Abstract
Synapses are the basic unit of neuronal communication and their disruption is associated with many neurological disorders. Significant progress has been made towards understanding the molecular and genetic regulation of synapse formation, modulation, and dysfunction, but the underlying cellular mechanisms remain incomplete. The actin cytoskeleton not only provides the structural foundation for synapses, but also regulates a diverse array of cellular activities underlying synaptic function. Here we will discuss the regulation of the actin cytoskeleton in dendritic spines, the postsynaptic compartment of excitatory synapses. We will focus on a select number of actin regulatory processes, highlighting recent advances, the complexity of crosstalk between different pathways, and the challenges of understanding their precise impact on the structure and function of synapses.
Keywords: Actin, spines, dendrites, synaptic plasticity, neurodegenerative disease, neuropsychiatric disorders, brain developmental disorders
Introduction
In the vertebrate brain, chemical synapses are the major form of synaptic connection comprised of paired pre- and post-synaptic structures for neurotransmitter release and reception, respectively. Chemical synapses are known to be plastic and undergo short- and long-term modifications during developmental refinement of neural circuits, as well as in learning and memory. On the postsynaptic side, synaptic modulation involves changes in the number of synaptic receptors on the cell surface via regulated trafficking [1–4]. Many neurological disorders have also been associated with alterations in synaptic connections [5]. Therefore, a better understanding of the molecular and cellular mechanisms underlying synaptic development and plasticity is of importance to our understanding of the complex brain functions underlying both physiological and pathological conditions.
Most of the excitatory synapses reside on dendritic spines, small actin-rich membrane protrusions that host neurotransmitter receptors, scaffolding proteins, signal transduction molecules, and cytoskeletal regulators. Dendritic spines come in different shapes and sizes including mushroom-shaped, stubby and thin, of which mushroom-shaped spines represent the mature form highlighted by an expanded head sitting on a thin neck. Spines undergo changes in shape and size during development and synaptic plasticity [6] and these structural changes have been considered as a hallmark of synaptic modifications associated with learning, aging, and neurodegenerative diseases [7–11]. In this review, we will discuss the regulation and function of the actin cytoskeleton in the development and modification of postsynaptic spine structure and function.
The actin cytoskeleton in spine development and modification
The actin cytoskeleton is built by a balancing act of filament assembly and disassembly, which is regulated by a wide range of accessory proteins [12]. Actin filaments are typically organized into distinct networks for different cellular functions by actin regulatory proteins. In vertebrate cells, over a hundred actin-binding proteins may exist, but it remains unknown how many of them are present and function in dendritic spines. Of these actin regulatory proteins, those involved in filament nucleation, severing, crosslinking, end capping, and monomer sequestering are believed to be crucial for the formation and dynamics of actin structures [12].
Nucleation of the actin filaments
Nucleation of actin filaments is the rate limiting step of filament formation and is thus tightly regulated. Two classic families of actin nucleating factors are well known for their role in the assembly of distinct F-actin networks: the actin-related protein 2/3 (Arp2/3) complex and formin molecules. The Arp2/3 complex is composed of seven subunits and it binds to the sides of existing actin filaments to nucleate new actin filaments with a 70-degree angle [12]. In dendritic spines, branched F-actin networks comprise the main cytoskeletal architecture in the spine head [13] and is required for the proper development and maintenance of dendritic spines [14–16]. The Arp2/3 complex can be activated by multiple nucleation-promoting factors (NPFs) [17], many of which have been shown to play an important in spine formation and plasticity [15,18,19]. Interestingly, the Arp2/3 activity can also be regulated at both the transcriptional and translational level in neurons. For example, downregulation of Arpc3, a component of the ARP2/3 complex, by miR-29a/b was shown to modulate dendritic spine shape and size [20]. Furthermore, translational control of three subunits of the Arp2/3 complex has been shown to mediate the memory loss in C. elegans [21]. Given that local protein translation represents an important mechanism for regulating synaptic function [22], future studies of the actin cytoskeleton in synaptic function and modification will need to consider the transcriptional and translational regulation of Arp2/3-mediated synaptic modification.
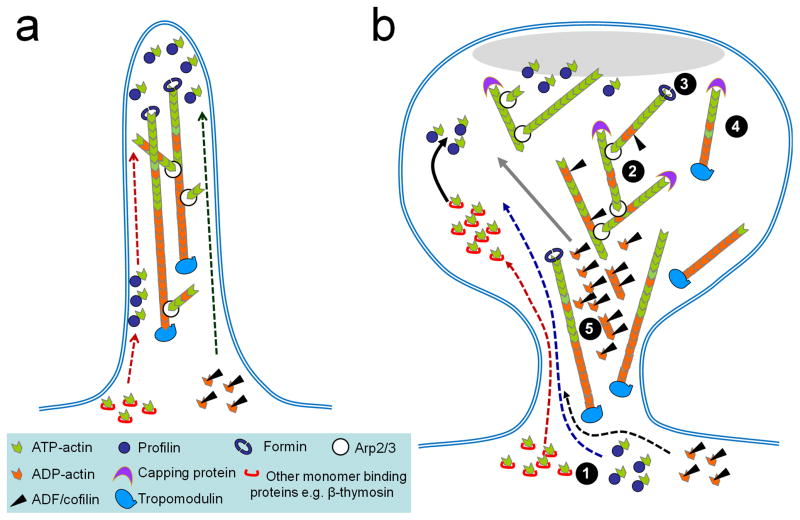
Formins represent a large family of actin regulatory proteins that are instrumental to the polymerization and rearrangement of the actin cytoskeleton [23]. The formins act as dimers to nucleate and elongate unbranched actin filaments by associating with the fast-growing barbed ends. The formin mDia2 was found to mediate the formation and elongation of dendritic filopodia, followed by Arp2/3-mediated actin polymerization for spine head expansion [14]. Overexpression of a dominant-active form of mDia2 resulted in an increase in dendritic filopodia and a reduction in mature spines [14], suggesting that the Arp2/3-mediated spine head expansion requires the down regulation of mDia2 activity. Similarly, the overexpression of another formin, Daam1, has been shown to cause a reduction in spine density [24]. Together, these results suggest that formins and Arp2/3 function at two distinct steps of spine formation: the former in generating linear F-actin mostly found in dendritic filopodia and the latter in nucleating branched F-actin networks for spine head expansion. It should be noted that formins and Arp2/3 do not function exclusively and may cooperate to generate specific dendritic protrusions. For example, branched actin filaments and Arp2/3 have been observed in dendritic filopodia [13]. Moreover, the mRNA level of another formin, formin-like 2(FMNL2), was found to be markedly elevated in adult mouse brain [25]. FMNL2 does not appear to nucleate but instead to polymerize F-actin in lamellipodial protrusion [26]. Finally, the formin Delphilin was found to co-localize with GluRδ2 at the postsynaptic side of the parallel fiber–Purkinje cell synapses, suggesting that formins may function in mature spines to regulate synaptic receptors [27]. It is exciting to speculate that different formin members could function at distinct stages of spine development, as well as in different aspects of regulation of the actin structure and dynamics in spines (see Figure 1).
Figure 1.
Schematic illustration depicting the actin mechanism underlying spine formation and modification. (a) Dendritic filopodia are considered to be the precursors of spines and their formation mainly depends on formin-based F-actin assembly, although branched actin filaments and Arp2/3 are present in filopodia. (b) Dendritic spines contain branched and unbranched actin filaments and their formation and modification depend on a combination of actin regulatory proteins. Only the factors involving monomer-binding, nucleation, polymerization, capping, and severing are shown for simplicity. Both solid and dashed lines with arrows indicate the potential source of actin monomers to be delivered for polymerization site. The PSD is depicted as the light gray oval. Circled numbers represent the essential steps of actin filaments formation and modification: (1) actin monomer binding and delivery to the polymerization site, (2) nucleation, (3) assembly, (4) capping, and (5) severing/disassembly.
Recent studies have identified a third family of actin nucleation molecules: the tandem-monomer binding nucleators consisting of Spire, Cordon-bleu (Cobl), Leiomodin (Lmod), JMY and adenomatous polyposis coli (APC). These molecules contain tandem repeats of Wiskott- Aldrich syndrome protein (WASP) homology 2 (WH2) domain that nucleate filaments by bringing actin monomers together [28–32]. Several of these tandem-monomer binding nucleators have been recently shown to play a general role in neuronal development and function [31,33–37]. Interestingly, some members of this family, e.g. Cobl, can be regulated by Ca2+/calmodulin (CaM), thus making them potential targets of neuronal activity [38]. However, more studies are needed to determine if these tandem monomer-binding nucleators play a role specifically in dendritic spines during synapse development and plasticity. Regardless, the nucleation process is complex and involves a diverse array of nucleation factors which are tightly spatiotemporally regulated. Future studies attempting to dissect the specific roles of these nucleators and their distinct effects on spine structure and function will require new advanced experimental approaches including nanoscale single molecule imaging. As an example, a recent study utilizing the single molecule tracking technique has identified two distinct sub-spine domains characterized by different types of actin nucleation: Arp2/3- mediated branched nucleation occurs in specific nano-domains at the PSD and formindependent actin polymerization drives the formation of finger-like protrusions from the sides of the spine [39]. These findings highlight the need to understand the nanoscale organization of the actin structure and subcellular compartments in dendritic spines.
End capping of actin filaments
Actin filaments are polarized with their barbed (plus) and pointed (minus) ends favoring polymerization and disassembly, respectively. Capping of the barbed end represents an important mechanism for regulating filament length and promoting Arp2/3-mediated nucleation and branched actin polymerization [40]. Capping protein (CP, also known as CapZ), is the best characterized barbed-end capping protein and is essential for actin-based motility [41]. In cultured hippocampal neurons, the loss of CP resulted in a substantial reduction in the number of mushroom-shaped dendritic spines with a concomitant increase in filopodia-like protrusions [42]. The loss of CP also resulted in the emergence of aberrant filopodia-like protrusions from the spine head. These results suggest that barbed end capping promotes Arp2/3-mediated growth of branched actin networks during spine head expansion.
Epidermal growth factor receptor pathway substrate 8 (Eps8), is a multifunctional protein that can cap barbed ends, bundle actin filaments, and activate the Rac signaling pathway [43]. Genetic knockout of Eps8 in mice leads to the formation of immature spines and impaired cognitive function [44], phenotypes shown to be mediated by the capping function of Eps8. Spines lacking Eps8 also showed increased actin polymerization [45], less stable PSD-95 dynamics [44], and reduced synaptic strength [44]. Together, these studies suggest that Eps8 regulates actin dynamics as well as the accumulation and/or clustering of PSD components during spine formation. It remains to be determined if Eps8 and CP function redundantly in spine development. However, the observation that the loss of Eps8 decreased the number of spine synapses, but increased the number of synapses on the dendritic shaft, suggests that Eps8 may exert distinct effects on synapse formation [44].
Barbed end capping of actin filaments may represent a key event that enables the modification of the spine actin structure during synaptic plasticity. Consistent with this notion, both CP and Eps8 are recruited into spines during LTP-inducing stimuli [44,46] and Eps8 capping activity is required for LTP-mediated spine formation [45] and synaptic strengthening [44]. However, the mechanism by which barbed end capping is regulated by synaptic activity is currently unclear. One possibility involves the actin severing protein Gelsolin, which can be regulated by Ca2+ and PIP2 [47] and may function in activity-dependent barbed capping of actin filaments in spines without filament severing [48]. Alternatively, adducins could function in activity-dependent barbed end capping as they cap the barbed end of actin filaments and cross-link them with the spectrin cytoskeleton [49]. β-adducin is localized to dendritic spines and mice lacking β-adducin display impaired synaptic plasticity, motor coordination and learning deficits [50–53]. Modulation and specificity of capping activity amongst these various classes of barbed end capping proteins is likely specified by binding partners, differential affinity for barbed ends, other actin-regulatory domains within the protein (such as bundling or severing) and post-translational modifications. Further studies will be needed to elucidate the mechanism by which barbed end capping proteins work in concert to regulate the actin cytoskeleton at the synapse.
Very little is currently known about pointed end capping and its function in dendritic spines. The pointed end can be capped by nucleation complexes (e.g. Arp2/3) as long as they remain bound to the complex [54]. Arp2/3-based branched actin networks are seen in spine head, but unbranched actin filaments are also present, especially in the spine neck region [13]. Therefore, pointed end capping of these actin filaments could substantially impact their stability. The tropomodulins (Tmods) are the best known pointed end capping proteins and both Tmods 1 and 2 are expressed in the nervous system [55]. Interestingly, knockout of Tmod2 was found to alter synaptic activity and cause defects in learning and memory [56]. However, no direct connection to pointed end capping of F-actin was demonstrated. The role of Tmods and the pointed end capping in synapse formation and plasticity remains an area for future research.
Actin monomer binding
F-actin is polymerized from actin monomers (globular actin: G-actin) with ATP bound (ATP-G-actin). While the critical concentration for polymerization at the barbed end is low (~0.1 μM), a large pool of G-actin is sequestered from spontaneous nucleation and polymerization by monomer-sequestering proteins [57]. As a result, the availability of polymerization-competent actin monomers at specific subcellular locations may be limited and could be rapidly depleted by polymerization unless it is quickly replenished [40]. Dendritic spines contain a limited space followed by a neck that has been shown to limit diffusion [58]. As such, an increase in F-actin for spine enlargement during development and synaptic potentiation requires the availability of a sufficient amount of G-actin that can be rapidly incorporated into the F-actin network. It is unknown if there is a pool of G-actin in spines that is sufficient to support the rapid F-actin polymerization during synaptic remodeling, or if G-actin is provided on-demand from outside the spine (e.g. dendritic shaft) through passive diffusion. Besides the cytosolic diffusible pool, G-actin also recycles from existing filaments by depolymerization at the pointed end through ADF/cofilin (Figure 1b) [12,40]. In this case, ADPG- actin generated via depolymerization is believed to diffuse to the polymerization site, being converted to ATP-actin through nucleotide exchange factors, and added to the barbed end for filament elongation by a number of binding proteins [57]. However, whether this recycling pathway plays a role in spine remodeling is unknown. Interestingly, it was found that the recycling pool of G-actin in motile lamellipodia is molecularly and functionally distinct from the cytosolic G-actin pool: the former is involved in homeostatic control of lamellipodial size whereas the latter localizes to the leading edge for F-actin polymerization underlying membrane protrusion [59]. Therefore, G-actin from recycling and cytosolic pools may contribute to distinct aspects of actin remodeling in spines.
Several families of G-actin binding proteins may contribute to G-actin localization, nucleotide state, and assembly [57]. For example, profilin binds G-actin, promotes its nucleotide exchange for polymerization-competent ATP-G-actin, and delivers ATP-G-actin to the growing barbed end for assembly [40,57,60]. Both profilin 1 and 2 have been shown to localize to dendritic spines through an activity-dependent manner [61–64] . Therefore, it is plausible that profilin and its translocation into spines may be responsible for delivering G-actin for actin remodeling underlying the structural modification of dendritic spines during synaptic plasticity. ADF/cofilin severs and depolymerizes aged F-actin at the rear part of the actin network, resulting in ADF/cofilin-bound ADP-G-actin that is recycled back to the leading edge for assembly [65–67]. Inhibition of cofilin has been shown to be important for increasing F-actin associated with spine enlargement during LTP [6], which appears to argue against this ADF/cofilin-mediated recycling model. However, it was shown that ADF/cofilin was first activated to increase actin turnover for AMPA receptor insertion, followed by inactivation for actin polymerization and spine enlargement [68]. Therefore, ADF/cofilin could potentially generate actin monomers needed to support actin polymerization underlying spine enlargement. Other families of actin monomer binding proteins could also be involved in spine development and remodeling [57], and future studies are needed to understand how the actin structure in spines is constructed, modified, and maintained during development and activity-dependent plasticity.
Activity-dependent regulation of the actin cytoskeleton in spines
One hallmark of dendritic spines is that they undergo synaptic activity-dependent modification: enlargement and shrinkage/elimination during long term synaptic potentiation (LTP) or depression (LTD), respectively. Increases in actin polymerization and depolymerization are known to underlie the corresponding changes in the spine structure [69], but how synaptic activities are linked to specific changes in the actin cytoskeleton in spines remains to be fully understood. Synaptic activities are known to elicit Ca2+ signals that can regulate the activity of a large number of actin regulatory proteins such as ADF/cofilin [65] and gelsolin [47]. Interestingly, Ca2+/calmodulin-dependent kinase II (CaMKII), the key molecule involved in LTP, has been found to gate the activity-dependent F-actin remodeling by controlling the access of actin regulatory proteins to F-actin [70]. Furthermore, a number of membrane lipids, especially phosphoinositides, are regulated by synaptic activities [71], which are known to play an important role in the regulation of actin polymerization near the plasma membrane [72]. Moreover, a recent study shows a novel activity-dependent mechanism of spine remodeling in which activity-dependent proteolytic cleavage of Neuroligin 1 generated a cytoplasmic C-terminal domain to activate LIM kinase 1, which inhibits ADF/cofilin leading to an increase in spine number and enhanced synaptic plasticity [73]. Finally, a number of actin-binding proteins have been shown to undergo activity-dependent translocation into dendritic spines during LTP, with specific sub-spine patterns and timing associated with distinct phases of spine remodeling [74]. Therefore, synaptic activities appear to regulate the actin remodeling in spines during synaptic plasticity through spatiotemporal targeting and coordinated control of the activities of a wide range of actin binding proteins.
Concluding remarks
Formation and remodeling of distinct F-actin structures involves the concerted efforts of a diverse array of actin regulatory proteins and activities. As illustrated in Figure 1, the essential steps include: 1) actin monomer binding and delivery to the polymerization site, 2) nucleation, 3) assembly, 4) capping, and 5) severing/disassembly. It is important to note that these steps are not discreet and that there is considerable overlap and crosstalk between the molecules involved at each step. In fact, due to the extensive crosstalk between different actin regulatory molecules/pathways, delineating the specific functions of individual actin binding molecules during spine formation and remodeling remains a huge challenge. Therefore, future studies are needed to understand how these actin regulatory molecules work in concert in response to synaptic signals, and how their functions overlap in distinct sub-spine regions. Importantly, many neurological disorders have been linked to disrupted actin mechanisms in neurons. For example, LIM kinase 1 (LIMK1) is a key regulator of ADF/cofilin molecules by phosphorylating and inhibiting ADF/cofilin. Its hemizygous deletion in Williams Syndrome patients is believed to contribute to behavioral and cognitive deficiencies [75]. Moreover, cofilin-actin enriched inclusions have also been observed in a number of neurodegenerative diseases [76]. Therefore, a better and full understanding of the actin regulation underlying synapse development and modification will help to delineate the molecular mechanisms underlying many of these devastating disorders.
Highlights.
Dendritic spines depend on the actin cytoskeleton and its remodeling for their structure and plasticity.
Actin nucleation factors enable the formation and growth of distinct F-actin networks in spines
Capping of actin filaments is a crucial step for the actin networks underlying spine formation and pasticity
Different actin regulatory proteins are spatiotemporally targeted and regulated in spines.
Disruption of the actin cytoskeleton underlies synaptic dysfunction in brain disorders.
Acknowledgments
The work is partially supported by research grants from National Institutes of Health to JQZ (GM083889 and MH104632), F32 fellowship to KRM (NS092342), and a F31 fellowship to OFO (NS092437).
Footnotes
Conflict of Interest Statement:
Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 2.Collingridge GL, Isaac JT, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5:952–962. doi: 10.1038/nrn1556. [DOI] [PubMed] [Google Scholar]
- 3.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 4.Song I, Huganir RL. Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 2002;25:578–588. doi: 10.1016/s0166-2236(02)02270-1. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni VA, Firestein BL. The dendritic tree and brain disorders. Mol Cell Neurosci. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segal M. Dendritic spines and long-term plasticity. Nat Rev Neurosci. 2005;6:277–284. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- 8.Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- 9.Geinisman Y. Structural synaptic modifications associated with hippocampal LTP and behavioral learning. Cereb Cortex. 2000;10:952–962. doi: 10.1093/cercor/10.10.952. [DOI] [PubMed] [Google Scholar]
- 10.Halpain S, Spencer K, Graber S. Dynamics and pathology of dendritic spines. Prog Brain Res. 2005;147:29–37. doi: 10.1016/S0079-6123(04)47003-4. [DOI] [PubMed] [Google Scholar]
- 11.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 12.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- **13.Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. This fluorescence and electron microscopy study provides a high-resolution view of the architecture of dendritic spines in neurons. In addition, this paper shows the fundamental architecture differences between the dendritic filopodia and other types of filopodia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hotulainen P, Llano O, Smirnov S, Tanhuanpaa K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim IH, Racz B, Wang H, Burianek L, Weinberg R, Yasuda R, Wetsel WC, Soderling SH. Disruption of Arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J Neurosci. 2013;33:6081–6092. doi: 10.1523/JNEUROSCI.0035-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 18.Hering H, Sheng M. Activity–dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci. 2003;23:11759–11769. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, Langeberg LK, Banker G, Raber J, Scott JD. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippi G, Steinert JR, Marczylo EL, D'Oro S, Fiore R, Forsythe ID, Schratt G, Zoli M, Nicotera P, Young KW. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Hadziselimovic N, Vukojevic V, Peter F, Milnik A, Fastenrath M, Fenyves BG, Hieber P, Demougin P, Vogler C, de Quervain DJ, et al. Forgetting Is Regulated via Musashi-Mediated Translational Control of the Arp2/3 Complex. Cell. 2014;156:1153–1166. doi: 10.1016/j.cell.2014.01.054. This report presents that the RNA-binding protein Musashi (MSI-1) can clear unwanted interneuron memories through the actin cytoskeleton by modulating the mRNA translation of Arp2/3 complex proteins in the Caenorhabditis elegans. [DOI] [PubMed] [Google Scholar]
- 22.Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomon SN, Haber M, Murai KK, Dunn RJ. Localization of the Diaphanous-related formin Daam1 to neuronal dendrites. Neurosci Lett. 2008;447:62–67. doi: 10.1016/j.neulet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 25.Dutta P, Maiti S. Expression of multiple formins in adult tissues and during developmental stages of mouse brain. Gene Expr Patterns. 2015;19:52–59. doi: 10.1016/j.gep.2015.07.003. [DOI] [PubMed] [Google Scholar]
- *26.Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. This paper provides the evidence that FMNL2 functions in actin polymerization, not nucleation, in membrane protrusion downstream of Cdc42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyagi Y, Yamashita T, Fukaya M, Sonoda T, Okuno T, Yamada K, Watanabe M, Nagashima Y, Aoki I, Okuda K, et al. Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J Neurosci. 2002;22:803–814. doi: 10.1523/JNEUROSCI.22-03-00803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. In this paper, the authors show that the vertebrate protein JMY, previously identified as transcriptional co-activator of p53, exhibits actin nucleating activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. This article reports a potentially new actin filament nucleator Leiomodin (Lmod), which is an actin-binding protein found in muscles. Furthermore, the Lmod cellular localization and nucleating activity is modulated by tropomyosin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **30.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. This important study shows that Drosophila Spire, which has a role in oocyte polarization, can nucleate actin filaments by a novel mechanism. The actin nucleation activity is mediated by four actin monomer-binding WH2 domains and a novel actin-binding motif. [DOI] [PubMed] [Google Scholar]
- **31.Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordonbleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. This ground-breaking paper uncovers a novel actin-nucleating protein Cordon-Bleu (cobl), which is enriched in brain and controls neuronal morphology and development. Cobl promotes formation of nonbundled, unbranched actin filaments more efficiently than formins via a mechanism involving all three WH2 domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **32.Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. This paper describes the novel actin-nucleation activity of adenomatous polyposis coli (APC). APC is possibly regulated by microtubules locally, and it is potentially very potent if synergized with formins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gates MA, Kannan R, Giniger E. A genome-wide analysis reveals that the Drosophila transcription factor Lola promotes axon growth in part by suppressing expression of the actin nucleation factor Spire. Neural Dev. 2011;6:37. doi: 10.1186/1749-8104-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira T, Ou Y, Li S, Giniger E, van Meyel DJ. Dendrite architecture organized by transcriptional control of the F-actin nucleator Spire. Development. 2014;141:650–660. doi: 10.1242/dev.099655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwintzer L, Koch N, Ahuja R, Grimm J, Kessels MM, Qualmann B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011;30:3147–3159. doi: 10.1038/emboj.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haag N, Schwintzer L, Ahuja R, Koch N, Grimm J, Heuer H, Qualmann B, Kessels MM. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J Neurosci. 2012;32:17842–17856. doi: 10.1523/JNEUROSCI.0843-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Firat-Karalar EN, Hsiue PP, Welch MD. The actin nucleation factor JMY is a negative regulator of neuritogenesis. Mol Biol Cell. 2011;22:4563–4574. doi: 10.1091/mbc.E11-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou W, Izadi M, Nemitz S, Haag N, Kessels MM, Qualmann B. The Actin Nucleator Cobl Is Controlled by Calcium and Calmodulin. PLoS Biol. 2015;13:e1002233. doi: 10.1371/journal.pbio.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chazeau A, Mehidi A, Nair D, Gautier JJ, Leduc C, Chamma I, Kage F, Kechkar A, Thoumine O, Rottner K, et al. Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 2014;33:2745–2764. doi: 10.15252/embj.201488837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- **41.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. This study provides evidence that capping protein gates actin monomers to the Arp2/3 complexpromoted filament nucleation by eliminating the competition from barbed ends, thus enhances actin filament nucleation by the Arp2/3 complex and actin-based motility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **42.Fan Y, Tang X, Vitriol E, Chen G, Zheng JQ. Actin capping protein is required for dendritic spine development and synapse formation. J Neurosci. 2011;31:10228–10233. doi: 10.1523/JNEUROSCI.0115-11.2011. This study provides the first evidence that the barbed end capping of actin filaments is essential for filopodia-to-spine transition, the formation of mushroom spines and funcational synapses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol. 2004;6:1180–1188. doi: 10.1038/ncb1199. [DOI] [PubMed] [Google Scholar]
- 44.Menna E, Zambetti S, Morini R, Donzelli A, Disanza A, Calvigioni D, Braida D, Nicolini C, Orlando M, Fossati G, et al. Eps8 controls dendritic spine density and synaptic plasticity through its actin-capping activity. EMBO J. 2013;32:1730–1744. doi: 10.1038/emboj.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatakou E, Marzo A, Gibb A, Salinas PC. Activity-dependent spine morphogenesis: a role for the actin-capping protein Eps8. J Neurosci. 2013;33:2661–2670. doi: 10.1523/JNEUROSCI.0998-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitanishi T, Sakai J, Kojima S, Saitoh Y, Inokuchi K, Fukaya M, Watanabe M, Matsuki N, Yamada MK. Activity-dependent localization in spines of the F-actin capping protein CapZ screened in a rat model of dementia. Genes Cells. 2010;15:737–747. doi: 10.1111/j.1365-2443.2010.01411.x. [DOI] [PubMed] [Google Scholar]
- 47.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 48.Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- 49.Matsuoka Y, Li X, Bennett V. Adducin: structure, function and regulation. Cell Mol Life Sci. 2000;57:884–895. doi: 10.1007/PL00000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porro F, Rosato-Siri M, Leone E, Costessi L, Iaconcig A, Tongiorgi E, Muro AF. beta-adducin (Add2) KO mice show synaptic plasticity, motor coordination and behavioral deficits accompanied by changes in the expression and phosphorylation levels of the alpha- and gamma-adducin subunits. Genes Brain Behav. 2010;9:84–96. doi: 10.1111/j.1601-183X.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 51.Rabenstein RL, Addy NA, Caldarone BJ, Asaka Y, Gruenbaum LM, Peters LL, Gilligan DM, Fitzsimonds RM, Picciotto MR. Impaired synaptic plasticity and learning in mice lacking beta-adducin, an actin-regulating protein. J Neurosci. 2005;25:2138–2145. doi: 10.1523/JNEUROSCI.3530-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52.Bednarek E, Caroni P. beta-Adducin is required for stable assembly of new synapses and improved memory upon environmental enrichment. Neuron. 2011;69:1132–1146. doi: 10.1016/j.neuron.2011.02.034. This study shows that beta-adducin, a protein that caps and stabilizes the growing ends of F-actins, is essential for the dynamic formation of synapses in the adult mouse hippocampus for augmented learning and long-term memory upon environmental enrichment. [DOI] [PubMed] [Google Scholar]
- 53.Engmann O, Giralt A, Gervasi N, Marion-Poll L, Gasmi L, Filhol O, Picciotto MR, Gilligan D, Greengard P, Nairn AC, et al. DARPP-32 interaction with adducin may mediate rapid environmental effects on striatal neurons. Nat Commun. 2015;6:10099. doi: 10.1038/ncomms10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamashiro S, Gokhin DS, Kimura S, Nowak RB, Fowler VM. Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton (Hoboken) 2012;69:337–370. doi: 10.1002/cm.21031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox PR, Fowler V, Xu B, Sweatt JD, Paylor R, Zoghbi HY. Mice lacking Tropomodulin-2 show enhanced long-term potentiation, hyperactivity, and deficits in learning and memory. Mol Cell Neurosci. 2003;23:1–12. doi: 10.1016/s1044-7431(03)00025-3. [DOI] [PubMed] [Google Scholar]
- 57.Paavilainen VO, Bertling E, Falck S, Lappalainen P. Regulation of cytoskeletal dynamics by actinmonomer- binding proteins. Trends Cell Biol. 2004;14:386–394. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- *58.Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 2005;310:866–869. doi: 10.1126/science.1114816. The authors establish a new principle which impacts on the fundamental understanding of synaptic plasticity. The new principle is that the necks of dendritic spines can act as neuronal activity-dependent diffusion barriers for molecules. [DOI] [PubMed] [Google Scholar]
- 59.Vitriol EA, McMillen LM, Kapustina M, Gomez SM, Vavylonis D, Zheng JQ. Two functionally distinct sources of actin monomers supply the leading edge of lamellipodia. Cell Rep. 2015;11:433–445. doi: 10.1016/j.celrep.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schluter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- *61.Pilo Boyl P, Di Nardo A, Mulle C, Sassoe-Pognetto M, Panzanelli P, Mele A, Kneussel M, Costantini V, Perlas E, Massimi M, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. Embo J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. This paper identifies profilin2’s role in brain through behavioral study in profilin2 knockout mice, which role cannot be compensated for by the ubiquitously expressed profilin1. This should provoke future studies on profilin2's role in modulating actin dynamics at the synapse. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neuhoff H, Sassoe-Pognetto M, Panzanelli P, Maas C, Witke W, Kneussel M. The actin-binding protein profilin I is localized at synaptic sites in an activity-regulated manner. Eur J Neurosci. 2005;21:15–25. doi: 10.1111/j.1460-9568.2004.03814.x. [DOI] [PubMed] [Google Scholar]
- 63.Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- 64.Lamprecht R, Farb CR, Rodrigues SM, LeDoux JE. Fear conditioning drives profilin into amygdala dendritic spines. Nat Neurosci. 2006;9:481–483. doi: 10.1038/nn1672. [DOI] [PubMed] [Google Scholar]
- 65.Bernstein BW, Bamburg JR. ADF/cofilin a functional node in cell biology. Trends in Cell Biology. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. U http://www.ncbi.nlm.nih.gov/pubmed/20133134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Troys M, Huyck L, Leyman S, Dhaese S, Vandekerkhove J, Ampe C. Ins and outs of ADF/cofilin activity and regulation. Eur J Cell Biol. 2008;87:649–667. doi: 10.1016/j.ejcb.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 67.Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. Journal of Neurobiology. 2004;58:103–117. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- *68.Gu J, Lee CW, Fan Y, Komlos D, Tang X, Sun C, Yu K, Hartzell HC, Chen G, Bamburg JR, et al. ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat Neurosci. 2010;13:1208–1215. doi: 10.1038/nn.2634. This study provides evidence that temporally separated ADF/cofilin activation and inactivation mediate synaptic receptor insertion and spine enlargement, respectively. Therefore, a single actin regulatory protein can regulate both the functional and structural aspects of synaptic plasticity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- *70.Kim K, Lakhanpal G, Lu HE, Khan M, Suzuki A, Hayashi MK, Narayanan R, Luyben TT, Matsuda T, Nagai T, et al. A Temporary Gating of Actin Remodeling during Synaptic Plasticity Consists of the Interplay between the Kinase and Structural Functions of CaMKII. Neuron. 2015;87:813– 826. doi: 10.1016/j.neuron.2015.07.023. This paper provides evidence for a novel gating mechanism for actin remodeling by CaMKII. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dotti CG, Esteban JA, Ledesma MD. Lipid dynamics at dendritic spines. Frontiers in Neuroanatomy. 2014;8 doi: 10.3389/fnana.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 73.Liu A, Zhou Z, Dang R, Zhu Y, Qi J, He G, Leung C, Pak D, Jia Z, Xie W. Neuroligin 1 regulates spines and synaptic plasticity via LIMK1/cofilin-mediated actin reorganization. J Cell Biol. 2016;212:449–463. doi: 10.1083/jcb.201509023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **74.Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82:444–459. doi: 10.1016/j.neuron.2014.03.021. In this study, the authors provided evidence for translocation of a number of actin regulatory proteins into spines with specific patterns and sequential phases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoogenraad CC, Akhmanova A, Galjart N, De Zeeuw CI. LIMK1 and CLIP-115: linking cytoskeletal defects to Williams syndrome. Bioessays. 2004;26:141–150. doi: 10.1002/bies.10402. [DOI] [PubMed] [Google Scholar]
- 76.Bamburg JR, Bloom GS. Cytoskeletal pathologies of Alzheimer disease. Cell Motil Cytoskeleton. 2009;66:635–649. doi: 10.1002/cm.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]