction data were collected at APS 24-ID-C at 100K and were processed using HKL-3000.25 The structure was solved by Fourier synthesis using the previously reported T790M/WZ-4002 structure (PDB 3IKA) without the compound, water molecules and ions as the starting model. The structure was then refined using Phenix and Coot.26, 27 The inhibitor was modeled into the closely fitting positive Fo-Fc electron density and then included in the following refinement and fitting cycles. Topology and parameter files for the inhibitor were generated using PRODRG.28 Data collection and structure refinement statistics were listed in Supplemental Table 2.
BTK and MNK2 docking
Docking QL-X-138 to BTK and MNK2 was performed with Autodock 4.0. PDB 3GEN and 2HW7 were chosen as the model structures for BTK and MNK2 respectively. QL-X-138 was constructed using online demo: CORINA. Then the grip map was set with the 56×56×56 points and the space of 0.375 angstrom. The other parameters were used with the default values. Finally, the docket conformations were sorted by the binding energy.
IP kinase assay (BTK, MNK, mTORC1)
1. BTK IP kinase assay
1). Generating BTK stable cell lines
Retroviral expression vectors were constructed by inserting wild-type (wt) and C481S mutant BTK coding regions into a pMSCV-puro vector (Clontech) that was modified by inserting a 3XFLAG sequence at the 3′ end of the multi-cloning sites. Retroviruses of BTK-WT-FLAG and BTK-MT-FLAG were packaged by transfecting the plasmids with two helper plasmids into 293T cells using Fugene 6 (Roche). After 48 hours, the medium of 293T cells was replaced with DMEM medium containing 1 μg/ml of puromycin for 48 hours. Then cells were maintained in DMEM medium containing 1 μg/ml of puromycin.
2). Kinase assay
The serially diluted QL-X-138 was added to the BTK kinase. After 30 min incubation at RT, ATP (final concentration: 20μM) was added and incubated for 20 min at 37° C. The reactions were stopped by addition of 5X sample buffer. Then the phosphorylation of BTK was detected by western-blot using a pBTK Y551 antibody (BD).
2. MNK IP kinase assay
1). Generating MNK1/2 stable cell lines
Retroviral expression vectors were constructed by inserting full-length wt and MNK1(C190S), MNK2(C225S) coding regions into a pMSCV-puro vector (Clontech) that was modified by inserting a 3XFLAG sequence at the 3′ end of the multi-cloning sites. Retroviruses of MNK1/2-WT-FLAG and MNK1/2-MT-FLAG were packaged by transfecting the plasmids with two helper plasmids into 293T cells using Fugene 6 (Roche). After 48 hours, the medium of 293T cells was replaced with DMEM medium containing 1 μg/ml of puromycin for 48 hours. Cells were then maintained in DMEM medium containing 1 μg/ml of puromycin.
2). Expression of eIF4E
The eIF4E cDNA fragment encoding the full length of eIF4E was cloned into a pTriEx vector (Novagen), in frame with an N-terminal histidine tag for bacterial expression. The protein was expressed in E. Coli cells with an overall yield of 2–5 mg/liter.
3). Kinase assay
The reaction system contained MNK wt or mutant and eIF4E. Serial dilutions of QL-X-138 were added to the protein. After 30 min incubation at room temperature (RT), ATP (final concentration: 20μM) was added and incubated for 30 min at 37°C. The reactions were stopped by addition of 5X sample buffer. Then, the phosphorylation of eIF4E was detected by western-blot using peIF4E Ser209 antibody (CST).
3. mTORC1 IP kinase assay
The HEK293T-Raptor cell line, which stably expresses Raptor-FLAG fusion protein, was cultured in DMEM (Corning, USA). After 48 hours, the cells were collected and lysed, and Raptor –mTORC1 complex was purified by ANTI-FLAG M2 Affinity Gel (Sigma). The substrate 4EBP1was expressed in E. coli with 6XHis tag and purified by Ni-Resin (GE). The kinase reaction was performed by incubation of serially diluted QL-X-138 with mTORC1 and 4EBP1. After a 30 min incubation at RT, ATP was added and incubated for another 30 min at 37° C. The reactions were stopped by addition of 5X sample buffer. Then the phosphorylation of 4EBP1 was detected by western-blot using P-4EBP1 Thr37/46 antibody (CST).
Cell culture and anti-proliferation assay
RV-1, HCT116, HeLa, and DU145 cell lines were grown in DMEM (Corning, USA) and all other cell lines used in this paper were grown in RPMI 1640 (Corning, USA), both containing 10% fetal bovine serum (Gibco, USA), 1% penicillin-streptomycin (Gibco, BRL). Cells were grown in 96-well culture plates (2500–3000/well) for 12h before compounds of various concentrations were added. Cell proliferation was determined after treatment with compounds for 72 hours. Cell Titer-Glo assays were performed according to the manufacturer’s instructions; and luminescence was measured in a multi-label reader (Envision, PerkinElmer, USA). Data were normalized to control groups (DMSO) and represented by the mean of three independent measurements with standard error <20%. GI50 values were calculated using Prism 5.0 (GraphPad Software, San Diego, CA).
Signaling pathway effect examination
Ramos cells were cultured in 10% FBS-containing RPMI and OCI-AML3 cells were cultured in 10% FBS-containing alpha-MEM medium. The serially diluted QL-X-138 was added to cells for 4 hours. The cells were collected and lysed, and cell lysates were analyzed by western blotting.
Apoptosis
Ramos and OCI-AML3 cells were cultured in 10% FBS-containing alpha-MEM medium. The serially diluted QL-X-138 was added to cells for 16/24/48 hours. Then, apoptosis of OCI-AML3 cells and Ramos cells were detected by western-blot using PARP and Caspase 3 antibodies (CST).
Cell cycle analysis
Ramos cells were cultured in 10% FBS-containing RPMI and OCI-AML3 cells were cultured in 10% FBS-containing alpha-MEM medium with QL-X-138 at concentrations of 0.5μM and 1μM. After 24, 48, and 72 hours, cells were collected and stained with PI/RNase staining buffer (BD Pharmingen) and cell cycle progression of cells was analyzed using FACS Calibur (BD).
CLL and AML primary patient cells: anti-proliferation assay
Mononuclear cells were isolated from CLL and AML patients. Cells were tested in liquid culture (DMEM, supplemented with 20% FBS) in the presence of different concentrations of QL-X-138 or PCI-32765. The trypan blue exclusion assay was used for quantification of cells for seeding for CellTiter Glo assays (Promega, Madison, WI) and for proliferation studies as indicated. The CellTiter Glo assay was used for proliferation studies and carried out according to manufacturer instructions. Cell viability is reported as percentages of control (untreated) cells.
Results
Rational design of BTK/MNK dual inhibitor QL-X-138
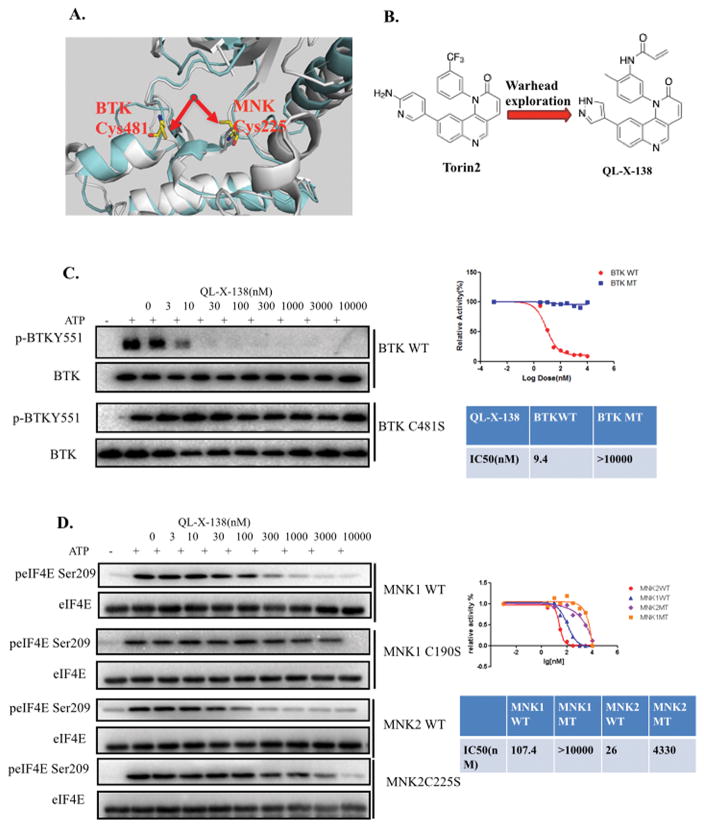
Based on inspection of the available X-ray structures of BTK (PDB ID: 3GEN) and MNK1/2 (PDB ID: 2HW6/2HW7) kinases, we envisioned that a suitably designed inhibitor could potentially target two distinct cysteine residues in the ATP-binding sites of both kinases. BTK kinase possesses a cysteine (C481) in the hinge binding area that is covalently modified by irreversible inhibitors such as PCI-32765 and AVL-292.29, 30 MNK1/2 possesses a cysteine (C190/C225), which is located before the highly conserved DFG(D) motif that marks the start of the kinase activation loop.31 We aimed to develop an inhibitor bearing a mobile acrylamide electrophile that could form a covalent bond with either cysteine residue. (Fig. 1A)
Fig. 1. Discovery of the BTK/MNK dual kinase inhibitor QL-X-138.
(A) Illustration of the rational design concept in X-ray structure of BTK and MNK kinase, BTK(cynate: PDB ID: 3GEN), MNK2(gray, PDB ID:2HW7) (B) Chemical structure of Torin2 and QL-X-138. (C) IP kinase assay of BTK wt and BTKC481S against QL-X-138. (D) IP kinase assay of MNK1/2 wt and MNK1 (C190S), MNK2(C225S) against QL-X-138.
In order to explore this possibility we searched our kinase inhibitor database to identify a scaffold that could provide a suitable platform to incorporate an electrophilic “warhead,” which led to our previously-developed tricyclic mTOR inhibitor.32 Torin2 is an exceptionally potent biochemical inhibitor of mTOR (IC50 = 250 pM) but also less potently inhibits MNK2 (IC50 = 620 nM), with no apparent activity against MNK1 and BTK kinases. Molecular modeling suggested that replacing the CF3 group with an acrylamide group would provide an inhibitor with the ability to potentially form a covalent bond with the cysteine in BTK or MNK1 and MNK2. We replaced the aminopyrimidine side chain of Torin2 with a pyrazole that previous results suggested would greatly diminish mTOR inhibitory potency (Fig. 1B) An IP kinase assay with BTK kinase demonstrated that QL-X-138 exhibits an IC50 of 9.4 nM in a fixed time-point assay with no apparent inhibitory activity against BTKC481S, which indicates that QL-X-138 might be an irreversible inhibitor of BTK kinase. (Fig. 1C) In order to further confirm this irreversible binding mode, we made a reversible version of QL-X-138 by replacing the acrylamide with a propionamide (QL-X-138R). (Supplemental Fig. 1A) An IP kinase assay demonstrated that QL-X-138R completely lost its inhibitory activity against both the wt BTK and BTK C481S mutant, which further supports the notion that QL-X-138 is a covalent inhibitor of BTK kinase. (Supplemental Fig. 1B) In addition, we performed mass spectrum study and detected the site specific labeling of BTK Cys481 by QL-X-138 which confirmed this covalent binding. (Supplemental Fig. 2)
An IP kinase assay with MNK1 and MNK2 demonstrates that QL-X-138 can inhibit these kinases with an IC50 of 107.4 and 26 nM, respectively, which can be rescued up to a concentration of 3μM using mutated MNK1(C190S) and MNK2(C225S) kinases. (Fig. 1D) However, the non-covalent analog, QL-X-138R, is only two-fold less potent as a biochemical inhibitor of MNK1 and MNK2 relative to QL-X-138, demonstrating that this scaffold has excellent non-covalent affinity. (Supplemental Fig. 3A) To further investigate whether QL-X-138 is a covalent MNK2 inhibitor, we performed cellular ‘washout’ experiments and evaluated the kinetics of the recovery of eIF4E Ser209 phosphorylation. The results showed that eIF4E phosphorylation is restored quickly. (Supplemental Fig. 3B) Given the fact that reversible inhibitor binding activity usually is more sensitive to ATP concentrations, we then conducted an ATP competition IP kinase assay with MNK2 kinase. The result showed that with higher ATP concentrations, the inhibitory activity of QL-X-138 against MNK2 got significantly decreased (over 30 fold change comparing 500 μM and 10 μM of ATP). (Supplemental Fig. 3C) Collectively these experiments suggest that QL-X-138 is a non-covalent inhibitor of MNK1/2 kinases. These results demonstrated that we have achieved the development of a potent dual BTK/MNK kinase inhibitor containing dual binding modes, which targets BTK kinase through covalent binding and targets MNK1/2 kinase using reversible binding.
Selectivity and binding mode illustration of QL-X-138
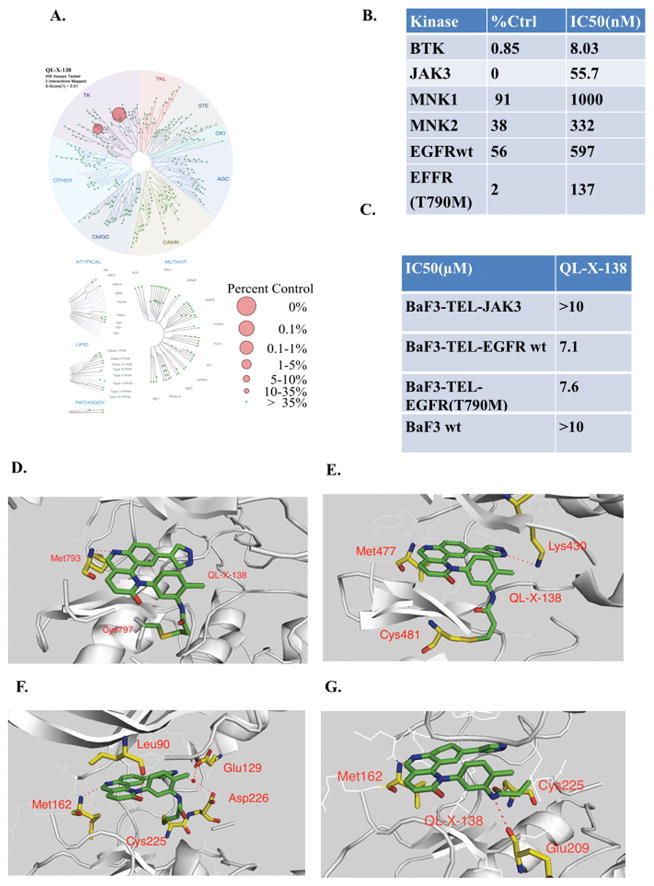
In order to further characterize the selectivity profile of QL-X-138 in the kinome wide, we then subjected QL-X-138 to a kinome selectivity analysis using DiscoveRx’s KinomeScan™ technology at a concentration of 1 μM. QL-X-138 exhibits good overall kinase selectivity with a Score (1) of 0.01.33 QL-X-138 exhibited strong binding to BTK and JAK3 kinases (0.85 and 0 percent activity remaining). (Fig. 2A and Supplemental Table 1) Enzymatic assays (Invitrogen SelectScreen®) confirmed potent inhibition of BTK (IC50 of 8 nM) and JAK3 (IC50 of 55.7 nM). However, further analysis with Tel-fused JAK3-Ba/F3 cells showed that QL-X-138 did not strongly inhibit the JAK3 kinase in a cellular context (GI50>10 μM), which suggests that the biochemical JAK3 inhibitory activity does not predict activity in a cellular assay for this compound series (Fig. 2B,C) Considering EGFR also share a similar cysteine residue in the hinge binding area we also examined QL-X-138 against EGFR and EGFR T790M mutant although it did not show strong binding according to the KinomeScan™ profiling data (56 and 2 percent activity remaining, Supplemental Table 1). It exhibited 597 nM and 137 nM IC50 against EGFR wt and EGFR (T790M) mutant in a biochemical assay, however, these potencies did not translate into the cellular assay again when using TEL transfusion kinase BaF3 isogenic cell lines. (Fig. 2C) Since QL-X-138 was originally derived from an mTOR inhibitor, we further tested QL-X-138 in an mTORC1 IP kinase assay as well as in a cellular assay using HeLa cells. Results validated that QL-X-138 lacks mTOR inhibitory activity, both in vitro and in the cellular assay at concentrations below 1 μM. (Supplemental Fig. 4 A, B) Interestingly, QL-X-138 did not strongly target MNKs in KinomeScan™ panels, which is not consistent with the IP in vitro kinase assay. It is possible that this was due to limitations of the binding assay format or special features of the MNK kinase that favors DFD-out conformation.14
Fig. 2. Demonstrating of binding mode of QL-X-138 and selectivity profiling.
(A) TreeSpot™ demonstration of QL-X-138 selectivity against a panel of 452 kinases with DiscoveRx KinomeScan™ technology with a S score =1. (B) Invitrogen biochemical IC50 of BTK, JAK3, MNK1/2 kinases. (C)Anti-proliferation effect of QL-X-138 against TEL-fused JAK3-BaF3 cells. (D)X-ray crystal structure of QL-X-138 with EGFR (T790M) (PDB ID: 4WD5). (E) Molecular modeling of the binding mode of QL-X-138 with BTK kinase (PDB ID: 3GEN). (F) Molecular modeling demonstration of the designed irreversible binding mode of QL-X-138 with MNK2 kinase (PDB ID: 2HW7). (G) Reversible binding mode analysis of QL-X-138 with MNK2 kinase (PDB ID: 2HW7).
In order to further elucidate the binding mode of QL-X-138, we attempted to crystalize the inhibitor in complex with the targeted kinases. Although we were unable to obtain an X-ray crystal of QL-X-138 bound to BTK or MNK, we instead obtained an EGFR (T790M) protein complex with the inhibitor (PDB ID: 4WD5 and Supplemental Table 2). (Fig. 2D) The structure shows that QL-X-138 forms a hinge binding with Met793 and the acrylamide forms a covalent binding with Cys797, which is located in a position equivalent to Cys481 in the BTK kinase.34 A molecular modeling study with the BTK kinase X-ray structure (PDB: 3GEN) showed that QL-X-138 can form a hinge binding with Met477 and a hydrogen bond with Lys430, and can form a covalent bond with Cys481 with its warhead acrylamide. (Fig. 2E) A docking study of QL-X-138 with MNK2 kinase X-ray structure (PDB ID: 2HW7) suggests that the inhibitor may form a hinge hydrogen bond with Met162 and a salt-bridged hydrogen bond through its pyrazole ring with Gly129 and Asp226. Our initial design involved projecting the acrylamide “warhead” to rotate toward the inside of the ATP binding pocket and forming a covalent bond with Cys225. (Fig. 2F) However, the biological data suggests that we have not achieved covalent binding to Cys225. A detailed molecular modeling study suggests that Glu209 may make a hydrogen bond with amide NH in the “warhead” moiety, which helps to project the carbonyl group toward Cys225 and form another hydrogen bond. (Fig. 2G) These two hydrogen bonds force the double bond to rotate outward against Cys225 and hence prevent the covalent bond formation.
QL-X-138 exhibits greater anti-proliferation activity against lymphoma and leukemia cell lines
We surveyed the anti-proliferative activity of QL-X-138 against a panel of different cancer cell lines. The results showed that QL-X-138 is generally more potent than the BTK kinase inhibitor PCI-32765 and MNK kinase inhibitor cercosporamide. (Table 1) For the cell lines that do not express BTK kinase, such as RV-1, DU-145, HCT-116, HeLa and K562, QL-X-138 did not exhibit apparent anti-proliferation activity at concentrations below 10 μM. For those cell lines that have been validated for expression of BTK kinase, such as Burkitt lymphoma cell line (Ramos), DLBCL cell line (U2932), AML cell lines (MOLM14, OCI-AML3, SKM-1, NB4, HEL, NOMO-1 and U937) and an ALL cell line (NALM6), QL-X-138 exhibited a GI50 of around 1 μM. However, neither the BTK kinase inhibitor, PCI-32765, nor the putatively selective MNK inhibitor, cercosporamide, exhibited apparent inhibitory effects even at a concentration of 10μM. This indicates that dual inhibition of MNK and BTK kinase may translate into higher anti-proliferative activity as compared to inhibition of each individual kinase alone. This was further validated by an experiment showing that a close analogue of QL-X-138, the previously published BMX kinase inhibitor (BMX-IN-1), which possesses similar BTK kinase activity with QL-X-138 but no apparent MNK kinase activity, exhibits weaker anti-proliferation activity in comparison to QL-X-138.35 (Supplemental Table 3) We also conducted a combination experiment to examine the proliferation inhibitory activity of dual inhibition of BTK and MNK kinase activity. Combining the BTK kinase inhibitor, PCI-32765, and MNK kinase inhibitor, cercosporamide, indeed exhibited combinatorial efficacy against the AML cell lines, NB4 and U937. (Supplemental Fig. 5) Interestingly, the TMD8 cell line, which has been reported to be highly sensitive to BTK inhibitor PCI-32765 (GI50: 0.002 μM), did not exhibit high responsiveness against QL-X-138 (GI50: 0.31 μM). This might be due to the polypharmacology of PCI-32765 which requires further detailed exploration.36, 37, 38
Table 1.
QL-X-138 anti-proliferation effect on a panel of cancer cell lines
| Cell line | Cell type | QL-X-138 | PCI-32765 | Cercosporamide |
|---|---|---|---|---|
| (GI50:μM) | (GI50:μM) | (GI50:μM) | ||
| RV-1 | Prostate | >10 | >10 | >10 |
| DU-145 | Prostate | >10 | >10 | >10 |
| HCT-116 | Colon | >10 | >10 | >10 |
| HeLa | Cervical | >10 | >10 | >10 |
| K562 | CML | >10 | >10 | >10 |
| TMD8 | DLBCL | 0.31 | 0.002 | >10 |
| U2932 | DLBCL | 1.2 | >10 | >10 |
| Ramos | B-lymphoma | 0.49 | 3.9 | >10 |
| OCI-AML3 | AML | 1.4 | >10 | >10 |
| SKM-1 | AML | 0.4 | 3.9 | >10 |
| NOMO-1 | AML | 0.23 | >10 | >10 |
| NB4 | AML | 0.95 | 7.5 | >10 |
| HEL | AML | 1.2 | 4 | >10 |
| U937 | AML | 1.4 | 6.4 | 3.9 |
| NALM6 | ALL | 0.23 | 2.8 | >10 |
| MEC-1 | CLL | 1.3 | >10 | >10 |
| MEC-2 | CLL | 0.93 | 4.6 | >10 |
| Hs 505.T | CLL | 1 | >10 | >10 |
| REC-1 | MCL | 2.4 | 3.3 | >10 |
QL-X-138 blocks BTK- and MNK-mediated signaling
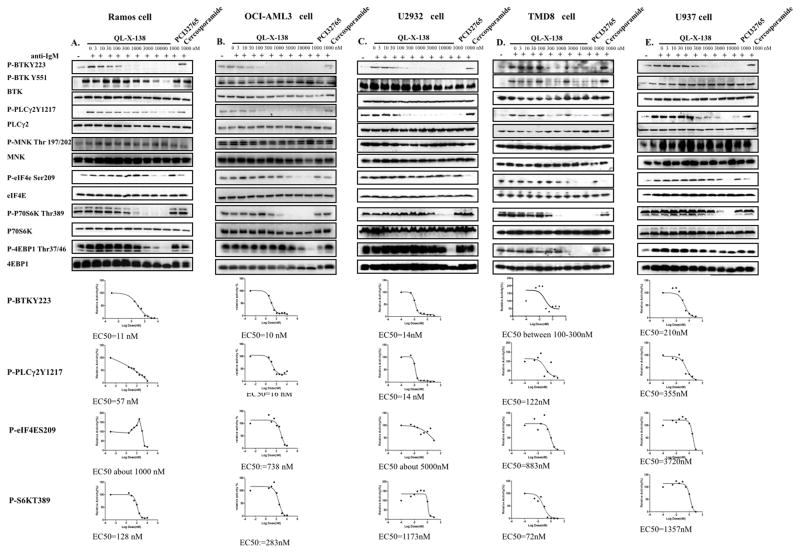
We next investigated the efficacy of QL-X-138 against the relevant signaling pathways in the cellular context. For the Burkitt lymphoma Ramos cell line, QL-X-138 significantly suppressed BTK auto-phosphorylation of Y223 (EC50: 11 nM), but did not affect Y551 as previously observed for PCI-32765. 34 (Fig. 3A) In addition, both QL-X-138 and PCI-32765 strongly blocked phosphorylation of the BTK downstream target PLCγ2 Y1217 (EC50: 57 nM). QL-X-138, PCI-32765 and cercosporamide did not affect MNK phosphorylation, which is consistent with what has been previously reported for cercosporamide.20 However, both QL-X-138 and cercosporamide suppressed the phosphorylation of the MNK downstream target eIF4E S209 at a concentration of 1 μM. QL-X-138 also affected mTOR signaling pathway mediators, S6K and 4EBP1, which is not consistent with the observed EC50 values observed from the non-BTK expressing cell line, HeLa (Supplemental Fig. 4B). This suggests that mTOR signaling is downstream of the BTK-mediated signaling pathway and the weak inhibitory activity of QL-X-138 against mTOR may have been strengthened in this case. Similar results were observed for these drugs against the AML cell lines (OCI-AML3 and U937) and DLBCL cell lines (U2932 and TMD8). This suggests that the dual inhibitory effect of QL-X-138 is not cell line-specific. (Fig. 3 B, C, D, E) Interestingly, for Ramos, OCI-AML-3 and U2932 cell lines, we observed hyper-activation of pMNK T197/202 at concentrations higher than 1μM, despite the down-regulation of peIF4E at the same time point; this suggests that a negative feedback mechanism might exist for the signaling as has been observed previously for mTOR inhibition and consequent re-activation of MNK kinase.39, 40 (Supplemental Fig. 6)
Fig. 3. Effect of QL-X-138 on BTK- and MNK-mediated signaling pathways.
(A) Effect of QL-X-138 on signaling in the Ramos cell line. (B) Effect of QL-X-138 on signaling in the OCI-AML3 cell line. (C) Effect of QL-X-138 on signaling in the U2932 cell line (D) Effect of QL-X-138 on signaling in the TMD8 cell line. (E) Effect of QL-X-138 on signaling in the U937 cell line.
QL-X-138 arrests cell cycle progression and strongly induces apoptosis
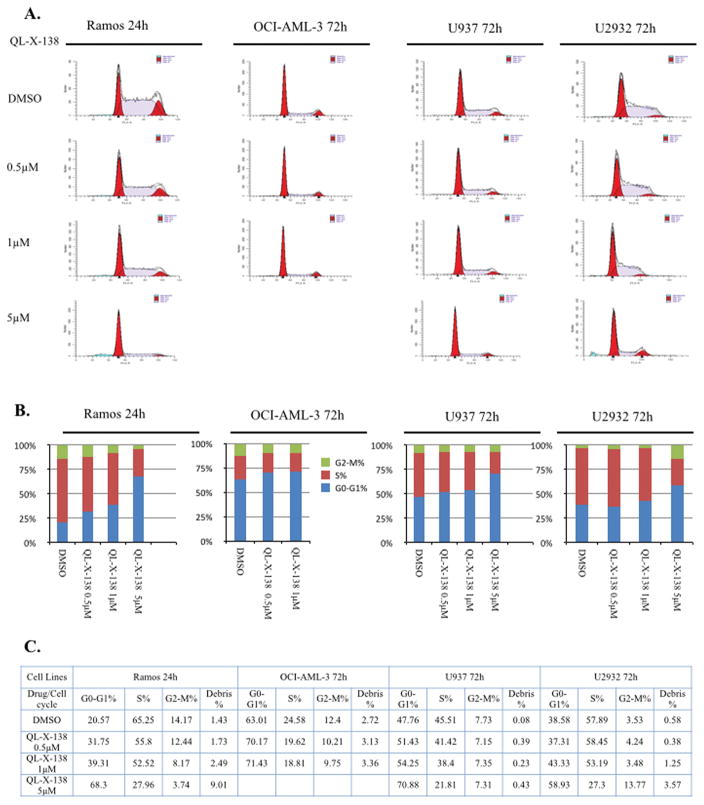
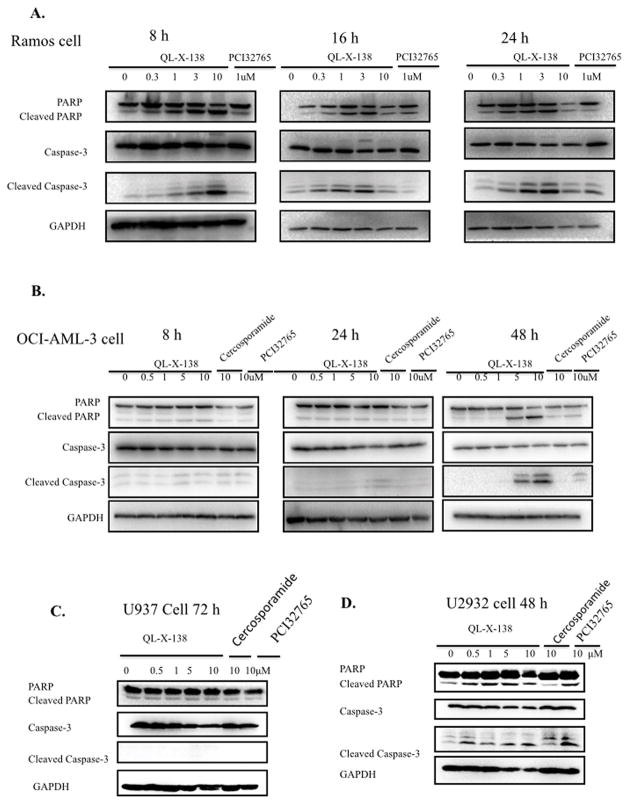
We then investigated the effects of QL-X-138 on the cell cycle progression. For the Ramos cell line, QL-X-138 significantly inhibited cell cycle progression at G0/G1 in a dose-dependent manner at 24h. (Fig. 4), while for AML cell (OCI-AML-3 and U937) and DLBCL cell U2932, QL-X-138 did not cause cell cycle arrest at concentrations up to 1 μM until after 72h of treatment. In Ramos cells, QL-X-138 strongly induced apoptosis at a concentration of 1 μM at 8h, while with a longer drug exposure (24 hours), a concentration of 300 nM was sufficient to induce apoptosis. (Fig. 5A) QL-X-138 induced apoptosis of OCI-AML3 cells at a higher concentration (5 μM) after 48 h of treatment. (Fig. 5B) Similarly, induction of apoptosis by QL-X-138 was observed following 48 h treatment, starting from the concentration of 500 nM. (Fig. 5D) However, even with 72h treatment, U937 cells did not undergo drug-induced apoptosis. (Fig. 5C) FACs analysis also revealed induction of apoptosis of Ramos and U2932 cells, however no apoptosis was observed for OCI-AML-3 and U937 cells; this is consistent with cell cycle results and debris observed at subG0 in these cell lines. (Supplemental Fig. 7)
Fig. 4. QL-X-138 arrest of cell cycle progression in a dose dependent manner.
(A) Flow cytometry illustration of the effect of QL-X-138 on cell cycle progression arrest in Ramos, OCI-AML-3, U937 and U2932 cells with different concentrations and time points. (B) Quantification of the cell cycle stage distribution (bar graph). (C) Effect of QL-X-138 on cell cycle progression (table).
Fig. 5. QL-X-138 demonstrates a time and dose-dependent effect on induction of apoptosis.
(A) QL-X-138 induction of apoptosis at different times and concentrations in Ramos cells. (B) QL-X-138 induction of apoptosis at different times and concentrations in OCI-AML3 cells. (C) QL-X-138 effect on induction of apoptosis at different concentrations in U937 cells. (D) QL-X-138 induction of apoptosis at different concentrations in U2932 cells.
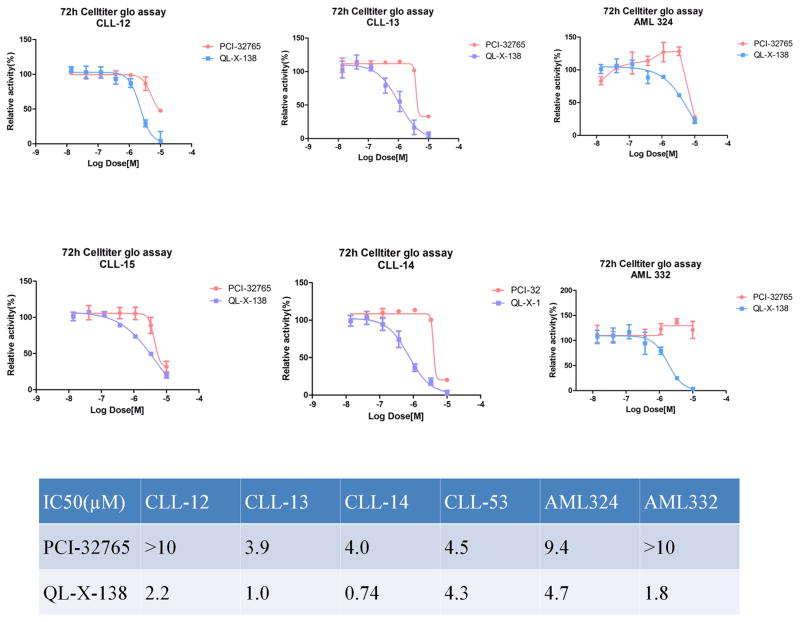
QL-X-138 inhibits the proliferation of primary CLL and AML patient cells
Both PCI-32765 and cercosporamide have shown moderate anti-proliferative activity against CLL and AML primary patient cells (Supplemental Table 4).13, 21 In order to similarly evaluate the potential clinical efficacy of QL-X-138, we tested the drug against a panel of CLL and AML patient cells. The results generally demonstrate a stronger effect of QL-X-138 over PCI-32765 and cercosporamide as single agents. (Fig. 6) Moreover, colony formation of normal bone marrow cells was not inhibited by QL-X-138 at a concentration of 1 μM, which suggests a moderate therapeutic window for the further drug development. (Supplemental Fig. 8)
Fig. 6.
QL-X-138 demonstrates anti-proliferation activity against CLL and AML primary patient cells.
All primary cells were cultured and treated for QL-X-138 for 72h before subject to Cell Titer-Glo analysis.
Discussion
The clinical success of PCI-32765 against CLL and MCL has proven BTK kinase to be a valuable drug discovery target for a variety of BCR constitutively activated B-cell malignances. In addition, an increasing amount of preclinical data has suggested that the BTK kinase inhibitor may be effective against AML and DLBCL.12 However, there has been some controversy regarding whether or not BTK plays a critical role in these diseases, like BCR-ABL does in CML, given the fact that there is no ubiquitous over-expression or gain-of-function mutation of BTK in BCR-mediated malignances.11 This is supported by the fact that a variety of BCR-mediated cancer cell lines do not strongly respond to PCI-32765. It is postulated that PCI-32765, a multi-targeted inhibitor, might also work through other targets and mechanisms, such as the tumor microenvironment, since the pharmacological inhibition of BTK induces molecular effects that cannot be explained by the canonical role of BTK in BCR signaling.4 Hsp90 inhibitors and other agents have been shown to potentiate the efficacy of BTK inhibitors and signaling pathway inhibitors such as those targeting JAK2.8, 41 These studies have made development of a multi-targeted BTK inhibitor an attractive approach for more effectively treating BCR-mediated cancers.42
Both BTK inhibitors and MNK inhibitors have been shown to be effective against leukemia cells. BTK is upstream of the PI3K/Akt/mTOR signaling pathway, and links to the NFκB-mediated transcription signaling pathway via PLCγ. MNK is downstream of the RAF-MEK-ERK signaling pathway and controls the transcription factor eIF4E, which is also linked to the mTOR signaling pathway.8 Therefore, combinatorial inhibition of BTK and MNK kinase activity seems a feasible way to obtain greater efficacy than individually.
The rational design of multiple targeted drugs is usually challenging considering the difficulty of achieving a high level of selectivity, however it is an attractive approach to achieving the improved efficacy against oncogene-driven diseases.43, 44, 45 Our study again exemplifies that it is feasible to achieve a selective multiple targeted inhibitor through a rational design approach. One clear challenge for multi-targeted kinase inhibitors is that it is much more difficult to ascribe the pharmacological effects of the inhibitor to a particular kinase target. Although we have characterized the effects of QL-X-138 as a dual inhibitor of BTK and MNK, we cannot exclude the possibility that other targets, including non-kinase targets, could contribute to the pharmacology exhibited by this inhibitor.
We have successfully developed the first highly potent and relatively selective dual BTK/MNK inhibitor, which exhibits greater anti-proliferative activity and induces apoptosis of a variety of lymphoma and leukemia cells as compared to the BTK inhibitor, PCI-32765, and the MNK inhibitor, cercosporamide. This effect is also observed in primary patient-derived AML and CLL cells. QL-X-138 can serve as a useful probe for further investigation of mechanisms underlying the development and progression of BTK- and MNK-mediated cancers, and provides a useful chemical starting point for developing future clinical candidates bearing this inhibitory profile.
Supplementary Material
Table 2.
Antiproliferation effect of QL-X-138 agansit a panel of CLL and AML patient primary cells
| GI50(μM) | CLL-12 | CLL-13 | CLL-14 | CLL-15 | AML324 | AML332 |
|---|---|---|---|---|---|---|
| PCI-32765 | >10 | 3.9 | 4 | 4.5 | 10 | >10 |
| QL-X-138 | 2.2 | 1 | 0.74 | 4.3 | 7.2 | 1.8 |
Acknowledgments
W. Wang, J. Liu and Q. Liu are supported by the grant of “Cross-disciplinary Collaborative Teams Program for Science, Technology and Innovation (2014–2016)” from Chinese Academy of Sciences. Z. Zhao is supported by Anhui Province Natural Science Foundation Annual Key Program (grant number: 1301023011). We want to thank China “Thousand Talents Program” support for Prof. Q. Liu and “Hundred Talents Program” of The Chinese Academy of Sciences support for Prof. J. Liu, and W. Wang. None of the authors included in this manuscript have a financial conflict of interest.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supplementary information is available at Leukemia’s website
References
- 1.Vetrie D, Vorechovsky I, Sideras P, Holland J, Davies A, Flinter F, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993 Jan 21;361(6409):226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 2.Tsukada S, Saffran DC, Rawlings DJ, Parolini O, Allen RC, Klisak I, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993 Jan 29;72(2):279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 3.Wiestner A. Targeting B-Cell receptor signaling for anticancer therapy: the Bruton’s tyrosine kinase inhibitor ibrutinib induces impressive responses in B-cell malignancies. J Clin Oncol. 2013 Jan 1;31(1):128–130. doi: 10.1200/JCO.2012.44.4281. [DOI] [PubMed] [Google Scholar]
- 4.Hendriks RW, Yuvaraj S, Kil LP. Targeting Bruton’s tyrosine kinase in B cell malignancies. Nat Rev Cancer. 2014 Apr;14(4):219–232. doi: 10.1038/nrc3702. [DOI] [PubMed] [Google Scholar]
- 5.Aoki Y, Isselbacher KJ, Pillai S. Bruton tyrosine kinase is tyrosine phosphorylated and activated in pre-B lymphocytes and receptor-ligated B cells. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10606–10609. doi: 10.1073/pnas.91.22.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurosaki T, Tsukada S. BLNK: connecting Syk and Btk to calcium signals. Immunity. 2000 Jan;12(1):1–5. doi: 10.1016/s1074-7613(00)80153-3. [DOI] [PubMed] [Google Scholar]
- 7.Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: regulators of T-helper-cell differentiation. Nat Rev Immunol. 2005 Apr;5(4):284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 8.D’Cruz OJ, Uckun FM. Novel Bruton’s tyrosine kinase inhibitors currently in development. Onco Targets Ther. 2013;6:161–176. doi: 10.2147/OTT.S33732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013 Jul 4;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013 Aug 8;369(6):507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woyach JA, Bojnik E, Ruppert AS, Stefanovski MR, Goettl VM, Smucker KA, et al. Bruton’s tyrosine kinase (BTK) function is important to the development and expansion of chronic lymphocytic leukemia (CLL) Blood. 2014 Feb 20;123(8):1207–1213. doi: 10.1182/blood-2013-07-515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akinleye A, Chen Y, Mukhi N, Song Y, Liu D. Ibrutinib and novel BTK inhibitors in clinical development. J Hematol Oncol. 2013;6:59. doi: 10.1186/1756-8722-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rushworth SA, Murray MY, Zaitseva L, Bowles KM, MacEwan DJ. Identification of Bruton’s tyrosine kinase as a therapeutic target in acute myeloid leukemia. Blood. 2014 Feb 20;123(8):1229–1238. doi: 10.1182/blood-2013-06-511154. [DOI] [PubMed] [Google Scholar]
- 14.Diab S, Kumarasiri M, Yu M, Teo T, Proud C, Milne R, et al. MAP kinase-interacting kinases-emerging targets against cancer. Chem Biol. 2014 Apr 24;21(4):441–452. doi: 10.1016/j.chembiol.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990 Jun 7;345(6275):544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 16.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007;2(2):e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008 Feb 1;68(3):631–634. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 18.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004 May;10(5):484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 19.Hou J, Lam F, Proud C, Wang S. Targeting Mnks for cancer therapy. Oncotarget. 2012 Feb;3(2):118–131. doi: 10.18632/oncotarget.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 2011 Mar 1;71(5):1849–1857. doi: 10.1158/0008-5472.CAN-10-3298. [DOI] [PubMed] [Google Scholar]
- 21.Altman JK, Szilard A, Konicek BW, Iversen PW, Kroczynska B, Glaser H, et al. Inhibition of Mnk kinase activity by cercosporamide and suppressive effects on acute myeloid leukemia precursors. Blood. 2013 May 2;121(18):3675–3681. doi: 10.1182/blood-2013-01-477216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dasmahapatra G, Patel H, Dent P, Fisher RI, Friedberg J, Grant S. The Bruton tyrosine kinase (BTK) inhibitor PCI-32765 synergistically increases proteasome inhibitor activity in diffuse large-B cell lymphoma (DLBCL) and mantle cell lymphoma (MCL) cells sensitive or resistant to bortezomib. Br J Haematol. 2013 Apr;161(1):43–56. doi: 10.1111/bjh.12206. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 23.Zhang M, Fu W, Prabhu S, Moore JC, Ko J, Kim JW, et al. Inhibition of polysome assembly enhances imatinib activity against chronic myelogenous leukemia and overcomes imatinib resistance. Mol Cell Biol. 2008 Oct;28(20):6496–6509. doi: 10.1128/MCB.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009 Dec 24;462(7276):1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006 Aug;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010 Feb;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010 Apr;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004 Aug;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 29.Pan Z, Scheerens H, Li SJ, Schultz BE, Sprengeler PA, Burrill LC, et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem. 2007 Jan;2(1):58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 30.Evans EK, Tester R, Aslanian S, Karp R, Sheets M, Labenski MT, et al. Inhibition of Btk with CC-292 provides early pharmacodynamic assessment of activity in mice and humans. J Pharmacol Exp Ther. 2013 Aug;346(2):219–228. doi: 10.1124/jpet.113.203489. [DOI] [PubMed] [Google Scholar]
- 31.Jauch R, Jakel S, Netter C, Schreiter K, Aicher B, Jackle H, et al. Crystal structures of the Mnk2 kinase domain reveal an inhibitory conformation and a zinc binding site. Structure. 2005 Oct;13(10):1559–1568. doi: 10.1016/j.str.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, Ahmed T, et al. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2( 1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J Med Chem. 2011 Mar 10;54(5):1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005 Mar;23(3):329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 34.Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010 Jul 20;107(29):13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F, Zhang X, Weisberg E, Chen S, Hur W, Wu H, et al. Discovery of a selective irreversible BMX inhibitor for prostate cancer. ACS Chem Biol. 2013 Jul 19;8(7):1423–1428. doi: 10.1021/cb4000629. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Shaffer AL, 3rd, Emre NC, Ceribelli M, Zhang M, Wright G, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012 Jun 12;21(6):723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010 Jul 20;107(29):13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao W, Wang M, Wang L, Lu H, Wu S, Dai B, et al. Selective antitumor activity of ibrutinib in EGFR-mutant non-small cell lung cancer cells. J Natl Cancer Inst. 2014 Sep;106(9) doi: 10.1093/jnci/dju204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cope CL, Gilley R, Balmanno K, Sale MJ, Howarth KD, Hampson M, et al. Adaptation to mTOR kinase inhibitors by amplification of eIF4E to maintain cap-dependent translation. J Cell Sci. 2014 Feb 15;127(Pt 4):788–800. doi: 10.1242/jcs.137588. [DOI] [PubMed] [Google Scholar]
- 40.Stead RL, Proud CG. Rapamycin enhances eIF4E phosphorylation by activating MAP kinase-interacting kinase 2a (Mnk2a) FEBS Lett. 2013 Aug 19;587(16):2623–2628. doi: 10.1016/j.febslet.2013.06.045. [DOI] [PubMed] [Google Scholar]
- 41.Axelrod M, Ou Z, Brett LK, Zhang L, Lopez ER, Tamayo AT, et al. Combinatorial drug screening identifies synergistic co-targeting of Bruton’s tyrosine kinase and the proteasome in mantle cell lymphoma. Leukemia. 2014 Feb;28(2):407–410. doi: 10.1038/leu.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kummar S, Chen HX, Wright J, Holbeck S, Millin MD, Tomaszewski J, et al. Utilizing targeted cancer therapeutic agents in combination: novel approaches and urgent requirements. Nat Rev Drug Discov. 2010 Nov;9(11):843–856. doi: 10.1038/nrd3216. [DOI] [PubMed] [Google Scholar]
- 43.Morphy R, Kay C, Rankovic Z. From magic bullets to designed multiple ligands. Drug Discov Today. 2004 Aug 1;9(15):641–651. doi: 10.1016/S1359-6446(04)03163-0. [DOI] [PubMed] [Google Scholar]
- 44.Schrattenholz A, Soskic V. What does systems biology mean for drug development? Curr Med Chem. 2008;15(15):1520–1528. doi: 10.2174/092986708784638843. [DOI] [PubMed] [Google Scholar]
- 45.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008 Nov;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.