Abstract
Previously, we identified the transcriptional co-activator CITED2 as a potential facilitator of bone metastasis using a murine mammary cancer model. Extending these studies to human breast cancer, it was observed that CITED2 mRNA expression was significantly elevated in patient specimens of metastatic breast cancer relative to primary tumors, with highest levels in metastasis to bone relative to non-bone sites. To further evaluate CITED2 functions in breast cancer metastasis, CITED2 expression was stably reduced in the human breast cancer cell lines MDA-MB-231 and MDA-MB-468, which are metastatic in animal models. While CITED2 knockdown had no effect on cell proliferation, cell migration and invasion were significantly reduced, as was the establishment of metastasis following intra-cardiac administration in athymic nude mice. To explore the mechanism behind these effects, gene expression following CITED2 knockdown in MDA-MB-231 cells by cDNA microarray was performed. As confirmed at the mRNA and protein levels in both MDA-MB-231 and MDA-MB-468 cells, expression of the NF-κB regulator IKKα was significantly reduced along with several NF-κB targets with known roles in metastasis (OPN, MMP9, uPA, SPARC, IL-11 and IL-1β). Further, ChIP assay revealed recruitment of CITED2 to the promoter of IKKα, indicating a direct role in regulating its expression. Consistent with reduced IKKα expression, CITED2 knockdown inhibited both canonical and non-canonical NF-κB signaling. Finally, restoration of IKKα expression following CITED2 knockdown in MDA-MB-231 and MDA-MB-468 cells rescued their invasive ability. Collectively, these data demonstrate that CITED2 modulates metastatic ability in human breast cancer cells, at least in part, through the regulation of IKKα.
Implications
The current study highlights the role of CITED2 in facilitating breast cancer metastasis, partly via regulation of IKKα.
Keywords: CITED2, IKKα, invasion, metastasis, cancer
Introduction
Breast cancer is the most frequently diagnosed cancer in women worldwide and the second most commonly occurring cancer overall. While primary tumors may be effectively treated when detected early, metastatic disease is largely incurable and represents the ultimate cause of mortality in breast cancer patients. It is estimated that ~6% of patients already have metastatic disease at the time of diagnosis while ~20–50% of patients who are initially diagnosed with early stage breast cancer will eventually develop metastasis (1). Sadly, the median survival time for patients with metastatic breast cancer is only 18–30 months. Despite recent research efforts, elucidation of the critical drivers of metastasis and their mechanism of action is lacking. Filling this knowledge gap is essential to the development of novel therapeutic modalities and improving the clinical management of this disease.
The Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain-2 (CITED2) is a non-DNA binding transcriptional co-activator that was originally discovered for its role in development (2–5). As a transcriptional co-activator, CITED2 interacts with several transcription factors such as p300/CBP, Lhx2, TFAP2, Smad2/Smad3, PPARγ and estrogen receptor, modulating their ability to activate gene transcription (6–11). Beyond its involvement in development, CITED2 has also been reported to play a role in cancer, including that of the skin, colon and lung (12–14). Recently, we identified CITED2 as a potential facilitator of breast cancer bone metastasis using a murine mammary cancer model (15). While our preliminary analysis of primary human breast tumor tissues revealed significantly higher levels of CITED2 mRNA relative to normal mammary epithelium (15), its expression pattern in metastatic lesions and functional contribution to human breast cancer metastasis remain unclear.
In this study, we investigated the role of CITED2 in human breast cancer metastasis. Here, we show that in breast cancer patients, CITED2 expression is significantly elevated in metastatic lesions relative to primary tumors, with highest levels in bone metastasis. Further, utilizing two highly invasive breast cancer cell lines, we show that stable knockdown of CITED2 significantly reduces tumor migration and invasion in vitro and the establishment of metastasis in vivo. Lastly, we provide evidence that CITED2 mediates metastatic ability in human breast cancer cells, at least in part, by regulating the expression of IKKα.
Materials and Methods
Cell lines, tissues and treatment
The human breast cancer cell lines MDA-MB-231 and MDA-MB-468 were obtained from American Type Culture Collection, Rockville, MD (2014) and were authenticated using DNA profiling and cytogenetic analysis by the cell bank. Cells were utilized for the experiments within 6 months from the time of resuscitation. MDA-MB-231 cells were maintained in RPMI medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals) and MDA-MB-468 cells were maintained in DMEM medium (Gibco) supplemented with 10% FBS and 1% L-Glutamine (Gibco). To identify genes that are regulated by the NF-κB pathway, cells were treated with 10 µM of PS1145 (Sigma-Aldrich) for 16 hours at 37°C.
Normal mammary epithelium samples, kindly provided by Dr. Saraswati Sukumar (Johns Hopkins University School of Medicine, Baltimore, MD), were prepared from reduction mammoplasty specimens of women with no breast abnormalities. Normal and tumor tissues were obtained from the Surgical Pathology Division of the Johns Hopkins Hospital following the approval of the institutional review board (IRB) of the Johns Hopkins University School of Medicine. For all specimens, required written informed patient consents were obtained as approved by the IRB.
Transfection
To study the effects of CITED2 in human breast cancer metastasis, MDA-MB-231 and MDA-MB-468 cells were infected with the lentiviral shRNA expression vector pLKO.1-puro (Addgene plasmid 8453) containing siRNA sequence specific for scrambled or CITED2. The CITED2 siRNA sequence has been described previously (9, 16). Stable cells were selected in the presence of 1 µg/ml puromycin (Sigma-Aldrich) for one week and utilized for subsequent experiments.
For experiments involving re-expression of IKKα, shCITED2-expressing cells were transiently transfected with a 3:1 ratio of Xtreme gene HP DNA transfection reagent (Roche) and pCR3.1-FLAG-IKKα vector [a kind gift from Hiroyasu Nakano (Addgene plasmid 15467) (17)] or empty vector in OPTI-MEM medium (Gibco) for 24, 48 or 72 hours.
Quantitative (q)RT-PCR
Total RNA from tissue samples and cell lines was extracted using Trizol (Invitrogen) and cDNA was generated using a reverse transcription system (Promega). The qRT-PCR parameters have been described previously (11). Amplification of 36B4 was used as an internal control. Relative expression between samples was calculated by the comparative CT method. The primer sequences used were: CITED2 (sense) 5’-ACCATCACCCTGCCCACC-3’, (antisense) CGTAGTGTATGTGCTCGCCCA; IKKα (sense) 5’-GGCTTCGGGAACGTCTGTC-3’, (antisense) 5’-TTTGGTACTTAGCTCTAGGCGA-3’; OPN (sense) 5’-GAAGTTTCGCAGACCTGACAT-3’, (antisense) 5’-GTATGCACCATTCAACTCCTCG-3’; MMP9 (sense) 5’-GGGACGCAGACATCGTCATC-3’, (antisense) 5’-TCGTCATCGTCGAAATGGGC-3’; uPA (sense) 5’-GGGAATGGTCACTTTTACCGAG-3’, (antisense) 5’-GGGCATGGTACGTTTGCTG-3’; SPARC (sense) 5’-AGCACCCCATTGACGGGTA-3’, (antisense) 5’-GGTCACAGGTCTCGAAAAAGC-3’; IL-11 (sense) 5’-CGAGCGGACCTACTGTCCTA-3’, (antisense) 5’-GCCCAGTCAAGTGTCAGGTG-3’; IL-1β (sense) 5’-ATGATGGCTTATTACAGTGGCAA-3’, (antisense) 5’-GTCGGAGATTCGTAGCTGGA-3’; 36B4 (sense) 5’-GAAGGCTGTGGTGCTGATGG-3’, (antisense) 5’-CCCCTGGAGATTTTAGTGGT-3’.
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissue sections were deparaffinized in xylene (Fisher Scientific) and rehydrated through a graded series of ethanol (Pharmco-AAPER). Sections were immersed in antigen retrieval solution (Dako) and heated in a steamer for 20 minutes. Cooled sections were washed with phosphate buffer saline (PBS, Gibco) and endogenous peroxidase activity was quenched by immersing sections in 3% hydrogen peroxide (Fischer Scientific) for 12 minutes and washed with PBS. Sections were blocked by incubation with protein block solution (Dako) for 30 minutes at room temperature and incubated at 4°C for 18 hours with goat anti-CITED2 (1:500; Everest Biotech). Sections were then sequentially incubated for 15 minutes at room temperature with streptavidin-biotin complex, Tyramide amplification reagent and streptavidin-horse radish peroxidase (HRP) from the DACO CSA Kit (Vector Laboratories). To visualize proteins, the chromogen 3, 3-diaminobenzamindine (DAB; Open Biosystems) was added for two minutes at room temperature. Sections were subsequently washed in water and counterstained with hematoxylin Gill No. 3 (Sigma-Aldrich).
Western analysis
Total protein extracts from cell lines were obtained as previously described (15). Cytoplasmic and nuclear extracts was processed using NE-PER cytoplasmic and nuclear extraction reagents (Thermo Scientific) according to manufacturer’s instructions. Conditioned medium was collected by maintaining the cells in serum free medium. Samples were resolved using SDS-PAGE, transferred to nitrocellulose membrane (Bio-Rad) and probed with sheep anti-CITED2 (1:250; R&D Systems), rabbit anti-IKKα, anti-IκBα, anti-p65, anti-RelB, anti-HDAC1 (1:1000; Cell Signaling Technology), mouse anti-GAPDH (1:10,000; kindly provided by Dr. Shanmugasundaram Ganapathy Kanniappan, Johns Hopkins University School of Medicine, Baltimore, MD) or Actin (1:1000; Sigma-Aldrich) antibodies. Membranes were incubated with horseradish peroxidase-conjugated antibody against sheep (1:2000; R&D Systems), rabbit or mouse (1:2000; GE HealthCare) IgG and binding was revealed by chemiluminescence detection (Millipore).
Proliferation assay
The in vitro proliferation of cells was determined by MTS assay using 0.2 mg/ml MTS reagent (Promega) as previously described (15). Data for each time point was obtained in triplicate per experimental condition.
Migration and invasion assay
A 24 well plate containing either 8.0 µm pore cell culture insert (BD Falcon) or 8.0 µm pore transwell-inserts pre-coated with 100 µl Matrigel (BD Falcon) was utilized for the migration and invasion assays, respectively. Tumor cells (2.5 × 104 cells) were seeded in the upper chamber in medium containing 0% FBS and the bottom chamber filled with medium containing either 0% or 20% FBS as the chemo-attractant. After 16 hours (in case of migration) or 48 hours (in case of invasion), cells in the upper chamber were removed with cotton swabs. Cells that migrated or invaded to the lower surface of the insert were fixed in 100% cold methanol (Fischer Scientific), washed in PBS and stained with 2% crystal violet (Harleco). Three representative images from each well were captured at 100× magnification by light microscopy and the total number of migrated or invaded cells per image was counted using ImageJ imaging software (National Institute of Health, Bethesda, MD). Data are representative of at least two independent experiments performed in triplicates per experimental condition.
In vivo assessment of breast cancer metastasis
Tumor cells (1 × 105 cells) from each group were injected into the left cardiac ventricle of five week old athymic nude mice (Taconic) [For MDA-MB-231 cells, n = 9 (scramble) and 10 (shCITED2); For MDA-MB-468 cells, n = 10 (scramble) and n = 7 (shCITED2)]. Two weeks later, the establishment of tumor-induced osteolysis in the bone was analyzed by obtaining digital radiographic images of the femur and tibia twice a week using a Faxitron MX-20 X-ray unit (Faxitron X-ray Corp.) until termination of the experiment. The experiment was terminated when animals became moribund. The osteolytic area in the radiographic images was measured using MetaMorph image analysis software (Meta Imaging Series version 6.1, Universal Imaging Corp.). Tumor lesions within the bone were analyzed by H&E staining of bone sections decalcified in 10% EDTA (Sigma-Aldrich). Brain, liver and lungs were harvested and maintained in Bouin’s fixative (RICCA chemical) for 24 hours and counted for the total number of macro-metastatic lesions.
All animal experiments were carried out in accordance with the National Research Council’s ‘‘Guide to the Care and Use of Laboratory Animals’’. Animal use was approved by the Johns Hopkins Animal Care and Use Committee, animal welfare assurance #A3272-01, protocol #MO10M450.
Microarray analysis
cDNA expression between scramble and shCITED2 MDA-MB-231 cells was compared using Agilent Human GE 4×44K v2 microarray (G4845A). Log2 transformed signal intensities, without background subtraction were imported into GeneSpring GX 10 software (Agilent Technologies) and (quantile) normalized within the sample type. Differentially expressed genes in shCITED2-expressing cells relative to scramble cells were identified based on ≥ two-fold change in gene expression. Quality assessment of samples and microarray analysis were conducted at the Sidney Kimmel Cancer Center Microarray Core Facility at Johns Hopkins University School of Medicine, Baltimore, MD (supported by NIH grant P30 CA006973 entitled Regional Oncology Research Center). Microarray data are deposited in the Array Express database www.ebi.ac.uk/arrayexpress under accession number E-MTAB-4267.
ELISA analysis
OPN and IL-11 ELISA immunoassay (R&D Systems) were performed on serum free tumor-conditioned media obtained from cell lines according to manufacturer’s instructions.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed on nuclear cell lysates using the LightShift EMSA and Chemiluminescent detection kit (Thermo Scientific) based on manufacturer’s instructions using NF-κB (5’-Biotin-AAGTTGAGGGGACTTTCCCAGGCT-3’ and 5’-Biotin-AGCCTGGGAAAGTCCCCTCAACTT-3’) oligonucleotides. Oct1 (5’-Biotin-TGTCGAATGCAAATCACTAGAA-3’ and 5’-Biotin-TTCTAGTGATTTGCATTCGACA-3’) was used as the loading control.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed on nuclear cell lysates using the SimpleChIP Enyzmatic Chromatin IP Kit (Cell Signaling Technology) based on manufacturer’s instructions. The promoter primer sequence used for IKKα was: (sense) 5’-GTGGTTCCGTTCAGCCCT-3’, (antisense) 5’-TGCTCGCGCGTCTTTG-3’.
Statistical analysis
Differences in the migratory and invasive ability, average tumor area and osteolytic area, and protein expression between experimental conditions were compared by unpaired Student’s t-test. Differences in the mRNA expression of pro-metastatic genes in the shCITED2-expressing cells relative to scramble cells normalized to 1.0, was compared by one sample t-test. CITED2 mRNA expression in tissues and the results of the invasion assay upon IKKα re-expression were analyzed by ANOVA and Tukey’s multiple comparison test. p-values below 0.05 were considered statistically significant. For all figures, (*) denotes p < 0.05, (**) denotes p < 0.01 and (***) denotes p < 0.001.
Results
CITED2 expression is elevated in breast cancer metastasis
Previously, we presented evidence that CITED2 expression is significantly elevated in primary human breast tumors relative to normal mammary epithelium, and is negatively correlated with survival (11, 15). Extending our analysis to metastatic lesions, CITED2 mRNA expression was significantly higher in human breast cancer metastases relative to primary breast tumors by qRT-PCR analysis (Fig. 1A). This difference appeared to be due to the fact that CITED2 levels in metastases were more frequently elevated beyond those observed in normal mammary epithelium as compared to primary tumors, many of which displayed CITED2 levels equivalent to that in normal. Consistent with mRNA results, this expression pattern was also appreciated at the protein level in a limited subset of samples by immunohistochemical analysis (Fig. 1B). Lastly, among metastases, higher expression of CITED2 mRNA was observed in metastasis to bone relative to non-bone sites. Taken together, these data demonstrate that CITED2 expression is frequently elevated in metastatic lesions of breast cancer patients, with highest levels in bone metastasis.
Figure 1.
CITED2 expression is elevated in human breast cancer metastasis. A, CITED2 mRNA expression was determined by qRT-PCR in human normal mammary epithelium (n=12), primary breast tumor tissues (invasive ductal carcinoma) from patients surviving greater than (n=11) and less than (n=8) five years from the time of diagnosis, and metastatic lesions obtained from non-bone (n=19) and bone (n=6) sites. (p* < 0.05; p** < 0.01). B, Immunohistochemical analysis was performed on paraffin-embedded tissue sections of normal mammary epithelium (n = 5), invasive ductal carcinoma (IDC) (n = 5) and breast cancer bone metastasis (BBM) (n = 8) using goat anti-CITED2 antibody. Protein was visualized using DAB (brown). Sections were counterstained with hematoxylin and visualized by light microscopy. N: Normal, T: Tumor and B: Bone. Magnification: 400×.
Down-regulation of CITED2 inhibits breast cancer metastasis
To explore the role of CITED2 in breast cancer metastasis, we utilized the human breast cancer cell lines MDA-MB-231 and MDA-MB-468. These cell lines are highly invasive in vitro, readily establish metastases following systemic administration in animal models (18–22), and as we have shown previously, express elevated levels of CITED2 relative to human mammary epithelial cells and breast cancer cell lines that are non-metastatic in animal models (15). MDA-MB-231 and MDA-MB-468 cells were stably infected with a lentiviral expression vector containing either shRNA specific for CITED2 (shCITED2) or scrambled shRNA (scramble) and levels of CITED2 were assessed at both the mRNA and protein levels (Fig. 2A). Stable expression of shCITED2 resulted in a greater than 75% reduction in CITED2 expression in both MDA-MB-231 and MDA-MB-468 cells by qRT-PCR and Western analysis. Prior to examining the effect of CITED2 on metastatic progression, we first examined whether reducing CITED2 expression altered the rate of cell proliferation in vitro. As determined by MTS assay, shCITED2 cells exhibited a similar growth rate to that of scramble cells, indicating that knockdown of CITED2 did not affect cell growth in either cell line (Fig. 2B). To begin exploring CITED2 involvement in metastatic progression, we evaluated the effects of CITED2 knockdown on the migratory and invasive ability of MDA-MB-231 and MDA-MB-468 cells using in vitro trans-well migration and invasion assays, respectively. CITED2 knockdown significantly reduced the migratory and invasive ability of both MDA-MB-231 and MDA-MB-468 cells (Fig. 2C and D). Next, we assessed the effects of CITED2 down-regulation on the establishment of metastasis following intra-cardiac administration of MDA-MB-231 and MDA-MB-468 cells stably expressing shCITED2 or scramble in athymic nude mice. While this model does not replicate the entire metastatic cascade, it effectively assesses the ability of tumor cells to extravasate from the vasculature, invade and colonize secondary organ sites, and establish vascularized metastatic lesions. Mice administered with shCITED2-expressing cells displayed significantly reduced metastasis to bone relative to the scramble group. As evidenced by digital radiography, osteolytic area was significantly lower in the shCITED2 group relative to the scramble group (Fig. 2E and F, top). Consistent with reduced osteolysis, the shCITED2 group also displayed a significant reduction in tumor area relative to the scramble group, as measured on histological sections (Fig. 2E and F, bottom). Additionally, mice injected with shCITED2-expressing MDA-MB-231 cells developed fewer brain metastases relative to those injected with scramble cells, although no differences were observed using MDA-MB-468 cells (Supplementary Fig. S1A). Further, metastasis to the lung and liver did not appear to be affected by shCITED2 expression in either MDA-MB-231 or MDA-MB-468 cells (Supplementary Fig. S1B and S1C). Taken together, these observations indicate that CITED2 may play a role in the establishment of breast cancer metastasis, particularly to the bone.
Figure 2.
CITED2 knockdown in breast cancer cells inhibits tumor migration and invasion in vitro and bone metastasis in vivo. A, CITED2 mRNA expression (left) was determined by qRT-PCR. Data are representative of triplicate experiments. CITED2 protein expression (right) was determined by Western analysis performed on equal amounts of protein from total cell lysates. GAPDH serves as the loading control. B, Cell proliferation was determined by MTS assay. C and D, The migratory and invasive capability of the tumors was determined by migration (C) and invasion (D) assays, respectively. E and F, scramble or shCITED2-expressing MDA-MB-231 (E) and MDA-MB-468 (F) cells were injected into the left cardiac ventricle of athymic nude mice. Top: Digital radiographic imaging of the femur and tibia showing areas of osteolysis marked by white circles. The adjacent histograms represent the average osteolytic area measured on the radiographic images between the experimental groups. Bottom: H&E staining of decalcified bone sections indicating presence of tumor lesions as shown by black rectangles and circles. The adjacent histograms represent the average tumor area measured on histological images between the experimental groups. (p* <0.05, p** < 0.01, p*** < 0.001).
Inhibition of CITED2 reduces expression of IKKα and pro-metastatic NF-κB target genes
To explore the mechanism through which CITED2 influences metastatic ability, we examined the effect of CITED2 knockdown on gene expression in MDA-MB-231 cells by cDNA microarray analysis (Supplementary Table 1). Notably, IKKα, a critical mediator of the NF-κB signaling cascade (23) was found to be down-regulated along with several downstream targets having reported roles in promoting metastasis, including osteopontin (OPN), matrix metalloproteinase 9 (MMP9), urokinase type plasminogen activator (uPA), secreted protein, acidic, cysteine-rich (SPARC), interleukin-11 (IL-11) and interleukin-1β (IL-1β). While the extracellular matrix protein SPARC and proteases MMP9 and uPA have been shown to promote tumor invasion (24–28), evidence suggests that the cytokines IL-11 and IL-1β facilitate the establishment of osteolytic metastasis (18, 29). Additionally, the multi-functional protein OPN reportedly affects numerous steps in the metastatic cascade (30). Regulation of these genes by NF-κB signaling was confirmed in MDA-MB-231 cells, wherein treatment with the IKK inhibitor PS1145, which prevents IκBα degradation (31), reduced both basal NF-κB signaling by Western analysis and expression of OPN, MMP9, uPA, SPARC, IL-11 and IL-1β by qRT-PCR (Supplementary Fig. S2A and S2B). Moreover, using qRT-PCR and Western/ELISA analyses, we confirmed down-regulation of IKKα along with the aforementioned NF-κB target genes in shCITED2-expressing MDA-MB-231 and MDA-MB-468 cells compared to scramble cells at both the mRNA and protein levels (Fig. 3A–F). Further, by ChIP assay in MDA-MB-231 cells, CITED2 was found to localize to the promoter of IKKα indicating a potentially direct role for CITED2 in the regulation of its expression. (Fig. 3G). Collectively, these data indicate that CITED2 knockdown reduces the expression of IKKα and several downstream pro-metastatic genes in breast cancer cells.
Figure 3.
CITED2 knockdown in breast cancer cells reduces expression of IKKα and pro-metastatic NF-κB target genes. A and D, mRNA expression was determined by qRT-PCR. Data are representative of triplicate experiments. B and E, Protein expression of IKKα was determined by Western analysis of total cell lysates (CE). GAPDH serves as the loading control. Protein expression of MMP9 and uPA was determined by Western analysis of serum free condition medium (CM). C and F, protein expression of IL-11 and OPN (MDA-MB-231 only) was analyzed by ELISA. G, Localization of CITED2 or IgG to the IKKα promoter was assessed by ChIP assay using anti-sheep CITED2 or sheep IgG antibodies in wild-type MDA-MB-231 cells. (p* <0.05, p** < 0.01, p*** < 0.001).
Down-regulation of CITED2 attenuates NF-κB signaling
NF-κB signaling is constitutively active in breast cancer (32) and occurs via both canonical and non-canonical pathways, each of which involves IKKα. In the canonical pathway the trimeric IKKα/β/γ complex phosphorylates the NF-κB inhibitor IκBα, inducing its degradation and concomitantly releasing p65/p50 transcription factors to translocate into the nucleus for regulation of gene expression (33). In the non-canonical pathway, phosphorylation of p100 by IKKα triggers nuclear translocation of the RelB/p52 transcription factors for regulating gene expression (33). Since CITED2 knockdown resulted in the down-regulation of IKKα expression along with several downstream targets of NF-κB, we next investigated the effect of CITED2 on basal NF-κB signaling in breast cancer cells. Examining the effects of CITED2 knockdown on the expression levels and localization of NF-κB signaling factors by Western analysis revealed increased levels of IκBα and reduced nuclear levels of p65 and RelB in shCITED2-expressing MDA-MB-231 and MDA-MB-468 cells relative to scramble cells (Fig. 4A), indicating that both the canonical and non-canonical pathways were affected. Consistent with reduced nuclear p65 and RelB levels, NF-κB DNA binding activity of the p65/p50 heterodimer was also markedly reduced in shCITED2-expressing MDA-MB-231 and MDA-MB-468 cells relative to scramble cells as determined by EMSA (Fig. 4B). Together, these data demonstrate the ability of CITED2 to influence NF-κB signaling.
Figure 4.
CITED2 regulates NF-κB signaling. A, Western analysis of cytoplasmic IκBα and nuclear p65 and RelB proteins was performed on equal amounts of protein obtained from cytoplasmic (CE) and nuclear (NE) cell lysates. GAPDH and HDAC1 serve as the cytoplasmic and nuclear loading controls, respectively. B, Non-radioactive EMSA analysis was performed on equal amounts of nuclear cell lysate using NF-κB and Oct-1 oligonucleotides. Oct-1 serves as the loading control. (p* <0.05, p** < 0.01).
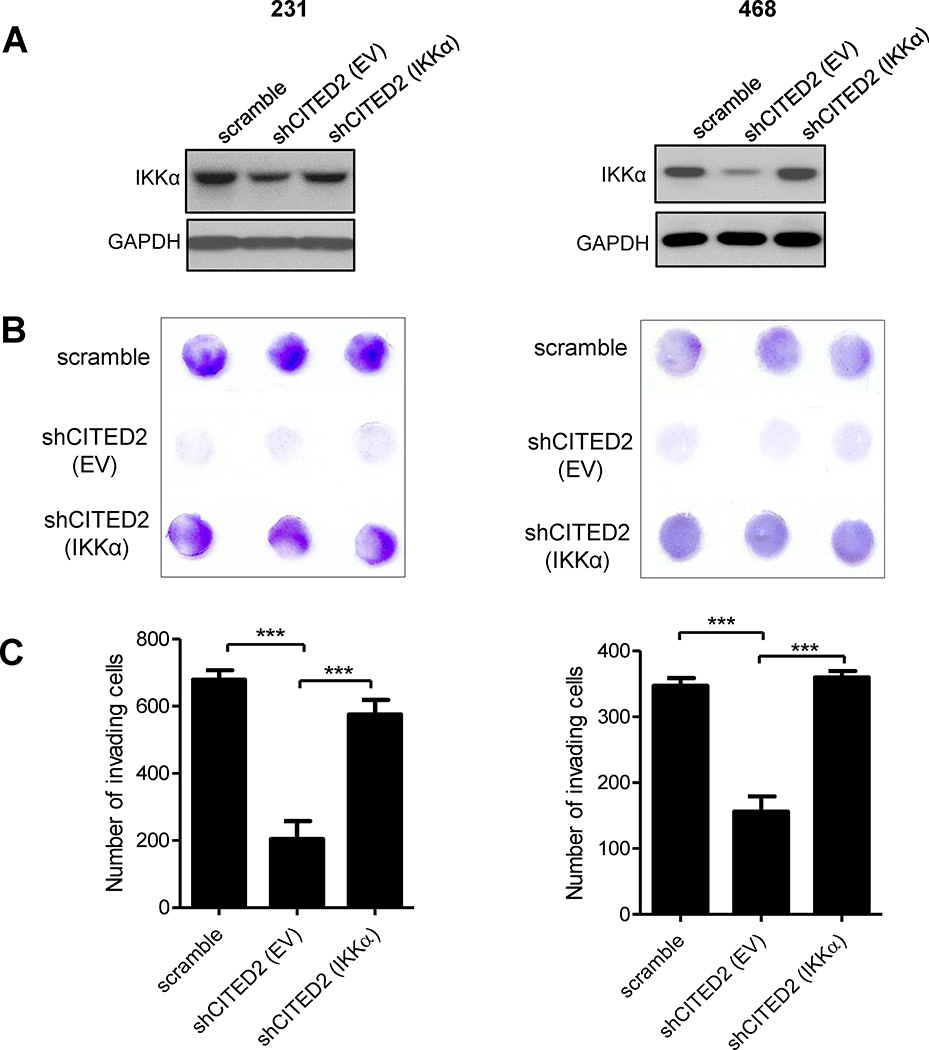
Restoration of IKKα expression reverses effects of CITED2 knockdown on breast cancer cell invasion
Since CITED2 knockdown reduced levels of IKKα along with numerous downstream factors with reported roles in promoting metastatic dissemination (Fig. 3), we next investigated the possibility that the reduced metastatic ability of shCITED2-expressing breast cancer cells was related to the down-regulation of IKKα expression. To address this question, we transiently restored IKKα expression in shCITED2-expressing MDA-MB-231 and MDA-MB-468 cells (Fig. 5A) and examined invasive ability by in vitro trans-well invasion assay. Notably, restoring IKKα expression in shCITED2-expressing cells significantly increased invasive ability relative to empty vector-transfected shCITED2 cells, returning invasiveness to levels commensurate with those observed in scramble cells (Fig. 5B and C). These data support a role for IKKα in mediating the effects of CITED2 on the metastatic ability of breast cancer cells.
Figure 5.
Restoration of IKKα expression following CITED2 knockdown in breast cancer cells rescues cell invasiveness. A, IKKα expression was determined by Western analysis of equal amounts of total protein lysates obtained from scramble cells or shCITED2-expressing cells transfected with either empty vector (EV) or IKKα. GAPDH serves as the loading control. B and C, Invasive ability of scramble cells and shCITED2 cells transfected with either empty vector (EV) or IKKα was determined by in vitro trans-well invasion assay. (B) Representative images of trans-well inserts following staining of invading cells with crystal violet (purple). (C) Quantification of invading cells showing the average number per experimental condition. (p*** < 0.001).
Discussion
Despite current treatment efforts, the vast majority of patients diagnosed with metastatic breast cancer ultimately succumb to this disease, highlighting the need for clearer understanding of the drivers of metastasis and their mechanism of action. In this study, we have shown that expression of the non-DNA binding transcriptional co-activator CITED2 is significantly elevated in metastatic lesions of breast cancer patients relative to primary tumors. Further, we have demonstrated that down-regulation of CITED2 significantly attenuates invasive and metastatic ability in two human breast cancer cell lines (MDA-MB-231 and MDA-MB-468). Lastly, we provide evidence that the effects of CITED2 on metastatic ability are mediated, at least in part, by controlling expression of the NF-κB regulator IKKα, the levels of which have been shown to negatively correlate with relapse-free survival in breast cancer patients (Supplementary Fig. S3).
While elevated levels of CITED2 were found in patient samples of metastatic disease relative to primary tumors, highest levels were noted in metastases to bone (Fig. 1A). Further, the inhibition of metastatic colonization observed following knockdown of CITED2 in breast cancer cell lines (Fig. 2E and F; Supplementary Fig. S1) was largely limited to skeletal disease. These findings are in agreement with our previous data demonstrating that reducing CITED2 expression in the murine mammary tumor cell line NT2.5 significantly reduces bone metastasis in vivo (15), highlighting the potential importance of CITED2 as a critical mediator of bone metastasis in breast cancer. Although the mechanism by which CITED2 mediates this effect remains unclear, CITED2 knockdown in MDA-MB-231 and MDA-MB-468 cells resulted in reduced expression of IL-11 and IL-1β, both of which are reported mediators of bone metastasis and osteolysis (18, 29), thus warranting further investigation into their potential contribution towards the metastatic effects of CITED2.
In addition to the impairment of bone metastasis, CITED2 knockdown in MDA-MB-231 cells also inhibited the establishment of brain metastasis. This effect was not observed in MDA-MB-468 cells, possibly due to the fact that this cell line colonized the brain less frequently than MDA-MB-231 in the control condition. While the lesser ability of MDA-MB-468 cells to colonize the brain could be due to various differences between these two cell lines, it is interesting to note that canonical TGF-β signaling is absent in MDA-MB-468 cells due to the lack of Smad4 in this cell line (34). Combined with the reported role of CITED2 in regulating TGF-β signaling through Smad interactions (9), it is tempting to speculate that the divergent ability of MDA-MB-231 and MDA-MB-468 cells to establish brain metastasis may be related to TGF-β responsiveness. Moreover, it should be noted that the lack of canonical TGF-β signaling in MDA-MB-468 cells did not impact the effect of CITED2 knockdown on invasion or the establishment of bone metastasis. This indicates that these effects were mediated in a TGF-β-independent manner, further supporting a role for NF-κB signaling, which was attenuated in both the MDA-MB-231 and MDA-MB-468 cell lines.
Despite the ability of CITED2 to directly regulate expression of the NF-κB pathway regulator IKKα, it is not yet clear how CITED2 modulates NF-κB signaling and transcriptional activity in breast cancer cells. Although we did not observe changes in the mRNA expression of NF-κB signaling intermediates downstream of IKKα, upstream mediators also exist whose expression could be impacted by CITED2. Additionally, CITED2 is known to interact with CREB-binding protein (CBP) and p300, reported co-activators of p65-mediated gene transcription (35), suggesting that CITED2 could also regulate the activity of NF-κB as part of the transcriptional complex. Although the ability of IKKα to restore NF-κB signaling (data not shown) and rescue tumor invasion in MDA-MB-231 and MDA-MB-468 cells following CITED2 knockdown (Fig. 5B and C) implicates involvement of the NF-κB pathway, it should be noted that IKKα can also exert NF-κB-independent effects (36). Thus, further investigation is required not only to assess the mechanism whereby CITED2 modulates NF-κB activity, but also to determine the extent of its contribution to the pro-metastatic effects of CITED2, as well as the ultimate effectors of its action. Addressing these questions will not only further our understanding of CITED2 action in breast cancer, but may also provide new avenues for the prevention and treatment of metastatic spread.
Supplementary Material
Acknowledgments
Financial support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA157687. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Lu J, Steeg PS, Price JE, Krishnamurthy S, Mani SA, Reuben J, et al. Breast Cancer Metastasis: Challenges and Opportunities. Cancer Res. 2009;69:4951–4953. doi: 10.1158/0008-5472.CAN-09-0099. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Doughman YQ, Gu S, Jarrell A, Aota S, Cvekl A, et al. Cited2 is required for the proper formation of the hyaloid vasculature and for lens morphogenesis. Development. 2008;135:2939–2948. doi: 10.1242/dev.021097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu X, Lam E, Doughman YQ, Chen Y, Chou YT, Lam M, et al. Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J. 2007;26:4445–4456. doi: 10.1038/sj.emboj.7601883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B, Qu X, Gu S, Doughman YQ, Watanabe M, Dunwoodie SL, et al. Cited2 is required for fetal lung maturation. Dev. Biol. 2008;317:95–105. doi: 10.1016/j.ydbio.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin Z, Haynie J, Yang X, Han S, Kiatchoosakun S, Restivo J, et al. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10488–10493. doi: 10.1073/pnas.162371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya S, Michels CL, Leung MK, Arany ZP, Kung AL, Livingston DM. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glenn DJ, Maurer RA. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J. Biol. Chem. 1999;274:36159–36167. doi: 10.1074/jbc.274.51.36159. [DOI] [PubMed] [Google Scholar]
- 8.Bragança J, Eloranta JJ, Bamforth SD, Ibbitt JC, Hurst HC, Bhattacharya S. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J. Biol. Chem. 2003;278:16021–16029. doi: 10.1074/jbc.M208144200. [DOI] [PubMed] [Google Scholar]
- 9.Chou YT, Want H, Chen Y, Danielpour D, Yang YC. Cited2 modulates TGFbeta-mediated upregulation of MMP9. Oncogene. 2006;25:5547–5560. doi: 10.1038/sj.onc.1209552. [DOI] [PubMed] [Google Scholar]
- 10.Tien ES, Davis JW, Vanden Heuvel JP. Identification of the CREB-binding protein/p300-interacting protein CITED2 as a peroxisome proliferator activated receptor alpha coregulator. J. Biol. Chem. 2004;279:24053–24063. doi: 10.1074/jbc.M401489200. [DOI] [PubMed] [Google Scholar]
- 11.Lau WM, Doucet M, Huang D, Weber KL, Kominsky SL. CITED2 modulates estrogen receptor transcriptional activity in breast cancer cells. Biochem. Biophys. Res. Commun. 2013;437:261–266. doi: 10.1016/j.bbrc.2013.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun HB, Zhu YX, Yin T, Sledge G, Yang YC. MRG1, the product of a melanocyte-specific gene related gene, is a cytokine-inducible transcription factor with transformation activity. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13555–13560. doi: 10.1073/pnas.95.23.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai L, Merchant JL. A role for CITED2, a CBP/p300 interacting protein, in colon cancer cell invasion. FEBS Lett. 2007;581:5904–5910. doi: 10.1016/j.febslet.2007.11.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou YT, Hsieh CH, Chiou SH, Hsu CF, Kao YR, Lee CC, et al. CITED2 functions as a molecular switch of cytokine-induced proliferation and quiescence. Cell Death Differ. 2012;19:2015–2028. doi: 10.1038/cdd.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau WM, Weber KL, Doucet M, Chou YT, Brady K, Kowalski J, et al. Identification of prospective factors promoting osteotropism in breast cancer: a potential role for CITED2. Int J Cancer. 2010;126:876–884. doi: 10.1002/ijc.24780. [DOI] [PubMed] [Google Scholar]
- 16.Chou YT, Yang YC. Post-transcriptional control of CITED2 by transforming growth factor beta. Regulation via Smads and CITED2 coding region. J Biol. Chem. 2006;281:18451–18462. doi: 10.1074/jbc.M601720200. [DOI] [PubMed] [Google Scholar]
- 17.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, et al. Differential regulation of IkappaB kinase alpha and beta by two upstream kinases, NF-kappaB-inducing kinase and mitogen-activated protein kinase/ERK kinase kinase-1. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordόn-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 19.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau WM, Doucet M, Stadel R, Huang D, Weber KL, Kominsky SL. Enpp1: a potential facilitator of breast cancer bone metastasis. PLoS One. 2013;8:e66752. doi: 10.1371/journal.pone.0066752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vantyghem SA, Allan AL, Postenka CO, Al-Katib W, Keeney M, Tuck AB, et al. A new model for lymphatic metastasis: development of a variant of the MDA-MB-468 human breast cancer cell line that aggressively metastasize to lymph nodes. Clin Exp Metastasis. 2005;22:351–361. doi: 10.1007/s10585-005-0745-1. [DOI] [PubMed] [Google Scholar]
- 23.Adli M, Merkhofer E, Cogswell P, Baldwin AS. IKKalpha and IKKbeta each function to regulate NF-kappaB activation in the TNF-induced/canonical pathway. PLoS One. 2010;5(e9428) doi: 10.1371/journal.pone.0009428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seno T, Harada H, Kohno S, Teraoka M, Inoue A, Ohnishi T. Downregulation of SPARC expression inhibits cell migration and invasion in malignant gliomas. Int J Dev Neurosci. 1999;17:463–472. doi: 10.3892/ijo_00000197. [DOI] [PubMed] [Google Scholar]
- 25.Golembieski WA, Ge S, Nelson K, Mikkelsen T, Rempel SA. Increased SPARC expression promotes U87 glioblastoma invasion in vitro. Int J Oncol. 2009;34:707–715. doi: 10.1016/s0736-5748(99)00009-x. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Wang M, Xi B, Xue J, He D, Zhang J, et al. SPARC is a key regulator of proliferation, apoptosis and invasion in human ovarian cancer. PLoS One. 2012;7:e42413. doi: 10.1371/journal.pone.0042413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehner C, Hockla A, Miller E, Ran S, Radisky DC, Radisky ES. Tumor-cell produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget. 2014;5:2736–2749. doi: 10.18632/oncotarget.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang L, Han X. The urokinase plasminogen activator system in breast cancer invasion and metastasis. Biomed Pharmacother. 2013;67:179–182. doi: 10.1016/j.biopha.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Russell MR, Shahriari K, Jernigan DL, Lioni MI, Garcia FU, et al. Interleukin-1β promotes skeletal colonization and progression of metastatic prostate cancer cells with neuroendocrine features. Cancer Res. 2013;73:1–9. doi: 10.1158/0008-5472.CAN-12-3970. [DOI] [PubMed] [Google Scholar]
- 30.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biology. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yemelyanov A, Gasparian A, Lindholm P, Dang L, Pierce JW, Kisselijov F, et al. Effects of IKK inhibitor PS1145 on NF-κB function, proliferation, apoptosis and invasion activity in prostate carcinoma cells. Oncogene. 2006;25:387–398. doi: 10.1038/sj.onc.1209066. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi N, Ito T, Azuma S, Ito E, Honma R, Yanagisawa Y, et al. Constitutive activation of nuclear factor-kappaB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer Sci. 2009;100:1668–1674. doi: 10.1111/j.1349-7006.2009.01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schutte M, Firuban RH, Hedrick L, Cho KR, Nadasdy GM, Weinstein CL, et al. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 35.Gerritsen ME, William AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang WC, Hung MC. Beyond NF-κB activation: nuclear functions of IκB kinase α. J Biomed Sci. 2013;20:3. doi: 10.1186/1423-0127-20-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.