Summary
The tumor microenvironment contributes important information in gene expression signatures but may be susceptible to sampling variance. Mesenchymal signatures in particular may be influenced by sampling of non-representative regions with high stromal content. Appropriate pathology quality control is required to ensure reproducibility of gene expression signatures.
In this issue of Clinical Cancer Research, Dunne and colleagues explore the impact of intratumoral heterogeneity on gene expression signatures in colorectal cancer (1). They utilize macrodissected invasive front and tumor center for multi-region transcriptome analyses in 25 patients, and demonstrate a higher stromal microenvironment signature in the invasive front. A list of 30 genes are identified that are differentially expressed between the invasive front and tumor center, and they show that this signature is stromally derived. When macrodissected invasive front specimens are applied to a classifier, they identify that the classifier assigns most of the samples into a stem cell/mesenchymal subtype, in contrast to the expected results when the whole tumor or tumor center are utilized for profiling. With this, they call into question the robustness of these gene expression assays for patient classification, specifically the subset defined by a mesenchymal signature.
This work builds on an increasing body of literature in colorectal cancer that has identified molecularly distinct subtypes of colorectal cancer by gene expression, similar to the well-recognized efforts in lymphoma and breast cancer. These studies in colorectal cancers have demonstrated anywhere between two and six subtypes of colorectal cancer. Recent collaboration with many of the research teams who have published in this area resulted in a consensus molecular subtypes (CMS) classification system (2). These efforts identified four primary consensus subtypes, termed CMS1-4. A common feature of many of the individual efforts was a subtype defined by a stem cell or mesenchymal phenotype. This was labeled as CMS4 in the consensus framework, and was associated with a substantially worse recurrence free and overall survival in early-stage colon cancer. Therefore there is clear potential for translation of this subtyping into clinical management.
This finding is a useful addition to the literature, albeit not surprising as intratumoral heterogeneity has been well described for a variety of tumor characteristics, with many studies evaluating differences between the invasive margin and tumor center (3, 4). Several markers of prognostic and biologic importance, such MACC1, E-cadherin, and ZEB2, have been shown to vary in expression between invasive margin and center of the tumor (5–7). Indeed, several of the discriminatory genes identified by Dunne et al. (1), including matrix metallopeptidases and Wnt ligands, have been shown to play important functional roles in the invasive and proliferative characteristics of the associated tumors. The invasive front is common defined as the region incorporating the tumor-stromal interface, which may include regions of tumor budding, in contrast to the tumor margin. Given the extension of the tumor compartment into the stroma in the invasive margin, this typically incorporates greater stromal content than the tumor center (Figure 1). An invasive tumor border, with a greater degree of tumor infiltrating stromal and less well defined border, have been long shown to be a poor-prognostic marker (8). As a result, many studies macrodissecting (as opposed to microdissecting) the invasive margin will obtain a sample with higher amounts of stromal infiltrate, including the work by Dunne et al., and results should be interpreted with this in mind.
Figure 1.
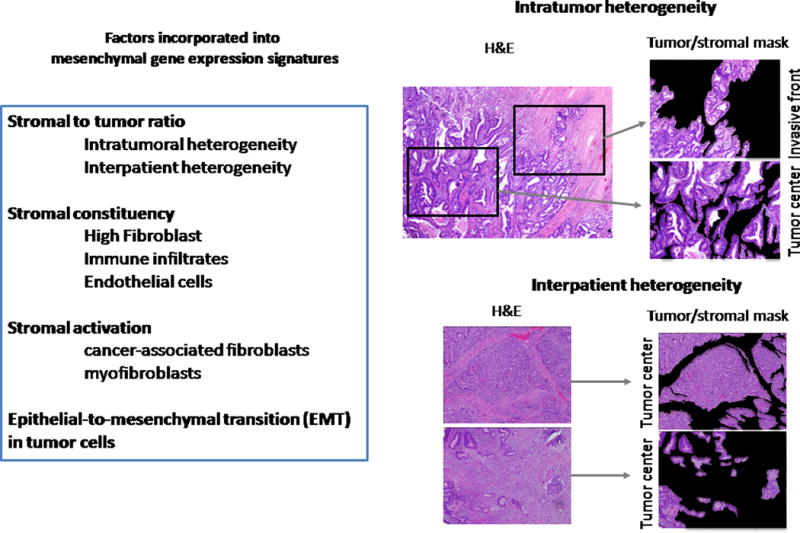
A non-exhaustive list of factors influencing the gene expression profiles from clinical samples, emphasizing impact on mesenchymal signatures. Stromal content is one such factor, as outlined by Dunne et al. (1), as shown in the comparison of the intra- and inter-patient variability of stromal content.
Multiple studies have demonstrated that stromal genes are critical for the classification of the mesenchymal (CMS4-associated) subgroup (9, 10). After microdissection, Calon et al. found that the key prognostic genes in early stage CRC were significantly upregulated in stromal components, including CAFs, endothelial cells, and leukocytes, and CAFs are known to be critical components of tumor microenvironment promoting tumor growth and invasion, and their signatures prognostic in CRC (9). Restriction to gene expression in the epithelial compartment resulted in mesenchymal subgroup patients misclassified into good-prognosis subtypes. We and others have found that when applying the CMS classifier designed for primary tumors to patient-derived xenografts (PDX), we see CMS1-3 proportions similar to the clinical population, but no CMS4 due to differences between murine and human stroma (10). It is clear that tumor microenvironment plays a significant role in the biology underlying the CMS4 subgroup, with a fundamental interaction with the tumor cells producing its characteristic behavior. For example, Calon et al. showed that TGF-β expression in the presence of fibroblasts led to a 200-fold increase in tumor-initiating cells (TICs) while TGF-β expression or presence of fibroblasts alone led to only a 5-fold increase.
Dunne and colleagues’ work (1) does reiterate an important concept of nomenclature, as the distinctions between mesenchymal and EMT signatures are commonly blurred in the gene expression literature. The former is the preferred term in most cases, as it appropriately incorporates many of the features of the underlying microenvironment including fibroblast activation, stromal admixture, and mesenchymal features of tumor cells. In contrast, the use of the term EMT implies the well described transition of the tumor cell compartment from epithelial to mesenchymal biology, which is commonly not possible to discern with the macrodissected source material used in most gene expression studies. As shown in the Figure, multiple potentially biologically important components are incorporated in a classification of a tumor from a mesenchymal subtype, and care should be taken to ensure the proper terminology is used to describe the observations.
Critically, the presence of mesenchymal features from stroma doesn’t preclude contributions from tumor cells with EMT or stem-cell like features. One of the earliest studies to demonstrate the role of epithelial-to-mesenchymal transition in the invasive front of primary tumors identified loss of E-cadherin expression and localization of β-catenin to the nucleus, in contrast to the tumor center in many primary tumors (5). Assessment of EMT features by immunohistochemistry has the added advantage of being able to localize the changes to the tumor compartment, and has similarly shown prognostic features in several settings (6).
A mesenchymal subtype is a recurrent feature of a subset of colorectal cancers with strong and independent prognostic information, and arguably represents critical biology and not solely confounding noise. If the CMS4/mesenchymal subtype was simply an artifact of sampling more stroma, one would not expect to see a persistent association with poor prognosis. Importantly, high collagen levels or other immunohistochemical assessments of stromal content are not a negative prognostic factor in isolation (11). The presence of a poor prognosis mesenchymal/stem-cell subgroup is arguably one of the few unifying conclusions in the gene expression classification literature in colorectal cancer. Similarly, mesenchymal signatures have been identified in multiple tumor types, with a diversity of stromal infiltrates and primary tumor locations, identifying this as a recurring theme in cancer biology (12).
Future gene expression studies should continue to incorporate close pathology evaluation of the source material. Full representation of the entire tumor-bearing region (including invasive margin, tumor center, tumor budding) would most closely match completed gene expression studies. As the stromal content changes with metastases, a related issue that remains to be addressed is the ability to classify gene expression subtypes based on sampling from regions other than the primary tumor. It is possible that deconvolution of the stromal content in the samples may further improve the discriminatory effects of gene expression classifications, for example by reclassifying tumor subtypes with high rates of recurrence but low stromal content into a mesenchymal subgroup. The goal of these gene expression classification efforts is to observe and classify the most global and recurrent patters of biology to improve patient care, and awareness of the limitations and opportunities are clearly needed.
Acknowledgments
Grant Support: J.S. Morris was supported by the NCI of the NIH under award number CA178744 and the National Science Foundation (1550088). S. Kopetz was supported by the NCI of the NIH under award numbers CA172670 and CA184843.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Dunne PD, McArt DG, Bradley CA, O’Reilly PG, Barrett HL, Cummins R, et al. Challenging the cancer molecular stratification dogma: Intratumoral heterogeneity undermines consensus molecular subtypes and potential diagnostic value in colorectal cancer. Clin Cancer Res. 2016 May 5; doi: 10.1158/1078-0432.CCR-16-0032. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karamitopoulou E, Zlobec I, Panayiotides I, Patsouris ES, Peros G, Rallis G, et al. Systematic analysis of proteins from different signaling pathways in the tumor center and the invasive front of colorectal cancer. Human Pathol. 2011;42:1888–96. doi: 10.1016/j.humpath.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Kim TM, Jung SH, An CH, Lee SH, Baek IP, Kim MS, et al. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin Cancer Res. 2015;21:4461–72. doi: 10.1158/1078-0432.CCR-14-2413. [DOI] [PubMed] [Google Scholar]
- 5.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, et al. Variable β-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci. 2001;98:10356–61. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahlert C, Lahes S, Radhakrishnan P, Dutta S, Mogler C, Herpel E, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin Cancer Res. 2011;17:7654–63. doi: 10.1158/1078-0432.CCR-10-2816. [DOI] [PubMed] [Google Scholar]
- 7.Koelzer VH, Herrmann P, Zlobec I, Karamitopoulou E, Lugli A, Stein U. Heterogeneity analysis of Metastasis Associated in Colon Cancer 1 (MACC1) for survival prognosis of colorectal cancer patients: a retrospective cohort study. BMC Cancer. 2015;15:160. doi: 10.1186/s12885-015-1150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halvorsen TB, Seim E. Association between invasiveness, inflammatory reaction, desmoplasia and survival in colorectal cancer. J Clin Pathol. 1989;42:162–6. doi: 10.1136/jcp.42.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–9. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 10.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–9. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 11.Offerhaus GJ, Giardiello FM, Bruijn JA, Stijnen T, Molyvas EN, Fleuren GJ. The value of immunohistochemistry for collagen IV expression in colorectal carcinomas. Cancer. 1991;67:99–105. doi: 10.1002/1097-0142(19910101)67:1<99::aid-cncr2820670119>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 12.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22:609–20. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]


