Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) infections cause significant morbidity and mortality in neonatal intensive care units (NICUs). We characterized the clinical and molecular epidemiology of MRSA strains colonizing NICU patients. Nasal MRSA isolates (n=250, from 96 NICU patients) recovered through active surveillance from 2009-2014 were characterized with Staphylococcal cassette chromosome mec (SCCmec) typing and detection of mupA (marker of high-level mupirocin resistance) and qacA/B (marker associated with chlorhexidine resistance). Factors associated with community-associated (CA-) or healthcare-associated (HA-) MRSA were evaluated. The overall prevalence of MRSA nasal colonization was 3.9%. Of 96 neonates in our retrospective cohort, 60 (63%) were colonized with CA-MRSA strains and 35 (36%) were colonized with HA-MRSA strains. Patients colonized with HA-MRSA were more likely to develop MRSA infections than patients colonized with CA-MRSA (13/35 [37%] vs. 8/60 [13%], p=0.007), although the interval from colonization to infection was shorter in CA-MRSA-colonized infants (0 days [range −1 to 4] versus HA-MRSA-colonized infants, 7 days [−1 to 43], p=0.005). Maternal peripartum antibiotics were associated with CA-MRSA colonization (adjusted odds ratio [aOR] 8.7; 95% confidence interval [CI] 1.7, 45.0); intubation and surgical procedures were associated with HA-MRSA colonization (aOR 7.8; 95% CI 1.3, 47.6 and aOR 6.0; 95% CI 1.4, 24.4, respectively). Mupirocin- and chlorhexidine-resistant MRSA was isolated from 4 and 8 patients, respectively; carriage of a mupirocin-resistant strain precluded decolonization. CA-MRSA strains are prominent in the NICU and associated with distinct risk factors. Given community reservoirs for MRSA acquisition and transmission, novel infection prevention strategies are needed.
INTRODUCTION
From its emergence in 1961 [1, 2] until the late 1990s [3], methicillin-resistant Staphylococcus aureus (MRSA) was largely a nosocomial pathogen, affecting patients undergoing surgery or dialysis, receiving prolonged courses of antibiotics, residing in long-term care facilities, or requiring indwelling catheters or percutaneous medical devices [4]. During the late 1990s, new MRSA strains emerged in the community, affecting otherwise healthy adults and children. These community-associated (CA) MRSA strains are distinct, both clinically and genetically, from traditional healthcare-associated (HA) MRSA strains [5]. The enhanced virulence properties of CA-MRSA strains frequently result in hospitalization, thus introducing these clones into healthcare settings. The predominant strain types in many United States (U.S.) healthcare settings, including neonatal intensive care units (NICUs), have shifted from HA-MRSA to CA-MRSA [6-9], presenting new challenges for clinicians and infection prevention specialists. Additionally, our knowledge of factors associated with CA-MRSA acquisition and transmission in healthcare settings is limited.
Critically ill neonates are exposed to myriad factors which render them vulnerable to MRSA colonization and infection, subjecting them to increased morbidity and mortality as well as prolonged hospitalizations [7, 10-12]. Neonatal MRSA colonization is a demonstrated risk factor for invasive MRSA infection, and patients colonized with MRSA serve as reservoirs for transmission to other patients [9, 13, 14]. This underscores the importance of effective MRSA infection prevention measures in the NICU. To this end, many centers conduct active surveillance to detect MRSA colonization; some centers employ decolonization protocols which may include intranasal mupirocin and occasionally chlorhexidine baths for colonized patients [9, 15]. However, the efficacy of these decolonization measures in preventing MRSA infections is unclear, and many neonates may become recolonized over time [15]. Additionally, widespread use of these topical antimicrobials confers a risk of emerging resistance [16-18], although the prevalence of mupirocin- and chlorhexidine-resistant strains in NICU settings is largely unknown.
To inform infection prevention strategies, the objectives of our study were to measure the prevalence of MRSA colonization in our NICU based on active surveillance; determine the clinical and molecular epidemiology of MRSA strains recovered from NICU patients, specifically to identify factors associated with CA-MRSA versus HA-MRSA colonization; and measure the prevalence of mupirocin and chlorhexidine resistance in NICU MRSA isolates.
PATIENTS AND METHODS
Study Population
This retrospective cohort study was conducted from July 2009 to April 2014 in the St. Louis Children's Hospital (SLCH) Level IV NICU (70 beds: 36 in private rooms and 34 in 2 open bays). The study was approved by the Washington University School of Medicine Institutional Review Board with waiver of consent for the infants and mothers.
An MRSA active surveillance program was implemented in the SLCH NICU in 2004, whereby MRSA surveillance cultures are obtained from the nares of each patient upon admission and weekly thereafter throughout their NICU stay. Infants with positive MRSA cultures are considered colonized and are placed in contact isolation (requiring healthcare workers to wear gowns and gloves when handling the infants) for the remainder of their hospitalization. A standard decolonization protocol was introduced in 2006 for MRSA colonized NICU patients consisting of intranasal 2% mupirocin ointment twice daily for 7 days plus, for infants greater than 30 weeks gestation, a 1-time bath from the neck down with 2% chlorhexidine gluconate cloths.
Infants included in this study were NICU patients who were colonized with MRSA, as detected through active surveillance, and whose isolates were stored by the SLCH Clinical Microbiology Laboratory. We excluded MRSA-colonized neonates whose isolates were not stored or those who were discharged home from a hospital prior to SLCH NICU admission (and thus had exposures outside the hospital environment). The following infant-related data were collected: gender, race (self-reported), gestational age, birth weight, location of birth (patients born at our medical center [inborn] vs. born at an outside institution [outborn]), mode of delivery, presence of multiple gestations, underlying illnesses, surgical procedures, nutrition via nasogastric or orogastric tube, endotracheal intubation, number of ventilator days, presence of a central line, systemic antibiotic exposure, maternal skin-to-skin contact, exposure to maternal or donor breast milk, results of all MRSA surveillance cultures, and incidence of MRSA infections. Mothers’ charts were available for 60 infants and were reviewed for maternal antibiotic exposure prior to delivery.
Surveillance Cultures
MRSA surveillance swabs were collected from the anterior nares of each neonate by the NICU nursing staff upon admission and then weekly and submitted to the SLCH Clinical Microbiology Laboratory; swabs were inoculated onto MRSA chromogenic agar (BBL CHROMagar MRSA, Becton Dickinson [BD], Franklin Lakes, NJ from July 2009 to August 2011; chromID MRSA, bioMerieux, Durham, NC from August 2011 to April 2014). MRSA isolates were frozen and stored at −80°C prior to further analyses. Persistent colonization was defined as 3 or more consecutive positive surveillance MRSA cultures.
Antibiotic susceptibility testing
Disk diffusion testing on Mueller-Hinton agar (BBL, BD) was performed on all isolates to detect resistance to cefoxitin (as an indicator of methicillin resistance), erythromycin, clindamycin (and D-test determination), trimethoprim-sulfamethoxazole, rifampin, tetracycline, ciprofloxacin, linezolid, ceftaroline, and mupirocin according to Clinical and Laboratory Standards Institute guidelines [19]. Subsequently, multiplex polymerase chain reaction (PCR) was performed to detect the mupA and qacA/B genes, which confer high-level mupirocin and chlorhexidine resistance, respectively [20]. MRSA isolates possessing the qacA/B genes were subsequently characterized by repetitive-sequence polymerase chain reactions (repPCR) as described previously [21-23].
Staphylococcal Cassette Chromosome mec (SCCmec) Typing
All MRSA isolates underwent SCCmec genotyping by multiplex PCR testing to detect and differentiate types I through V, as described elsewhere [24]. Strains possessing SCCmec types I, II, or III were classified as HA-MRSA, and strains with SCCmec types IV or V were classified as CA-MRSA.
Statistical Analyses
Data were analyzed using SPSS 22 for Windows (IBM SPSS, Chicago, IL). Factors associated with HA- or CA-MRSA colonization were analyzed by the Mann-Whitney U test (for continuous data) and Pearson's chi-square test (for categorical data). All tests for significance were 2-tailed, and p-values <0.05 were considered significant. Multivariable analysis by backward stepwise logistic regression included variables with p≤0.1 in univariate analysis.
RESULTS
Patient characteristics
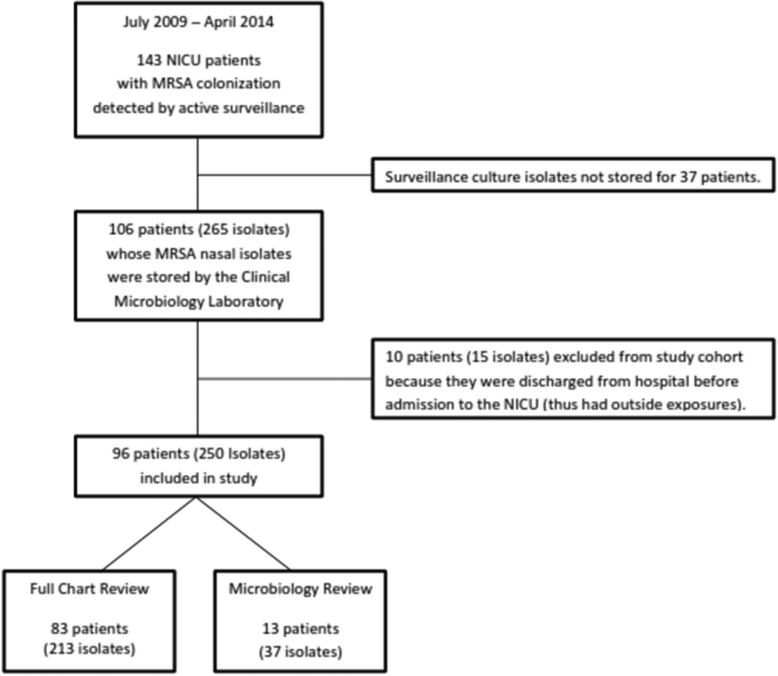
Between July 2009 and April 2014, there were 3,700 admissions to the SLCH NICU; 143 of these infants (3.9%) were found to be colonized with MRSA. From the 143 NICU patients colonized with MRSA over the study period, 265 MRSA isolates from 106 NICU patients had been stored by the SLCH Clinical Microbiology Laboratory and were available for antibiotic susceptibility testing and molecular analysis (Figure 1). Ten patients (15 isolates) were excluded from the study cohort because they had been discharged home from a hospital prior to SLCH NICU admission. Thirteen of the remaining 96 patients (37 isolates) were transferred from an outside hospital to the SLCH NICU more than 48 hours after birth; as complete epidemiologic data were not available from the transferring institution, these infants were included only in the microbial analysis. Of note, these 13 infants were colonized with MRSA at the time of transfer to our NICU. A thorough evaluation of the electronic medical record was conducted for the remaining 83 patients (213 isolates) in addition to microbial testing (Figure 1).
Figure 1.
Flow chart of participant selection. Patients transferred into the SLCH NICU from an outside hospital more than 48 hours after birth and colonized with methicillin-resistant S. aureus upon admission to the SLCH NICU were not included in the full chart review, but the microbiology data for their isolates was included in the analysis.
Within our cohort of 96 neonates, median gestational age was 30 weeks (range 23-41), 55/96 (57%) were male, 53/96 (55%) were very low or extremely low birth weight (<1500 g), and 53/96 (55%) were inborn. The median NICU length of stay was 57 days (range 5-455). Excluding neonates colonized at admission (14/96, 15%), the median time from admission to colonization was 18 days (range 4-133). Patients had a median of 9 colonization cultures obtained (range 1-67) during their NICU stay; the median number of positive MRSA cultures per patient was 2 (range 1-23) (Table 1). Fifteen patients in our study cohort were part of a multiple gestation pregnancy; of these 15 groups of multiples, in 2 sets of twins, both twins were colonized with MRSA. For the remaining MRSA-colonized multiples, their siblings were neither colonized nor infected with MRSA.
Table 1.
Comparison of characteristics of NICU patients with HA-MRSA vs. CA-MRSA nasal colonization cultures, univariate analysis
| Patient Characteristics (N=96 unless otherwise specified) | Total N=96 (%) | HA-MRSAa N=35 | CA-MRSAa N=60 | p-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 55 (57) | 19 (54) | 35 (58) | 0.70 |
| Female | 41 (43) | 16 (46) | 25 (42) | |
| Race | ||||
| Caucasian | 57 (59) | 26 (74) | 30 (50) | 0.02 |
| African-American and Otherb | 39 (41) | 9 (26) | 30 (50) | |
| Gestational age, weeks, median (range) | 29.9 (23.4-41.1) | 29.0 (23.4-41.1) | 30.0 (23.7-39.7) | 0.44 |
| Birth weight | ||||
| Extremely low, <1000g | 34 (35) | 16 (46) | 18 (30) | 0.24 |
| Very low, <1500g | 19 (20) | 4 (11) | 15 (25) | |
| Low, <2500g | 23 (24) | 7 (20) | 16 (27) | |
| Normal, ≥2500g | 20 (21) | 8 (23) | 11 (18) | |
| Location of birth | ||||
| Inbornc | 53 (55) | 14 (40) | 38 (63) | 0.03 |
| Outborn | 43 (45) | 21 (60) | 22 (37) | |
| Mode of delivery | ||||
| Cesarean | 61 (64) | 25 (71) | 36 (60) | 0.26 |
| Vaginal | 35 (37) | 10 (29) | 24 (40) | |
| Multiple gestation | 15 (16) | 7 (20) | 8 (13) | 0.39 |
| Length of NICU stay, median (range), daysd | 57 (5-455) | 82 (12-204) | 56 (5-455) | 0.53 |
| MRSA-colonized upon NICU admission | 14 (15) | 10 (29) | 4 (7) | 0.004 |
| Number of surveillance cultures, median (range) | 9 (1-67) | 10 (2-32) | 8.5 (1-67) | 0.39 |
| Number of positive surveillance cultures, median (range)e | 2 (1-23) | 2 (1-23) | 1.5 (1-18) | 0.22 |
| Persistent colonizationf | 30 (33) | 12 (36) | 17 (30) | 0.56 |
| MRSA infectiong | 22 (23) | 13 (37) | 8 (13) | 0.007 |
| Underlying illnessd,h | 73 (88) | 23 (92) | 50 (88) | 0.57 |
| Intubationd | 54 (65) | 21 (84) | 32 (56) | 0.02 |
| Length of intubation (if intubated, N=54)d | ||||
| >7 days | 28 (52) | 13 (62) | 14 (44) | 0.20 |
| ≤ 7 days | 26 (48) | 8 (38) | 18 (56) | |
| Received nutrition through gastric tubed | 76 (92) | 23 (92) | 52 (91) | 0.91 |
| Surgical procedured | 38 (46) | 15 (60) | 23 (40) | 0.10 |
| Central lined | 60 (72) | 19 (76) | 40 (70) | 0.59 |
| Received systemic antibioticsd | 79 (95) | 25 (100) | 53 (93) | 0.17 |
| Length of systemic antibiotic exposure before first | 6 (0-33) | 7 (1-33) | 6 (0-28) | 0.58 |
| positive culture (if given antibiotics, N=79), median (range), daysd | ||||
| Received clindamycin before first positive cultured | 10 (12) | 7 (28) | 3 (5) | 0.004 |
| Received vancomycin before first positive cultured | 24 (29) | 8 (32) | 16 (28) | 0.72 |
| Maternal peripartum antibiotics (N=60)d,i | 37 (62) | 8 (42) | 28 (70) | 0.04 |
| Skin-to-skin contact with mother (N=75)d | 44 (59) | 18 (75) | 26 (52) | 0.06 |
| Received mother's milkd | 75 (90) | 24 (96) | 50 (88) | 0.25 |
| Received donor milk (N=74)d | 15 (20) | 7 (29) | 8 (16) | 0.20 |
| Time to MRSA colonization, median (range), days | 16 (0-133) | 11 (0-133) | 17 (0-72) | 0.15 |
| Excluding patients colonized at admission, N=82 | 18 (4-133) | 18 (7-133) | 18 (4-72) | 0.66 |
| Received any decolonization measuresd | 73 (88) | 23 (92) | 50 (88) | 0.57 |
| Received both intranasal mupirocin and chlorhexidine bathd | 52 (63) | 18 (72) | 34 (60) | 0.29 |
| Colonized with chlorhexidine-resistant strain | 8 (8) | 1 (3) | 7 (12) | 0.14 |
| Colonized with mupirocin-resistant strain | 4 (4) | 2 (6) | 2 (3) | 0.58 |
Abbreviations: NICU, neonatal intensive care unit; HA-MRSA, healthcare-associated methicillin-resistant Staphylococcus aureus; CA-MRSA, community-associated MRSA.
Staphylococcal cassette chromosome mec (SCCmec) type II (HA-MRSA) or IV/V (CA-MRSA) of first recovered MRSA isolate, N=95 (SCCmec type not able to be determined for 1 isolate).
Other race: 1 biracial (Caucasian/African-American), 1 Asian, and 1 American Indian.
Born at our medical center.
N=83 (13 patients transferred into the SLCH NICU from an outside hospital >48 hours after birth and colonized upon admission to the SLCH NICU were included in microbial analyses only as epidemiologic and clinical data were not available).
401 total positive cultures; 250 isolates (62%) available in laboratory and included in analysis.
Colonized with MRSA at 3 consecutive nasal surveillance cultures; 3 patients with <3 cultures excluded, N=90.
Patients with at least 1 positive MRSA culture that was not a surveillance colonization culture: 15 tracheal aspirate, 2 blood, 2 both tracheal aspirate and blood, 1 urine, 1 peritoneal fluid, 1 eye drainage.
Underlying illness includes congenital heart disease, bronchopulmonary dysplasia, omphalocele, gastroschisis, retinopathy of prematurity, or other significant underlying disorder.
Antibiotics administered to mothers (N=37) included: penicillin (13), ampicillin (12), cefazolin (10), azithromycin (4), amoxicillin (3), amoxicillin-clavulanate (1), ceftriaxone (1), cephalexin (1), erythromycin (1), gentamicin (1), and levofloxacin (1).
Intranasal mupirocin administration was documented in 73/83 (88%) colonized neonates; 52/83 (63%) received both intranasal mupirocin and a chlorhexidine bath. Persistent colonization was seen in 30/90 (33%) patients. Patients who received decolonization measures were less likely to be persistently colonized compared to those who did not, though this did not reach statistical significance (28% [20/71] vs. 56% [5/9], p=0.10).
Molecular Epidemiology
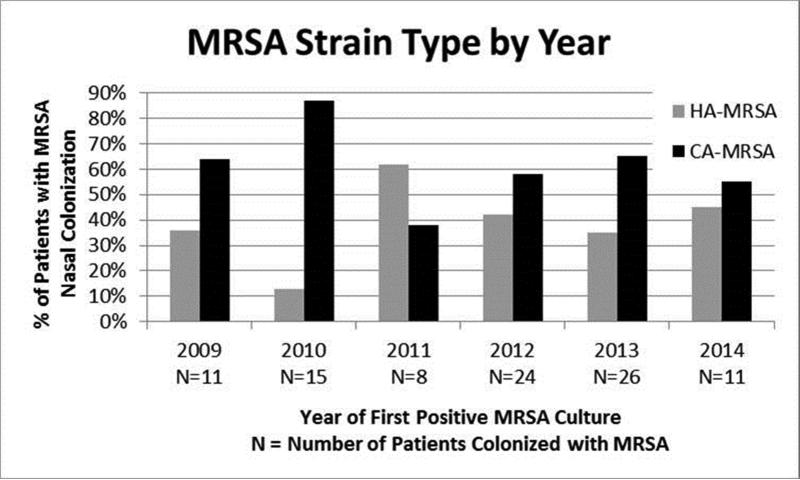
CA-MRSA strains were recovered from 60/96 (63%) patients (SCCmec type IV: 59 patients, 128/250 isolates [51%]; SCCmec type V: 1 patient, 1/250 isolates [0.4%]) (Figure 2). HA-MRSA strains (SCCmec type II) were recovered from 35/96 patients (36%; 120/250 isolates [48%]). One patient's isolate was not typable by SCCmec PCR. Approximately half of the patients (51/96) had multiple positive MRSA surveillance cultures and a third (31/96) had 2 or more MRSA isolates in our collection. When multiple MRSA isolates were recovered from the same patient, all isolates were of the same SCCmec type. Significant factors associated with CA-MRSA colonization (compared to neonates colonized with HA-MRSA) included being inborn (63% [38/60] vs. 40% [14/35], p=0.03) and maternal peripartum antibiotic exposure (70% [28/40] vs. 42% [8/19], p=0.04) (Table 1). Factors associated with HA-MRSA colonization (compared to neonates colonized with CA-MRSA) included Caucasian race (74% [26/35] vs. 50% [30/60], p=0.02), MRSA colonization at the time of admission (29% [10/35] vs. 7% [4/60], p=0.004), endotracheal intubation (84% [21/25] vs. 56% [32/57], p=0.02), and previous exposure to IV clindamycin (28% [7/25] vs. 5% [3/57], p=0.004). Time from NICU admission to MRSA colonization did not differ significantly between neonates colonized with CA- and HA-MRSA (Table 1).
Figure 2.
Proportion of patients colonized in the anterior nares with HA-MRSA (SCCmec type II) and CA-MRSA (SCCmec types IV and V) strains by year in the NICU.
In multivariable analysis, neonates whose mothers received peripartum antibiotics were more likely to be colonized with CA-MRSA (adjusted odds ratio [aOR] 8.7; 95% confidence interval [CI] 1.7, 45.0) than HA-MRSA. Neonates undergoing intubation or surgical procedures were more likely to be colonized with HA-MRSA (aOR 7.8; 95% CI 1.3, 47.6 and aOR 6.0; 95% CI 1.4, 24.4, respectively) than CA-MRSA.
MRSA Infection
MRSA infections were documented in 22 (23%) of the 96 colonized patients in our cohort: tracheitis/pneumonia (n=15), bacteremia (n=2), tracheitis/pneumonia and bacteremia (n=2), and urinary tract infection, peritonitis, and conjunctivitis (n=1 each). Five of the 14 (36%) infants who were colonized at the time of transfer to our NICU subsequently developed an MRSA infection, while 17 of the 82 (21%) infants not colonized at the time of NICU admission developed an MRSA infection (p=0.3). Patients colonized with HA-MRSA were more likely to develop an MRSA infection than patients colonized with CA-MRSA (13/35 [37%] vs. 8/60 [13%], p=0.007). The time from detection of MRSA nasal colonization to development of MRSA infection in the NICU ranged from −1 to 43 days (median 4 days). Of note, 10 of the 22 (45%) patients’ MRSA infections developed within 1 day of their first positive MRSA nasal culture. Patients colonized with CA-MRSA had a shorter time to MRSA infection (median 0 days, range −1 to 4) compared to patients colonized with HA-MRSA (median 7 days, range −1 to 43; p=0.005).
Mupirocin and Chlorhexidine Resistance
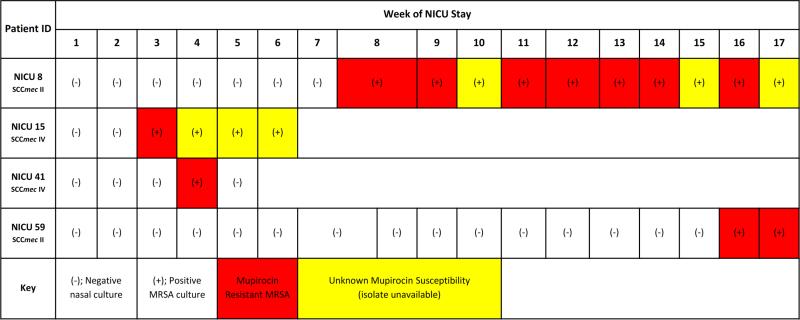
Four of 96 patients (4%; 11 of 250 isolates, 4%) were colonized with mupirocin-resistant (mupA positive) MRSA isolates. All 4 patients were colonized with mupirocin-resistant strains in their first positive surveillance culture, and none had overlapping NICU hospitalizations (Figure 3a). MRSA was eradicated from only 1 of 4 patients (25%) with a mupirocin-resistant strain, compared to 83% (76/92) of patients colonized with mupirocin-susceptible strains (p=0.005). HA-MRSA isolates were more likely to be mupirocin resistant than CA-MRSA strains (8% [9/120] vs. 2% [2/129], p=0.02) (Table 2).
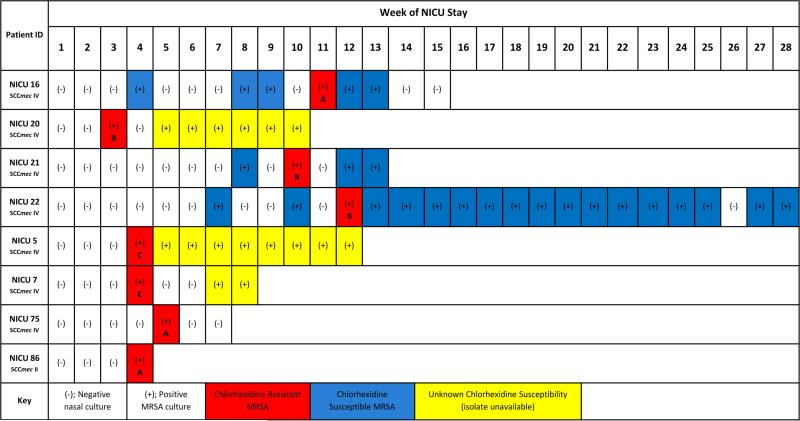
Figure 3.
a. Overview of surveillance cultures of patients colonized with MRSA strains exhibiting mupirocin resistance (possessing the mupA gene). 3b. Overview of surveillance cultures of patients colonized with MRSA strains exhibiting chlorhexidine resistance (possessing the qacA/B genes). Of the 8 chlorhexidine-resistant isolates, 3 distinct strain types were identified, designated as A, B, and C in the figure. Patient IDs NICU 21 and NICU 22 are twins.
Table 2.
Antimicrobial susceptibility profiles of MRSA colonizing isolates recovered from neonates, N=250
| % Susceptible |
||||||||
|---|---|---|---|---|---|---|---|---|
| Isolate Characteristics | CLIa | ERY | SXT | RIF | TET | CIP | MUP | CHG |
| Overall | 33 | 5 | 99 | 99 | 99 | 42 | 96 | 97 |
| Mupirocin | ||||||||
| Resistant (N=11) | 18 | 0 | 100 | 100 | 100 | 9c | N/A | 100 |
| Susceptible (N=239) | 34 | 5 | 99 | 99 | 99 | 43 | N/A | 97 |
| Chlorhexidine | ||||||||
| Resistant (N=8) | 38 | 0 | 100 | 88d | 100 | 63 | 100 | N/A |
| Susceptible (N=242) | 33 | 5 | 99 | 99 | 99 | 41 | 95 | N/A |
| SCCmec typeb | ||||||||
| HA-MRSA (N=120) | 1d | 0d | 100 | 99 | 100 | 12d | 93c | 99c |
| CA-MRSA (N=129) | 62 | 9 | 99 | 99 | 99 | 69 | 98 | 95 |
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; CLI, clindamycin; ERY, erythromycin; SXT, trimethoprim-sulfamethoxazole; RIF, rifampin; TET, tetracycline; CIP, ciprofloxacin; MUP, mupirocin; CHG, chlorhexidine; SCCmec, staphylococcal cassette chromosome mec; HA-MRSA, healthcare-associated MRSA; CA-MRSA, community-associated MRSA.
Note: All isolates were susceptible to linezolid and ceftaroline.
Clindamycin-susceptible isolates that were D-test positive (n=43) were considered clindamycin resistant.
HA-MRSA include SCCmec type II; CA-MRSA includes SCCmec types IV and V; 1 isolate was not typable by SCCmec testing (N=249).
p<0.05.
p≤0.001.
Eight of 96 patients (8%; 8 of 250 isolates, 3%) were colonized with chlorhexidine-resistant (qacA/B positive) MRSA strains (Figure 3b). Among the 8 chlorhexidine-resistant MRSA isolates, 3 distinct strain types were identified by repPCR (reducing the likelihood of clonal expansion of a single strain). Of interest, twins in adjacent rooms were colonized with identical, chlorhexidine-resistant, strains by repPCR (recovered within 6 days). MRSA eradication did not differ between patients colonized with chlorhexidine-resistant and chlorhexidine-susceptible MRSA strains. CA-MRSA isolates were more likely to be chlorhexidine resistant than HA-MRSA isolates (5% [7/129] vs. 1% [1/120], p=0.04) (Table 2). No patients carried both mupirocin- and chlorhexidine-resistant MRSA strains.
Antibiotic Susceptibility
The majority of isolates were susceptible to trimethoprim-sulfamethoxazole, rifampin, tetracycline, linezolid, and ceftaroline (Table 2). Only 33% (82/250) of isolates overall were clindamycin susceptible. HA-MRSA isolates were less likely to be susceptible than CA-MRSA isolates to clindamycin (1% [1/120] vs. 62% [80/129], p<0.001), erythromycin (0% [0/120] vs. 9% [11/129], p=0.001), and ciprofloxacin (12% [14/120] vs. 69% [89/129], p<0.001). Mupirocin-resistant strains were less likely to be susceptible to ciprofloxacin (9% [1/11] vs. 43% [103/239], p=0.025), and chlorhexidine-resistant strains were less likely to be susceptible to rifampin (88% [7/8] vs. 99.6% [241/242], p<0.001).
DISCUSSION
MRSA colonization represents a growing problem for critically ill neonates, posing risk for subsequent invasive MRSA infection, resulting in significant morbidity and mortality. Notwithstanding an active surveillance and isolation program and standardized decolonization protocol, nearly 4% of the infants in our NICU were colonized with MRSA over the study period, consistent with previous NICU studies in the U.S. [9, 25]. Within our study cohort, the predominant colonizing strains were CA-MRSA, and infants colonized with these CA-MRSA strains developed infections more quickly than infants colonized with traditional HA-MRSA strains. These findings are concerning given the insular and protected nature of the NICU environment; that is, these patients have not had prior exposure to the community. Healthcare workers and environmental surfaces have traditionally been considered reservoirs for MRSA transmission among hospitalized neonates. However, as MRSA is now disseminated throughout the community, we must also consider the role of family members [26] and other visitors as vectors for MRSA acquisition among NICU patients, and perhaps incorporate them into our infection prevention strategies, while at the same time preserving the culture of the unit.
Endogenous MRSA colonization poses a 20-fold increased risk for subsequent invasive MRSA infection in neonates [8, 9]. This risk, as well as the potential for colonized neonates to serve as reservoirs for transmission to other vulnerable patients within the NICU, has prompted a focus on infection prevention measures to decrease MRSA colonization rates [9, 13, 14]. While cohorting, isolation, and contact precautions are effective in reducing MRSA prevalence rates [25], many U.S. centers, upon identifying an MRSA colonized patient, employ topical antimicrobials in an effort to eradicate MRSA carriage and thereby prevent MRSA transmission and infection [27, 28]. Among adult patients, the practice of universal chlorhexidine bathing has yielded success in reducing the incidence of MRSA infection in ICUs compared to surveillance and isolation alone [29]. However, this practice may not be a feasible intervention in NICUs given the potential for toxicity in this patient population [30]. At present, there is a paucity of data from randomized trials to inform decolonization practices among critically ill neonates. With the application of mupirocin for decolonization, several centers have reported a reduction, albeit not complete elimination, in the incidence of S. aureus infections in neonates [14, 31]. In the present study, we were encouraged that patients receiving topical antimicrobials were less likely to be persistently colonized with MRSA compared to those not receiving decolonization (28% vs. 56%, respectively); while this finding did not reach statistical significance (p=0.10), it is clinically significant and supports the practice of decolonization in this setting. However, ongoing surveillance is essential as patients may reacquire MRSA colonization, likely due to ongoing exposure to transmission reservoirs [15], such as colonized family members or contaminated environmental surfaces in the healthcare setting.
A potential downside to the widespread use of topical antimicrobials is the emergence of resistance. In this study of MRSA isolates recovered from critically ill neonates, it was reassuring that the overall prevalence of resistance to the topical agents evaluated was low. Consistent with prior investigations of healthy children with skin and soft tissue infections [20], in the present study, carriage of a mupirocin-resistant MRSA strain precluded decolonization efforts, and thus, mupirocin resistance should be considered in patients persistently colonized with MRSA. McNeil et al. examined S. aureus isolates recovered from compromised pediatric patients (specifically those with malignancy and congenital heart disease) at Texas Children's Hospital, revealing a notable rise in the prevalence of chlorhexidine resistance, as high as 45%, coincident with the increased use of chlorhexidine bathing in these populations [32, 33]. Thus, when considering implementation of a decolonization program, the potential benefit of decreasing the incidence of infection must be weighed against the risk of emergence of resistant strains.
The strengths of this study include detailed clinical analysis and molecular characterization of the isolates for both strain typing and resistance to topical anti-infective agents. There are several limitations to this study. The MRSA isolates available for analysis represented a convenience sample of isolates stored in our clinical microbiology laboratory. Moreover, these isolates were collected from patients in a Level IV NICU at a large referral center in the U.S., and may not be generalizable to other institutions or settings. Active surveillance cultures were collected exclusively from the anterior nares, which is less sensitive for detecting MRSA colonization compared to swabbing multiple anatomic sites [34]. Given the nature of retrospective medical record review, we may have underestimated the proportion of infants treated with topical antimicrobials for the purposes of decolonization. Additionally, outside records were not available for all of the outborn patients, and thus these patients could not be included in the epidemiologic analyses. Finally, the small number of isolates carrying the mupA or qacA/B genes limited our ability to identify factors associated with mupirocin and chlorhexidine resistance.
Due to the risk MRSA poses to critically ill neonates, determining viable solutions to preventing infections is essential. To date, infection prevention practices have focused largely on identifying MRSA-colonized neonates and instituting isolation and targeted decolonization for these patients. To more effectively protect these fragile infants, we must expand our scope to fully understand MRSA reservoirs for acquisition, as well as the transmission dynamics of this pathogen, within the NICU. Logistical factors and social barriers have prohibited us from understanding the colonization dynamics of healthcare workers, as detecting colonization in these individuals raises a conundrum surrounding their involvement in patient care. Screening family members and other visitors entering the NICU environment for MRSA colonization raises similar questions. Finally, the role of the NICU environment in harboring and transmitting harmful microorganisms is relatively unexplored. Thus, future studies are needed to understand the interplay among NICU patients, healthcare workers, environmental sources, and community reservoirs. As we are nearing the limit of traditional infection prevention measures to prevent MRSA transmission and infection, this knowledge will inform future strategies, accounting for factors both internal and external to the hospital setting to prevent MRSA acquisition among neonates.
ACKNOWLEDGEMENTS
We would like to thank the employees of the SLCH clinical microbiology laboratory for help with isolate accrual and storage, and Christopher Brenner RN, BSN for assistance with data abstraction. We appreciate the thoughtful reviews of this manuscript by David Hunstad, MD and Jason Newland, MD, MEd.
Financial support Funding for this project was provided by the Children's Discovery Institute of Washington University and St. Louis Children's Hospital; National Institutes of Health grants K23-AI091690 and UL1-TR000448; and grants R01-HS021736 and R01-HS024269 from the Agency for Healthcare Research and Quality. These funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Agency for Healthcare Research and Quality.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential conflicts of interest All authors report no conflicts of interest relevant to this article.
This work was presented in part at IDWeek in San Diego, CA in October 2015.
REFERENCES
- 1.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci USA. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett FF, McGehee RF, Jr., Finland M. Methicillin-resistant Staphylococcus aureus at Boston City Hospital. Bacteriologic and epidemiologic observations. N Engl J Med. 1968;279:441–8. doi: 10.1056/NEJM196808292790901. [DOI] [PubMed] [Google Scholar]
- 3.Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–8. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 4.Klevens RM, Morrison MA, Fridkin SK, Reingold A, Petit S, Gershman K, et al. Community-associated methicillin-resistant Staphylococcus aureus and healthcare risk factors. Emerg Infect Dis. 2006;12:1991–3. doi: 10.3201/eid1212.060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290:2976–84. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 6.Diekema DJ, Richter SS, Heilmann KP, Dohrn CL, Riahi F, Tendolkar S, et al. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol. 2014;35:285–92. doi: 10.1086/675283. [DOI] [PubMed] [Google Scholar]
- 7.Healy CM, Hulten KG, Palazzi DL, Campbell JR, Baker CJ. Emergence of new strains of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Clin Infect Dis. 2004;39:1460–6. doi: 10.1086/425321. [DOI] [PubMed] [Google Scholar]
- 8.Carey AJ, Della-Latta P, Huard R, Wu F, Graham PL, 3rd, Carp D, et al. Changes in the molecular epidemiological characteristics of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2010;31:613–9. doi: 10.1086/652526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seybold U, Halvosa JS, White N, Voris V, Ray SM, Blumberg HM. Emergence of and risk factors for methicillin-resistant Staphylococcus aureus of community origin in intensive care nurseries. Pediatrics. 2008;122:1039–46. doi: 10.1542/peds.2007-3161. [DOI] [PubMed] [Google Scholar]
- 10.Song X, Perencevich E, Campos J, Short BL, Singh N. Clinical and economic impact of methicillin-resistant Staphylococcus aureus colonization or infection on neonates in intensive care units. Infect Control Hosp Epidemiol. 2010;31:177–82. doi: 10.1086/649797. [DOI] [PubMed] [Google Scholar]
- 11.Nelson MU, Gallagher PG. Methicillin-resistant Staphylococcus aureus in the neonatal intensive care unit. Semin Perinatol. 2012;36:424–30. doi: 10.1053/j.semperi.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shane AL, Hansen NI, Stoll BJ, Bell EF, Sanchez PJ, Shankaran S, et al. Methicillin-resistant and susceptible Staphylococcus aureus bacteremia and meningitis in preterm infants. Pediatrics. 2012;129:e914–22. doi: 10.1542/peds.2011-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang YC, Chou YH, Su LH, Lien RI, Lin TY. Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics. 2006;118:469–74. doi: 10.1542/peds.2006-0254. [DOI] [PubMed] [Google Scholar]
- 14.Delaney HM, Wang E, Melish M. Comprehensive strategy including prophylactic mupirocin to reduce Staphylococcus aureus colonization and infection in high-risk neonates. J Perinatol. 2013;33:313–8. doi: 10.1038/jp.2012.102. [DOI] [PubMed] [Google Scholar]
- 15.Popoola VO, Budd A, Wittig SM, Ross T, Aucott SW, Perl TM, et al. Methicillin-resistant Staphylococcus aureus transmission and infections in a neonatal intensive care unit despite active surveillance cultures and decolonization: challenges for infection prevention. Infect Control Hosp Epidemiol. 2014;35:412–8. doi: 10.1086/675594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Upton A, Lang S, Heffernan H. Mupirocin and Staphylococcus aureus: a recent paradigm of emerging antibiotic resistance. J Antimicrob Chemother. 2003;51:613–7. doi: 10.1093/jac/dkg127. [DOI] [PubMed] [Google Scholar]
- 17.Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth JD. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis. 2010;50:210–7. doi: 10.1086/648717. [DOI] [PubMed] [Google Scholar]
- 18.Antonov NK, Garzon MC, Morel KD, Whittier S, Planet PJ, Lauren CT. High prevalence of mupirocin resistance in Staphylococcus aureus isolates from a pediatric population. Antimicrob Agents Chemother. 2015;59:3350–6. doi: 10.1128/AAC.00079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute . Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data. Fourth Edition Wayne, PA: 2014. pp. M39–A4. [Google Scholar]
- 20.Fritz SA, Hogan PG, Camins BC, Ainsworth AJ, Patrick C, Martin MS, et al. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother. 2013;57:559–68. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Feghaly RE, Stamm JE, Fritz SA, Burnham CA. Presence of the bla(z) beta-lactamase gene in isolates of Staphylococcus aureus that appear penicillin susceptible by conventional phenotypic methods. Diagn Microbiol Infect Dis. 2012;74:388–93. doi: 10.1016/j.diagmicrobio.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Kang HP, Dunne WM. Stability of repetitive-sequence PCR patterns with respect to culture age and subculture frequency. J Clin Microbiol. 2003;41:2694–6. doi: 10.1128/JCM.41.6.2694-2696.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez M, Hogan PG, Satola SW, Crispell E, Wylie T, Gao H, et al. Discriminatory indices of typing methods for epidemiologic analysis of contemporary Staphylococcus aureus strains. Medicine. 2015;94:e1534. doi: 10.1097/MD.0000000000001534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boye K, Bartels MD, Andersen IS, Moller JA, Westh H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin Microbiol Infect. 2007;13:725–7. doi: 10.1111/j.1469-0691.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- 25.Nelson MU, Bizzarro MJ, Baltimore RS, Dembry LM, Gallagher PG. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in the decade following implementation of an active detection and isolation program. J Clin Microbiol. 2015;53:2492–501. doi: 10.1128/JCM.00470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milstone AM, Koontz DW, Voskertchian A, Popoola VO, Harrelson K, Ross T, et al. Treating parents to reduce NICU transmission of Staphylococcus aureus (TREAT PARENTS) trial: protocol of a multisite randomised, double-blind, placebo-controlled trial. BMJ Open. 2015;5:e009274. doi: 10.1136/bmjopen-2015-009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milstone AM, Song X, Coffin S, Elward A. Identification and eradication of methicillin-resistant Staphylococcus aureus colonization in the neonatal intensive care unit: results of a national survey. Infect Control Hosp Epidemiol. 2010;31:766–8. doi: 10.1086/653615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerber SI, Jones RC, Scott MV, Price JS, Dworkin MS, Filippell MB, et al. Management of outbreaks of methicillin-resistant Staphylococcus aureus infection in the neonatal intensive care unit: a consensus statement. Infect Control Hosp Epidemiol. 2006;27:139–45. doi: 10.1086/501216. [DOI] [PubMed] [Google Scholar]
- 29.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, et al. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med. 2013;368:533–42. doi: 10.1056/NEJMoa1113849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman AK, Aucott SW, Milstone AM. Safety of chlorhexidine gluconate used for skin antisepsis in the preterm infant. J Perinatol. 2012;32:4–9. doi: 10.1038/jp.2011.148. [DOI] [PubMed] [Google Scholar]
- 31.Milstone AM, Budd A, Shepard JW, Ross T, Aucott S, Carroll KC, et al. Role of decolonization in a comprehensive strategy to reduce methicillin-resistant Staphylococcus aureus infections in the neonatal intensive care unit: an observational cohort study. Infect Control Hosp Epidemiol. 2010;31:558–60. doi: 10.1086/652449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNeil JC, Ligon JA, Hulten KG, Dreyer WJ, Heinle JS, Mason EO, et al. Staphylococcus aureus infections in children with congenital heart disease. J Ped Infect Dis. 2013;2:337–44. doi: 10.1093/jpids/pit037. [DOI] [PubMed] [Google Scholar]
- 33.McNeil JC, Hulten KG, Kaplan SL, Mahoney DH, Mason EO. Staphylococcus aureus infections in pediatric oncology patients: high rates of antimicrobial resistance, antiseptic tolerance and complications. Pediatr Infect Dis J. 2013;32:124–8. doi: 10.1097/INF.0b013e318271c4e0. [DOI] [PubMed] [Google Scholar]
- 34.Fritz SA, Hogan PG, Hayek G, Eisenstein KA, Rodriguez M, Krauss M, et al. Staphylococcus aureus colonization in children with community-associated Staphylococcus aureus skin infections and their household contacts. Arch Pediatr Adolesc Med. 2012;166:551–7. doi: 10.1001/archpediatrics.2011.900. [DOI] [PMC free article] [PubMed] [Google Scholar]