Abstract
Time-lapse imaging of biological samples is important for understanding complex (patho)physiological processes. A growing number of point-of-care biomedical assays rely on real-time imaging of flowing or migrating cells. However, the cost and complexity of integrating experimental models simulating physiologically relevant microenvironments with bulky imaging systems that offer sufficient spatiotemporal resolution limit the use of time-lapse assays in research and clinical settings. This paper introduces a compact and affordable lens-free imaging (LFI) device based on the principle of coherent in-line, digital holography for time-lapse cell migration assays. The LFI device combines single-cell resolution (1.2 μm) with a large field of view (6.4 × 4.6 mm2), thus rendering it ideal for high-throughput applications and removing the need for expensive and bulky programmable motorized stages. The set-up is so compact that it can be housed in a standard cell culture incubator, thereby avoiding custom-built stage top incubators. LFI is thoroughly benchmarked against conventional live-cell phase contrast microscopy for random cell motility on two-dimensional (2D) surfaces and confined migration on 1D-microprinted lines and in microchannels using breast adenocarcinoma cells. The quality of the results obtained by the two imaging systems is comparable, and they reveal that cells migrate more efficiently upon increasing confinement. Interestingly, assays of confined migration more readily distinguish the migratory potential of metastatic MDA-MB-231 cells from non-metastatic MCF7 cells relative to traditional 2D migration assays. Altogether, this single-cell migration study establishes LFI as an elegant and useful tool for live-cell imaging.
Introduction
Scientists have imaged living organisms since the first observations of motile cells by Leeuwenhoek in the 17th century.1 Time-lapse analysis of biological specimens is essential for understanding dynamic and complex (patho)physiological processes and has been growing in use and importance 1-5 with applications in basic science research 6, 7, drug screening assays 8, and point-of-care tests 9. However, combining in vitro experimental models with complex imaging systems capable of generating data at the required temporal and spatial resolution presents technical challenges which limit the use of such assays in some research and most clinical settings. Therefore, an affordable alternative live-cell imaging platform is needed that can be extensively miniaturized and incorporated into (single-use) point-of-care tests.
One promising imaging technique, lens-free imaging (LFI), takes advantage of the principle of digital holography. 10 Holography was introduced by Dennis Gabor in 1948 11 and evolved into digital holography after direct recording of the Fresnel holograms with charge coupled devices by Schnars and Jüptner in the early 1990s.12 In these systems, objectives are replaced with mathematical calculations, greatly simplifying the optical set-up. Next to the cost and space reduction implied by this, the greatest advantage is that a lens-free image essentially is a 3D image that can be reconstructed at any focal depth. Focus becomes a digital parameter and is thus a data-processing parameter instead of a physical action (Movie S1), avoiding stage drift, which is a common issue in time-lapse phase contrast microscopy (Movie S2). In a typical LFI device, the field of view and resolution are determined by the size of the imaging sensor chip and pixel pitch, i.e. pixel interval spacing, which are constantly improving, with larger chip sizes, smaller pixel pitch and decreasing costs as a consequence of the rapidly evolving imager and semi-conductor industries. The large field of view allows inspection of large surfaces without the need for mechanical scanning while keeping sufficient resolution via digital zoom. The simple optical path enables LFI systems to be compact and housed in common cell culture incubators. Further miniaturization of all the system components, e.g. by incorporation of waveguides, will render the LFI even more compact and suited for point-of-care applications. In sum, these factors result in a robust, compact, and inexpensive technology attainable in both research and clinical settings.
In this study, we benchmark the lens-free imaging technique, with current state of the art field of view and resolution, as a time-lapse live-cell imaging tool. Three widely used motility assays are imaged side-by-side using both LFI and conventional phase contrast microscopy: random motility on 2D unconfined surfaces, confined motility on 1D microprinted lines, and confined motility in 3D microchannels. The motility assays are chosen in such a way that they account for 1D, 2D, and 3D cell migration, taking into account the importance of microenvironmental topography on cell response.13 Data are generated using two breast adenocarcinoma cell lines: non-metastatic MCF7 cells and metastatic MDA-MB-231 cells. 14-16 The quality of the results obtained by the two imaging systems is comparable and the high-throughput nature of LFI experiments further enables us to study how metastatic and non-metastatic cells respond to physical confinement. Importantly, we demonstrate that confined migration on microprinted lines or in microchannels more readily distinguishes the enhanced migratory potential of metastatic MDA-MB-231 cells versus non-metastatic MCF7 cells relative to traditional 2D migration assays. Overall, this study establishes the utility of LFI for live-cell imaging applications and provides important information on microenvironmental regulation of tumor cell behavior.
Results & discussion
Lens-free imaging via coherent in-line digital holography is suited for biological samples
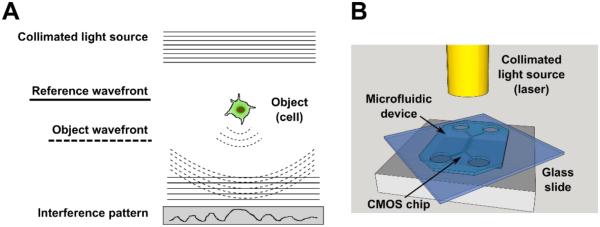
The LFI technique is based on the principle of in-line digital holography. A schematic representation of the principle and set-up is shown in Fig. 1A. In-line holography is very elegant for cell migration assays because the imaged objects are transparent and in this way the more complex optics needed to maintain a separate reference beam can be avoided. The biological sample is placed on top of the CMOS imaging sensor chip and illuminated with a laser (Fig. 1B). The sample is thus imaged in transmission mode, similar to inverted phase contrast microscopy. The imager records the interference pattern of the reference wavefront (corresponding to the laser light, full lines in Fig. 1A) and the object wavefront (corresponding to laser light scattered by the object, dotted lines in Fig. 1A). Subsequently, the imaged object is digitally reconstructed using the interference pattern, reference wavefront and focal depth, i.e. distance between imager and object, as inputs (Fig. S1). As the position of the sample is much closer to the imager than the light source, there is a one to one correspondence between the real cell size and the size of the reconstructed cell.17 Reconstruction is based on the Fresnel-Kirchhoff integral.18 To eliminate the distorting twin image inherently produced by in-line holography 19, iterative phase retrieval is used as described previously by Stahl et al..20 Use of coherent illumination enabled sub-cellular resolution and rapid image reconstruction. Live preview images could be obtained using rapid reconstructions of one-wavelength holograms for accurate sample positioning and real-time monitoring of the sample. The LFI set-up was assembled and operated in a standard cell culture incubator (37°C, 5% CO2; Fig. S2), circumventing the cost of custom-built stage-top environments.
Fig. 1. Lens-free imaging (LFI) principle of operation and schematic.
(A) Principle of operation of holographic in-line LFI. A collimated light source sent through a pinhole encounters a transparent object. The non-interacting reference wavefront and the object wavefront travel to a sensor (in this case, a CMOS chip), creating an interference pattern that is read by the sensor. The interference pattern is then reconstructed to create an image of the object. (B) Schematic of the LFI platform. Laser light travels through a fiber optic cable to illuminate the sample, here represented by a transparent microfluidic chip. The reference wavefront and object wavefront are detected using a CMOS sensor.
Images acquired by LFI were compared to inverted phase contrast microscopy images in Fig. S3-4. As the goal of time-lapse imaging is to track the paths of cells over time, they should remain in the field of view over the course of an experiment, and images should be taken at prescribed intervals. In our systems, the field of view for LFI was 6.41 × 4.58 mm2, and that of phase contrast (using a 10x, 0.45 NA objective) was 0.90 × 0.67 mm2, approximately a 50-fold difference. LFI images were taken every 10 min, resulting in stacked images showing migrating cells over large distances (Movie S3). However, imaging speeds of as high as 150 frames per second can be obtained. Although the resolution of the phase contrast system with this objective and a halogen lamp is marginally better than that of LFI (0.92 μm compared to 1.2 μm), single cell morphology is clearly visible for both imaging techniques. LFI resolution was determined using an US Air Force resolution test chart (Fig. S5). Of note, time-lapse LFI did not affect cell viability (Fig S6). Both phase contrast microscopy and LFI are thus suitable for imaging of migrating cells, with LFI being more favorable, as the large field of view allows cell tracking over much greater distances. Additionally, LFI needs markedly reduced handling steps and capital requirements and it has great potential for further miniaturization.
While a number of experimental approaches for lensless imaging have been developed, we specifically chose to use coherent illumination in a system without a pinhole or translatable stage for live-cell imaging. Lens-free LED platforms developed by Ozcan et al. 21, 22 that are similar to our set-up but use a LED light source in combination with a pinhole (~50 to 150 μm) to improve the spatial coherence of the LED light, achieve partially coherent digital in-line holography. This technique has been used for cell identification 23, 24 and semen analysis 25, to monitor adhering and dividing cells 26, 27, and to analyze random motility of fibroblasts on polyacrylamide gels 28, but fine cell morphology (~3-5 μm) was not resolved. While the resolution of partially coherent LED-based systems can be improved by using a synthetic aperture 29-37, this technique requires shifting the object or light source and recording a large number (~30-100) of sub-pixel shifted holograms, adding complexity to both the acquisition hardware and computational reconstruction scheme 38. Application of the synthetic aperture techniques for live-cell imaging to obtain sub-micron resolution is problematic, as it is important that the imaging speed is faster than that of the biological processes, hence there is not enough time to record a set of 100 holograms without any cell movement. As both the temporal and spatial coherence of lasers is much higher than that of LEDs, a single recorded hologram will contain more information, so fewer input holograms and less complex reconstruction algorithms are required. In this way image recording and reconstruction times are substantially decreased, enabling live views of the imaged sample 20. Next to this, the set-up design remains simple, as small and inexpensive semiconductor laser diodes are used and sub-pixel shifting is avoided. The presented LFI system uses four pigtailed laser diodes that are all coupled into one single mode optical fiber, avoiding alignment issues. The lasers are kept outside of the cell culture incubator and the light is guided into the incubator by the fiber to illuminate the sample and image sensor. An iterative phase retrieval technique is used based on 4 input holograms recorded using laser illumination at 4 different wavelengths 20, resulting in short image acquisition times (~6 ms) while correcting for the twin images inherently present when using digital in-line holography.
Although high-coherent laser light sources yield high quality holograms, they do introduce extra background signal. Speckle noise and multiple-reflection interference effects, generating extra background, are specific for laser illumination. Speckle noise was minimized by stabilizing the lasers through accurate temperature and power control. Next to this, the single mode optical fiber that is used to guide the light also serves as a spatial filter, severely reducing speckle noise. However, some speckle can still be observed in the LFI pictures obtained by our setup (Fig. S7A). Considering that the obtained picture quality is sufficient for the application at hand, no further action was taken to remove the remaining speckle noise using post-processing algorithms. Another issue with laser illumination is that, due to the high spatial coherence, interference occurs over larger distances. This means that the light diffracted by different cells, or cells and other structures in the sample, also adds to the complete interference pattern that is measured at the detector. This multi-interference issue is most relevant when cell motility in microchannels is imaged. Edge artefacts, e.g. dark areas and ring-like patterns, can be observed at the edges of the microchannels, and the repetitive pattern of channels causes shadow lines in between two consecutive channels (Fig. S7B). As a result, cells located close to the edges of the channels can be partially blurred (Fig. S7C). This issue can be minimized by designing the spacing of microstructures so that shadow lines do not fall at the positions of the cells and by adapting the image reconstruction software using the known interference created by the micropatterns. A more comprehensive comparison of the strengths and weaknesses of various holographic imaging systems is presented in the ESI. In summary, the LFI set-up presented in this work is ideally suited for live-cell imaging purposes as it combines short acquisition times (6 ms), large field of view (29 mm2), and good resolution (1.2 μm) with hardware void of moving stages or light sources. High imaging speeds can be combined with fine resolution as a result of the high-coherent laser illumination that enables high resolution hologram recording.
Cell migration assay results generated by LFI and phase contrast imaging are quantitatively compared
To quantitatively benchmark LFI for use in motility assays, we tracked cells migrating on planar surfaces, on 6 μm-wide microprinted lines, and in 6 μm-wide microchannels using both LFI and phase contrast microscopy. All substrates were coated with collagen type I. Metastatic MDA-MB-231 and tumorigenic but non-metastatic MCF7 breast adenocarcinoma cells were imaged at 10 min intervals for up to 2 h, although longer imaging periods were possible, as shown in Fig. S8. Quantitative descriptors of cell migration were calculated from cell trajectories tracked using the two imaging platforms and were directly compared to each other.
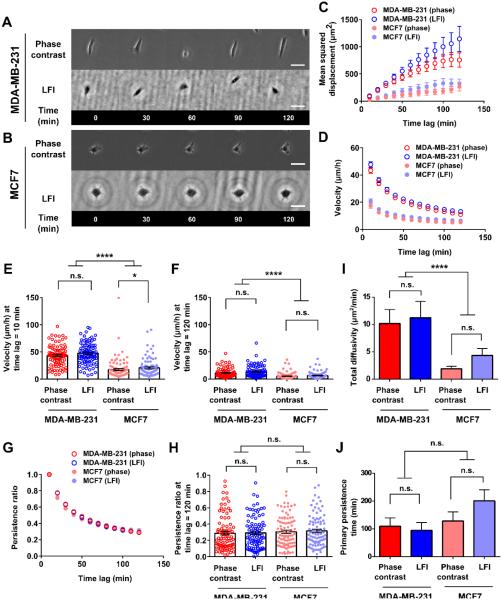
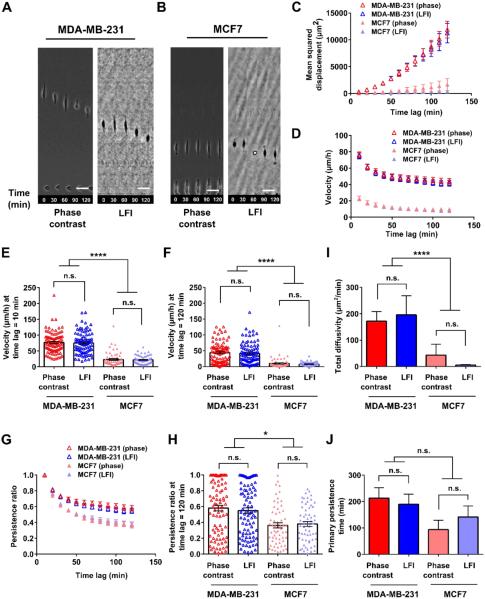
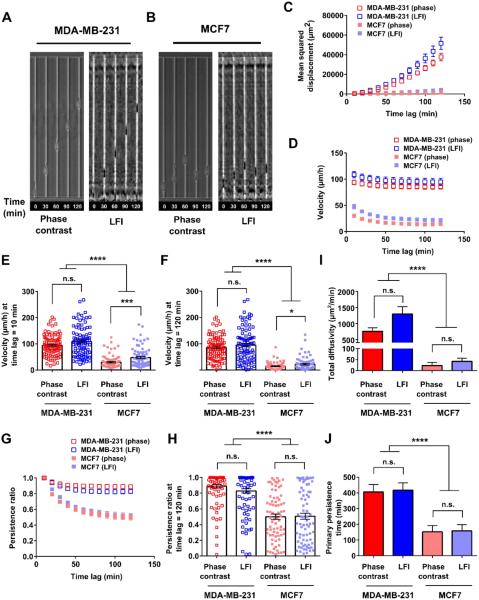
Representative images of cells migrating on planar substrates, on microlines, and in microchannels are shown in panels A and B of Fig. 2, 3, and 4 and in Movies S4-9. Confinement to microlines and microchannels elicited elongated cell morphologies similar to those observed in earlier studies. 14, 39-42 Mean squared displacement (MSD), velocity, and persistence were calculated for individual cells at different time lags and averaged across the entire cell population at any given time for each imaging platform (Fig. 2, 3, and 4, panels C, D, and G). MSD is a commonly used metric to illustrate the diffusion of a cell population over time. 43, 44 Velocity was calculated as the net displacement of a cell over a given time interval divided by that time period. Persistence was quantified by dividing the net cell displacement over a given time lag by the length of the total path traveled by the cell. 40, 45 These descriptors are further illustrated in Fig. S9. As shown in Fig. 2, 3, and 4C, the MSDs of metastatic MDA-MB-231 cells tracked by LFI versus phase contrast microscopy were similar in each physical microenvironment. Similarly, no statistical differences were detected between the two imaging platforms for non-metastatic MCF7 cells migrating on planar surfaces, on microprinted lines, or in microchannels. As expected, MSD increases with increasing time lag as cell displacement from initial position increases on average with time. Of note, the increase in MSD is more pronounced for the faster migrating MDA-MB-231 cells.
Fig. 2. Phase contrast and LFI imaging platforms generate similar results for random 2D motility assays.
Time-lapse images of (A) MDA-MB-231 and (B) MCF7 breast adenocarcinoma cells migrating on collagen type I-coated glass slides and imaged using either phase contrast microscopy (10x, 0.45 NA objective) or the LFI platform. Scale bars represent 50 μm. (C) Mean squared displacements observed for the two cell types with each imaging platform. (D) Cell velocity as a function of time lag. Velocities for time lags of (E) 10 min and (F) 120 min are also shown. (G) Persistence ratio as a function of time lag. The persistence ratio at a time lag of (H) 120 min is also shown. (I) Total diffusivity observed for the two cell types with each imaging platform. (J) Primary persistence time observed for the two cell types with each imaging platform. For all metrics, cell trajectories were tracked for the indicated time periods for up to 2 h. N=90 cells/condition, with 30 cells/experiment were analyzed from 3 independent experiments. Statistical significance between phase contrast and LFI imaging results was determined by an unpaired t test if cells passed the D’Agostino and Pearson omnibus normality test, or by Mann-Whitney test if they did not. Differences between MDA-MB-231 and MCF7 cells were assessed by Kruskal-Wallis test with Dunn’s multiple comparisons post-test. n.s., difference not statistically significant; *, p<0.05; ****, p<0.0001.
Fig. 3. Phase contrast and LFI imaging platforms generate similar results for microcontact printing migration assays.
Time-lapse images of (A) MDA-MB-231 and (B) MCF7 breast adenocarcinoma cells migrating on 6 μm-wide collagen type I printed lines and imaged using either phase contrast microscopy (10x, 0.45 NA objective) or the LFI platform. Scale bars represent 50 μm. (C) Mean squared displacements observed for the two cell types with each imaging platform. (D) Cell velocity as a function of time lag. Velocities for time lags of (E) 10 min and (F) 120 min are also shown. (G) Persistence ratio as a function of time lag. The persistence ratio at a time lag of (H) 120 min is also shown. (I) Total diffusivity observed for the two cell types with each imaging platform. (J) Primary persistence time observed for the two cell types with each imaging platform. For all metrics, cell trajectories were tracked for up to 2 h. For MDA-MB-231 cells, N=90 cells/condition, with 30 cells/experiment were analyzed from 3 independent experiments. For MCF7 cells, N=60 cells/condition, with 30 cells/experiment analyzed over 2 independent experiments. Statistical significance between phase contrast and LFI imaging results was analyzed by Mann-Whitney test. Differences between MDA-MB-231 and MCF7 cells were assessed by Kruskal-Wallis test with Dunn’s multiple comparisons post-test. n.s., difference not statistically significant; *, p<0.05; ****, p<0.0001.
Fig. 4. Phase contrast and LFI imaging platforms generate similar results for microchannel migration assays.
Time-lapse images of (A) MDA-MB-231 and (B) MCF7 breast adenocarcinoma cells migrating through 6 μm-wide, 10 μm-tall collagen type I-coated PDMS microchannels and imaged using either phase contrast microscopy (10x, 0.45 NA objective) or the LFI platform. (C) Mean squared displacements observed for the two cell types with each imaging platform. (D) Cell velocity as a function of time lag. Velocities for time lags of (E) 10 min and (F) 120 min are also shown. (G) Persistence ratio as a function of time lag. The persistence ratio at a time lag of (H) 120 min is also shown. (I) Total diffusivity observed for the two cell types with each imaging platform. (J) Primary persistence time observed for the two cell types with each imaging platform. For all metrics, cell trajectories were tracked for up to 2 h. For MDA-MB-231 cells, N=90 cells/condition, with 30 cells/experiment were analyzed from 3 independent experiments. For MCF7 cells, N=75 cells/condition, with 25 cells/experiment analyzed over 3 independent experiments. Statistical significance between phase contrast and LFI imaging results was analyzed by Mann-Whitney test. Differences between MDA-MB-231 and MCF7 cells were assessed by Kruskal-Wallis test with Dunn’s multiple comparisons post-test. n.s., difference not statistically significant; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.
Both velocity and persistence of metastatic MDA-MB-231 cells tracked by LFI versus phase contrast microscopy were similar in each physical microenvironment (Figs. 2-4, D and G). To more closely evaluate the two imaging platforms, velocities were also compared at the shortest (10 min) and longest (120 min) time periods recorded (Figs. 2-4, E-F). The velocity at a time lag of 10 min corresponds to the average instantaneous (point-to-point) speed of a cell over the course of tracking. At the longest time lag, velocity is the total displacement of the cell over the course of imaging divided by the time lag. No statistical differences were noted by the two imaging platforms for metastatic MDA-MB-231 cells tracked at short or long time periods. In contrast, the velocity of the slowly migrating non-metastatic MCF7 cells tracked by LFI was modestly higher than that of cells monitored by phase contrast microscopy for a subset of the cases examined. For instance, the velocity of randomly migrating MCF7 cells tracked by LFI was higher than that by phase contrast microscopy at 10 min but not at 120 min (Fig. 2, E-F). Similarly small differences were detected between the two imaging platforms for MCF7 cells migrating inside microchannels (Fig. 4, E-F), though no difference in cell velocity was noted in microprinted line migration assays (Fig. 3, E-F).
Typically, the velocity of a given cell decreases with increasing tracking times (Figs. 2-3, D) due to the non-linear path of a migrating cell on a planar surface (Fig. 2, A-B) or low persistence (Fig. 3, G-H). In contrast, no or small differences in velocities at short versus long times are observed for highly persistent cells moving unidirectionally inside narrow channels (Fig. 4D). It is noteworthy that the two imaging platforms yielded similar data for cell persistence in each of the three physical microenvironments examined in this work (Figs. 2-4, G-H).
Migration can be mathematically described by random walk models and persistent random walk models. Recently, Wu et al. developed a variation on the persistent random walk model, taking into account that certain cell types have a preferred direction of migration. 46, 47 The anisotropic random walk model (APRW) is well suited for our experiments, as migration on microprinted lines and in microchannels represents the extreme case of this model by exclusively allowing migration along one axis. Two descriptors of the APRW model, namely, primary persistence time and total diffusivity, were calculated here. No statistical differences were noted for total diffusivity and primary persistence when metastatic MDA-MB-231 cells were tracked by the two imaging platforms in each physical microenvironment (Figs. 2-4, I-J). Similar observations were made for non-metastatic MCF7 cells. Of note, many cells displayed primary persistence times longer than 120 min. The calculated persistence times represent extrapolations to best fit a mathematical model, and several cells had persistence times up to the user defined limit of parameter fit (1000 min). As the model was fit to cell tracks taken over 120 min intervals, longer persistence times have no physical meaning and should be interpreted as illustrating relative changes in speed and persistence between cell types and migration microenvironments instead of direct physical values.
Migration platforms demonstrate distinctive migratory potential of metastatic and non-metastatic breast adenocarcinoma cells
With LFI benchmarked for imaging of single-cell migration assays, we used this platform to compare the migratory potentials of metastatic and non-metastatic breast adenocarcinoma cells. For cells randomly migrating on a planar substrate, metastatic MDA-MB-231 cells displayed significantly higher MSDs, velocities, and total diffusivities than non-metastatic MCF7 cells (Fig. 2, C-F and I). MDA-MB-231 cells achieved on average about 9-fold greater displacements than MCF7 cells after 120 min. This finding is corroborated by detailed inspection of cell velocities at both short (10 min) and long (120 min) time lags and of total diffusivity calculated using the APRW model (p < 0.0001, Fig. 2, E-F, I). Interestingly, there was no significant difference in persistence between the two cell types (Fig. 2, G-H). Similarly, no difference was detected in the persistence time along the primary axis of migration calculated from the APRW model (Fig. 2J). Thus, on planar substrates, metastatic MDA-MB-231 cells achieve larger 2D displacements because they migrate faster, and not because they travel in a more persistent manner. Identical conclusions were reached when longer tracking times were used (Fig. S8).
On 6 μm-wide microprinted collagen I lines, MDA-MB-231 cells migrated with significantly greater velocities at all times (Fig. 3, D-F) and displayed a higher total diffusivity than MCF7 cells (Fig. 3I). Although the persistence of metastatic cells was higher than that of non-metastatic cells for t ≥ 30 min (e.g., at t = 120 min, p < 0.05) (Fig. 3, G-H), primary persistence times calculated from the APRW model (Fig. 3J) were similar for both cell types. Overall, the markedly higher velocities of metastatic MDA-MB-231 cells enabled them to achieve about 11-fold greater 1D displacements than MCF7 cells by the 120 min mark (Fig. 3C).
In contrast to cells migrating on planar surfaces or microprinted lines, metastatic MDA-MB-231 cells inside microchannels (W=6 μm, L= 600 μm, H=10 μm) not only displayed markedly higher MSD, velocity, and total diffusivity than non-metastatic MCF7 cells, but also augmented persistence (Fig. 4, C-J). The MDA-MB-231 cell’s larger 1D displacements (approximately 15x larger at the end of an experiment) are hence caused by faster and more persistent migration, as opposed to the two other migration assays in which these larger displacements were driven mainly by faster migration. Taken together, the three assays demonstrated the differences in migration behavior of metastatic MDA-MB-231 versus non-metastatic MCF7 breast cancer cells.
In order to resemble the natural environment of the cells more closely, the assays were coated with the extracellular matrix protein collagen type I. Prior work reveals that human osteosarcoma cells migrate with distinct efficiencies inside 6 μm-wide microchannels coated with different types of extracellular matrix proteins, such as collagen type I, laminin, hyaluronic acid, fibronectin, or collagen type IV 14. We thus anticipate that the speed of breast cancer cells will vary on different substrate coatings. Importantly, we expect that the elevated migratory potential of metastatic MDA-MB-231 relative to non-metastatic MCF7 cells would also be observed using extracellular matrix proteins other than collagen type I, especially in confined microenvironments.
Lateral confinement to microprinted lines and compressive confinement within microchannels differentially regulates cell motility
It has been postulated that cell behavior is influenced by the local microenvironment 13, 48 and that careful consideration of physical aspects of the in vivo microenvironment is required when designing experiments. In our case, the three experimental designs can be classified according to the following two parameters: i) allowed directions or freedom of migration, and ii) number of confining surfaces. A cell migrating in free space has six degrees of freedom (positive and negative x, y, and z). Cells on 2D surfaces can move in four directions and are confined only at their basal surface. On narrow 1D microprinted protein lines, cells can move in two directions but are confined by one surface only; lateral confinement merely occurs by cell-generated forces and is caused by adhesion of the cell to its substrate. 39 In 3D microchannel assays, cells are also restricted to move in only two directions but are confined by four surfaces.
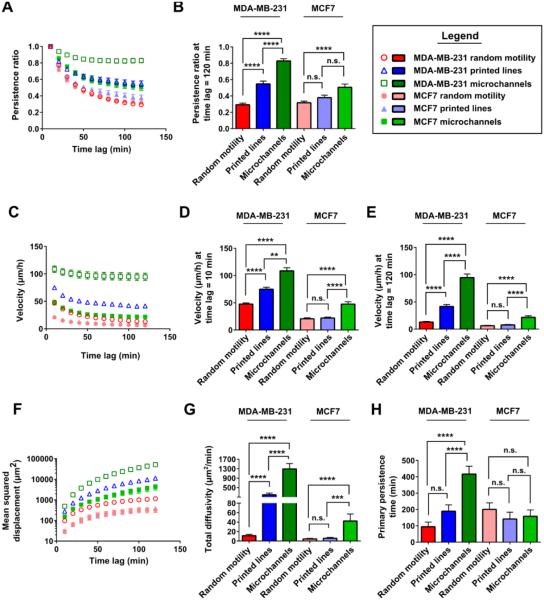
Comparison of cell behavior in the three different experimental designs demonstrated increasing displacement, diffusivity, velocity, and persistence for increased confinement (Fig. 5). This effect was most pronounced in microchannels, as they are the only case in which the cells are confined by four surfaces. For MDA-MB-231 cells, velocity and persistence are highest for microchannels, intermediate for microprinted lines, and lowest for planar random migration; this ranking proved to be of high statistical relevance (Fig. 5, A-E). For MCF7 cells, migration behavior is similar on surfaces with or without confinement by 1D microprinted lines. MCF7 cells migrating in microchannels show markedly higher diffusivity as well as velocity (Fig. 5, C-E and G), while results on persistence are mixed. Persistence ratios (time lag 120 min) are higher for microchannels than for the other experimental designs (Fig. 5B), but there are no relevant differences in primary persistence time for all assays (Fig 5H). Similar conclusions can be drawn from comparison of phase contrast imaging results (Fig. S10).
Fig. 5. Increasing confinement results in more efficient cell migration.
Data generated via LFI imaging across differing levels of physical cell confinement were compared. Persistence ratios were calculated (A) as a function of time lag and (B) at 120 min for both MDA-MB-231 and MCF7 cells in different physical microenvironments. Velocity was determined (C) as a function of time lag and at (D) 10 min and (E) 120 min for both MDA-MB-231 and MCF7 cells in each microenvironment. (F) Mean squared displacement, (G) total diffusivity, and (H) primary persistence time were calculated for MDA-MB-231 and MCF7 cells in each microenvironment. Cells were tracked by LFI for up to 120 min. For MDA-MB-231 cells, N=90 cells/condition, with 30 cells/experiment were analyzed from 3 independent experiments. For MCF7 cells, N=90 cells, with 30 cells/experiment were analyzed from 3 independent experiments from 2D assays; N=60 cells, with 30 cells/experiment were analyzed from 2 independent experiments for printed 1D lines; and N=75 cells, with 25 cells/experiment were analyzed from 3 independent experiments for microchannel results. Comparisons between microenvironments for a given cell type were made with Kruskal-Wallis test with Dunn’s multiple comparisons post-test. n.s., difference not statistically significant; **, p<0.01; ***, p<0.001; ****, p<0.0001.
In sum, microchannel assays amplified the differences in the migratory potential of MDA-MB-231 versus MCF7 breast cancer cells. It can be concluded that, for these breast adenocarcinoma cell lines, increasing confinement triggers larger cell displacements and higher diffusivity due to increasing cell velocity and persistence. This observation is more pronounced for faster moving metastatic MDA-MB-231 cells than for slow moving non-metastatic MCF7 cells.
Conclusions
Issues with time-lapse imaging involve maintenance of suitable microenvironments for both the organisms being imaged and the technology used to capture these images. Cells must be maintained in an environment with a controlled temperature and carbon dioxide (CO2) content, whereas phototoxicity and media evaporation must be minimized. 2, 3, 5 For this purpose, laboratories acquire expensive, custom-built stage top incubators that are designed to fit the specific optical tool and experimental conditions desired. Common problems with this approach include: damage to the microscope hardware by the humidity in the chamber necessary to avoid evaporation and changes in osmolarity 3; limited temporal resolution due to mechanical parts, e.g., motorized microscope stage translation 2; long times necessary to stabilize enclosure temperatures 3; stage drift caused by changes in room temperature 3-5 or heating of microscope components 49; and inflexible setups with optical tables overflowing with cables and tubes to component controllers, CO2 tanks, light sources, etc. Complex autofocus routines can help with stage drift, but they require additional illumination of the sample.5 The need for specialized microscopes, with expensive features and custom-built cell culture environments, hinders live-cell research, as the number of experiments being done is limited by the number of imaging tools available to the laboratory.
LFI using coherent lasers as light source was introduced in this study for time-lapse live-cell imaging applications as an alternative to the current complex and expensive microscopes. Our study employed various widely used assays of cell migration spanning different dimensions (i.e., 1D, 2D and 3D) on the order of those found in vivo 50, 51 to establish LFI as an alternative and reliable imaging technique for in vitro cell migration assays. The direct comparison of LFI with the conventional phase contrast method demonstrated the many advantages of the lens-free technique. Specifically, the LFI technique imaged a 50-fold larger area with 50-fold less expensive equipment, while it was housed in a conventional cell culture incubator common to cell biology laboratories. Importantly, because of the digital focus parameter, the sample could never move out of focus. Next to this, the large field of view avoided manual mechanical scanning. Together these features comprise a system that is inexpensive, easy to operate and well-suited for high-throughput applications.
In addition to benchmarking the LFI platform to phase contrast microscopy, the high-throughput nature of LFI experiments allowed assessment of how different migration microenvironments affect cell migration behavior, and how these behaviors are modulated when metastatic versus non-metastatic cell lines are assayed. For instance, although metastatic MDA-MB-231 cells and non-metastatic MCF7 cells migrated with essentially identical persistence on 2D surfaces, metastatic cells had a greater velocity. In contrast, the differences between metastatic and non-metastatic breast cancer cells were magnified in confinement. MDA-MB-231 cells were both faster and more persistent than MCF7 cells when confined to 1D migration. Interestingly, these parameters (velocity and persistence) were higher in microchannels than on microprinted lines, thereby illustrating the importance of 3D topography in modulating cell migration. These findings suggest that more complex microenvironments that better recapitulate aspects of the in vivo microenvironment can better distinguish the migratory potential of different cell types.
By increasing the throughput and decreasing the cost of in vitro experiments, LFI may enable a new generation of point-of-care single-cell inspection applications. This work demonstrates how LFI and microfluidic chips can be combined to render compact and user-friendly cell migration tests that show great potential for further translation to the clinic for point-of-care applications. Migratory potential of patient tumor cells could be investigated in this way and, by parallelization of the assay, the influence of various anti-cancer medication on cell (migration) behavior could be tested, enabling personalized medicine. For example, microfluidic point-of-care assays reliant on migration through PDMS microchannels are already in development to study the immune response from whole-blood samples.9, 52, 53 Next to this, the LFI platform is very well suited for high throughput screening, as many systems can be used parallel to each other due to their compactness and low cost. In future implementations, fluorescence microscopy techniques could be integrated with the lens-free apparatus 54, 55 to perform commercially available live-dead cell assays and obtain time-lapse images of fluorescently stained (e.g., CellTracker) or transgenic (e.g., LifeAct-GFP-expressing) cells. That said, fluorescence staining involves time-consuming protocols and often label-free assays are preferred. In that aspect, LFI of motility assays already visualizes dividing and dying cells clearly (Fig. S11), and information on the cell state can also be obtained from LFI phase images 26, 27, adding value to LFI as a label-free method. The results presented here demonstrate that LFI is an economical and reliable alternative to phase contrast imaging that would make live-cell imaging more translatable and economical.
Experimental
Study design
This study was designed as a controlled laboratory experiment in order to benchmark LFI against conventional phase-contrast microscopy for single-cell migration assays. For this, two types of breast adenocarcinoma cells were compared, i.e. MCF7 and MDA-MB-231 cell lines, and three different microenvironments were used, i.e. 2D random migration, 1D migration on microprinted collagen lines and 1D migration inside microchannels. For each combination of microenvironment and cell type three biologically independent repeats were performed. For each experiment, two identical samples were fabricated and seeded with cells originating from one culture flask. One sample was imaged using LFI, while the other sample was imaged using an inverted phase contrast microscope. Both samples were imaged every 10 min for 8 hours. Imaging was started and stopped at the same time for both samples. The imaged cells were tracked for as long they stayed in the field of view. Cells that were elongated on printed lines or fully within microchannels for less than 2 hours were not tracked. Tracks were discontinued if cells exited the microchannels or printed regions. Dividing cells were not tracked. After tracking, 30 cell tracks were randomly chosen per sample, resulting in a total of 90 cells per combination of microenvironment and cell type. There were two exceptions to this experimental design: (1) MCF7 microcontact printing experiments, where 30 cell tracks from two biologically independent experiments were grouped to give a total of 60 cell trajectories per condition; (2) MCF7 microchannel experiments, where 25 cell tracks from three biologically independent experiments were grouped to give a total of 75 cell trajectories per condition. These exceptions were made because MCF7 cells were less readily able to spread on confining, 6 μm-wide printed lines or enter microchannels. Experimental design was performed in this manner so that no single biological repeat would weigh the overall results if a large or small number of cells entered the microchannels or adhered to the printed lines. As the majority of cells didn’t adhere to the microprinted lines or stayed inside the microchannels for more than 2 hours, only the first 2 hours of the cell tracks were used for further data-analysis in order to consolidate the results. As all 2D randomly migrating cells were tracked for 8 hours, further data-analysis was performed on both 2h and 8h tracks for these cases. Quantitative migration descriptors were calculated for all chosen cells and used for further statistical analysis without identification or removal of outliers. Also, blinding was not used during this study.
Lens free imaging (LFI) setup
A schematic drawing of the LFI set-up that was built for live cell imaging is shown in Fig. 1. The set-up consists of a CMOS imaging sensor chip of 3840 × 2744 pixels with pixel pitch of 1.67 μm (Aptina MT9J003, ON Semiconductor, Phoenix, AZ, USA) and four lasers at multiple wavelengths in the range of 640 to 660 nm (Thorlabs, Newton, NJ, USA). The lasers are coupled into a single mode optical fiber that delivers the light on top of the imager chip. A USB interface provides connection to a PC with imaging software developed in Python. Samples were imaged every 10 minutes over a period of 8-10 hours.
Phase contrast microscopy setup
An inverted Nikon Eclipse Ti microscope (Nikon, Tokyo, Japan) with automated controls (NIS-Elements, Nikon) was equipped with a temperature- and CO2-controlled stage-top live cell incubator (Tokai Hit, Fujinomiya, Japan) mounted on a software-controlled motorized stage (NIS-Elements, Nikon). Migrating cells were visualized every 10 min with an Andor Clara DR-1261 camera head (Andor Technology, Belfast, UK) and a 10x, 0.45 NA objective for the duration of each live cell experiment (8-10 hours). In each experiment, the motorized stage was programmed to capture 10-15 fields of view.
Migration assay fabrication
Square glass slides were used as substrates for all experiments. These glass slides contained micromarkers to facilitate focusing of the lens-free images and reference point tracking. The micromarkers (10 nm titanium and 50 nm gold) were sputtered (Pfeiffer Spider 630, Pfeiffer Vacuum, Asslar, Germany) on glass wafers (Si-Mat Silicon Materials, Kaufering, Germany) using a standard lift-off process with LOR (Microchem, Westborough, MA, USA) and AZ® nLOF (Microchemicals GmbH, Ulm, Germany).
For 2D random migration assays, glass rings (Campus Glasblazerij KU Leuven, Leuven, Belgium) were glued on the glass slides with PDMS (Sylgard® 184 Silicone Elastomer Kit, Dow Corning, Midland, MI, USA). The surface of the dish was treated with 20 μg/ml collagen type I (BD Biosciences 354236, San Jose, CA, USA) for 1 hour at 37°C to facilitate cell adhesion to the surface.
For 3D microchannel migration assays, PDMS structures were bonded to the glass slides through treatment with oxygen plasma (Harrick PDC-32G plasma cleaner, Harrick Plasma, Ithaca, NY, USA). PDMS structures were fabricated using a standard soft lithography process described by Tong et al.14 Fig. S12 shows a schematic of the microfluidic chip. In short, silicon wafers (University Wafer, South Boston, MA. USA) were patterned with microchannels (W=6 μm, L= 600 μm, H=10 μm) and seeding channels (H=50 μm) using, respectively, SU-8 3010 and SU-8 3025 (Microchem). Masters were treated with fluorosilane ((tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane, Pfaltz & Bauer, Waterbury, CT, USA) to facilitate detachment of the PDMS devices. PDMS was cast against the silicon master to form microfluidic devices. PDMS prepolymer and crosslinker were mixed at weight ratio 10:1 and cured at 80°C overnight. Cured devices were carefully peeled from the mold and diced to size. Inlet ports were created using 4-6 mm-diameter biopsy punches (Harris Uni-Core, Electron Microscopy Sciences, Hatfield, PA, USA). After bonding to the glass slide, collagen type I was incubated in the microchannels for 1 hour at 37°C.
For 1D migration assays on microprinted lines, the PDMS microchannel structures were reversibly bonded to the glass surface in the dishes (i.e. glass rings glued to glass slides), primed with a 5% solution of Alconox (Alconox, Inc., White Plains, NY, USA) in DI water, and washed thoroughly with DPBS (Life Technologies Gibco, Carlsbad, CA, USA). Collagen type I was incubated in the microchannels for 1 hour at 37°C. After this, the microchannels were thoroughly washed with DPBS, emptied, and carefully peeled from the surface, leaving collagen patterned in a projection of the microchannel design on the glass surface. The dish was washed once more with DPBS, and incubated for 1 hour at room temperature with 2% Pluronic F127 (Sigma Aldrich P2443, St. Louis, MO, USA) to prevent cell adhesion to non-patterned regions of the glass.
Cell culture
MDA-MB-231 or MCF7 breast adenocarcinoma cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 4.5 g/l glucose, L-glutamine, and sodium pyruvate (Corning cellgro, Manassas, VA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (P/S; 100 U/ml-100 μg/ml; Gibco). Cells were passaged every 3-5 days and were used in experiments between passage 15 and passage 20. Prior to seeding for motility assays, cells were grown to 70-80% confluency, washed with versene (Gibco), detached with 0.05% trypsin-EDTA (Gibco), resuspendend in full growth medium, and spun down at 1000 rpm for 5 min. Cells were resuspended in growth medium to a final concentration of 400,000 cells/ml (random motility and microcontact printing assays) or 1×106 cells/ml (microchannel assays).
Cell migration experiments
For a given experiment, two migration substrates were prepared identically and a population of cells was distributed to them both, with one sample being imaged on a phase contrast microscope while the other was simultaneously imaged using the LFI platform.
For random and microprinted line migration assays, each sample was seeded with 40,000 cells. Cells were placed in an incubator at 37°C, 5% CO2 and given 1 hour to attach, except for MCF7 cells on microprinted lines; they were given 3-6 hours. After attachment and prior to time-lapse imaging, the dishes were washed with growth medium to remove non-adherent cells.
For migration assays in microchannels, 50,000 cells were added to the cell inlet port of the device (see Fig. S12). Cells were allowed to seed at the bases of the microchannels for 5-20 min. Following aspiration of the cell seeding solution, the device was washed to remove non-adherent cells. Growth medium was added to all wells of the device and time-lapse imaging started.
Cell tracking
Time lapse images were exported as image stacks to ImageJ (NIH, Bethesda, USA). For LFI experiments, phase reconstructions of the holographic images were created at each focal plane for the initial (time=0) image. The optimum focus depth was determined by visual inspection of the cells and LFI reference markers at each depth. Focal depth is a critical parameter for obtaining a high-resolution image (Movie S1). For random motility, microcontact printing, and microchannel migration assays, the centroid of cells was tracked for up to 8 hours using the MTrackJ plugin.56 Cells that were elongated on printed lines or fully within microchannels for less than 2 hours were not tracked. Tracks were discontinued if cells exited the microchannels or printed regions. Dividing cells were not tracked.
Data analysis
Calculations were made based on two-hour cell migration trajectories. Time (t) and time-dependent coordinate data (x(t) and y(t)) were used to calculate mean squared displacement (MSD), velocity (v) and persistence ratio (PR) as a function of time lag (τ) in the following way:
Where τ = n*dt with dt being the time between two consecutive frames and n = 1,2,3,…,12; <…> stands for time averaging.
The APRW model by Wu et al. 46, 47 was fitted to the MSD function, and the following parameters were obtained for each individual cell along the primary (p) and non-primary (np) axes of migration (identified by applying singular value decomposition to individual trajectories): persistence time (P), cell speed (S), and variance of observation noise in cell position. Parameter limits for the fits where: persistence time – 1000 min; speed – 20 μm/min; and variance of noise in cell position – 100 μm2. As 1D migration experiments represent the limit of anisotropic migration, only persistence time in the primary migration direction gives relevant information. Next to this, total diffusivity (D) represents displacement of the cell and is defined 46, 47 as
In the case of 2D random migration experiments, a 2 hour interval was selected out of 8 hour cell tracks for each cell. In order to assess whether this reduction introduces artefacts, the quantitative descriptors were additionally calculated for the 8 hour tracks. All calculations were performed in Matlab R2013a (The Mathworks Inc., Natick, MA, USA).
Statistical analysis
Results for a given cell type and microenvironment that were generated using LFI or phase contrast microscopy were compared using an unpaired t test or Mann-Whitney test after assessing the normality of the data via a D’Agostino & Pearson omnibus normality test. If populations were normally distributed but had significantly different variances, an unpaired t test with Welch’s correction was used. Results across cell types were compared using a Kruskal-Wallis test with Dunn’s multiple comparisons post-test. All statistical calculations were performed in Graphpad Prism 6 software (Graphpad Software Inc., La Jolla, CA, USA). These analyses are similar to those described previously. 41, 57
Cell viability
Samples were prepared similar to random migration assay experiments, however, 200,000 cells were seeded. One sample was imaged with LFI every 10 min for a minimum of 24 h, while another sample was kept in the same incubator without being imaged. Fluorescence staining was performed with 10 μg/mL Hoechst 33342 (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and 5 μg/mL Calcein AM (BD Biosciences) in PBS for 60 min and 1 μg/mL Propidium Iodide (Invitrogen) in PBS for 15 min in an incubator at 37°C, 5% CO2. Fluorescence imaging was performed immediately after staining using a confocal scanning microscope (LSM 780, Zeiss Microscopy GmbH, Jena, Germany). Cell counting was done in ImageJ. Cells were considered alive when the Hoechst nuclear stain co-located with the live cell stain (Calcein AM) and dead when the nuclear stain co-located with the dead cell stain (Propidium Iodide). Calcein AM and Propidium Iodide signals that did not co-locate with the nuclear stain were not counted, as well as cells exhibiting both stains.
Supplementary Material
Acknowledgments
E.M. is grateful for the financial support by IWT-Vlaanderen. C.D.P. acknowledges support from a National Science Foundation Graduate Research Fellowship. This work was partially supported by the European Research Council under the Consolidator Grant (SCALPEL; grant agreement no. 617312), and the National Institutes of Health R01 CA183804 and R01 CA186286.
References
- 1.Dunn GA, Jones GE. Nature reviews. Molecular cell biology. 2004;5:667–672. doi: 10.1038/nrm1439. [DOI] [PubMed] [Google Scholar]
- 2.Coutu DL, Schroeder T. Journal of cell science. 2013;126:3805–3815. doi: 10.1242/jcs.118349. [DOI] [PubMed] [Google Scholar]
- 3.Frigault MM, Lacoste J, Swift JL, Brown CM. Journal of cell science. 2009;122:753–767. doi: 10.1242/jcs.033837. [DOI] [PubMed] [Google Scholar]
- 4.Paddock S. BioTechniques. 2001;30:283–286. doi: 10.2144/01302bt01. 288-289. [DOI] [PubMed] [Google Scholar]
- 5.Stephens DJ, Allan VJ. Science (New York, N.Y.) 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 6.Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuze M, Takaki T, Voituriez R, Piel M. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Maiuri P, Rupprecht JF, Wieser S, Ruprecht V, Benichou O, Carpi N, Coppey M, De Beco S, Gov N, Heisenberg CP, Lage Crespo C, Lautenschlaeger F, Le Berre M, Lennon-Dumenil AM, Raab M, Thiam HR, Piel M, Sixt M, Voituriez R. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 8.van Roosmalen W, Le Devedec SE, Golani O, Smid M, Pulyakhina I, Timmermans AM, Look MP, Zi D, Pont C, de Graauw M, Naffar-Abu-Amara S, Kirsanova C, Rustici G, Hoen PA, Martens JW, Foekens JA, Geiger B, van de Water B. The Journal of clinical investigation. 2015;125:1648–1664. doi: 10.1172/JCI74440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones CN, Moore M, Dimisko L, Alexander A, Ibrahim A, Hassell BA, Warren HS, Tompkins RG, Fagan SP, Irimia D. PloS one. 2014;9:e114509. doi: 10.1371/journal.pone.0114509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isikman SO, Bishara W, Mavandadi S, Yu FW, Feng S, Lau R, Ozcan A. Proceedings of the National Academy of Sciences. 2011 doi: 10.1073/pnas.1015638108. DOI: 10.1073/pnas.1015638108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabor D. Nature. 1948;161:777–778. doi: 10.1038/161777a0. [DOI] [PubMed] [Google Scholar]
- 12.Schnars U, Jüptner W. Journal. 1994;33:179–181. doi: 10.1364/AO.33.000179. [DOI] [PubMed] [Google Scholar]
- 13.Stroka KM, Gu Z, Sun SX, Konstantopoulos K. Current opinion in cell biology. 2014;30:41–50. doi: 10.1016/j.ceb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong Z, Balzer EM, Dallas MR, Hung W-C, Stebe KJ, Konstantopoulos K. PLoS One. 2012;7:e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathak AP, Artemov D, Neeman M, Bhujwalla ZM. Cancer Research. 2006;66:5151–5158. doi: 10.1158/0008-5472.CAN-05-1788. [DOI] [PubMed] [Google Scholar]
- 16.Ziegler E, Hansen MT, Haase M, Emons G, Gründker C. Breast Cancer Research and Treatment. 2014;148:269–277. doi: 10.1007/s10549-014-3159-4. [DOI] [PubMed] [Google Scholar]
- 17.Kim SB, Bae H, Koo KI, Dokmeci MR, Ozcan A, Khademhosseini A. Journal of laboratory automation. 2012;17:43–49. doi: 10.1177/2211068211426695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulf S, Werner POJ. Measurement Science and Technology. 2002;13:R85. [Google Scholar]
- 19.Gabor D. Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences. 1949;197:454–487. [Google Scholar]
- 20.Stahl R, Vanmeerbeeck G, Lafruit G, Huys R, Reumers V, Lambrechts A, Liao C-K, Hsiao C-C, Yashiro M, Takemoto M, Nagata T, Gomi S, Hatabayashi K, Oshima Y, Ozaki S, Nishishita N, Kawamata S. The International Society for Optics and Photonics Proceedings. 2014;8947 doi: 10.1117/12.2037619. [Google Scholar]
- 21.Ozcan A, Demirci U. Lab on a chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 22.Mudanyali O, Tseng D, Oh C, Isikman SO, Sencan I, Bishara W, Oztoprak C, Seo S, Khademhosseini B, Ozcan A. Lab on a Chip. 2010;10:1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su TW, Seo S, Erlinger A, Ozcan A. Biotechnology and bioengineering. 2009;102:856–868. doi: 10.1002/bit.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo S, Su TW, Tseng DK, Erlinger A, Ozcan A. Lab on a chip. 2009;9:777–787. doi: 10.1039/b813943a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su TW, Erlinger A, Tseng D, Ozcan A. Analytical chemistry. 2010;82:8307–8312. doi: 10.1021/ac101845q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesavan SV, Navarro FP, Menneteau M, Mittler F, David-Watine B, Dubrulle N, Shorte SL, Chalmond B, Dinten JM, Allier CP. Journal of biomedical optics. 2014;19:36004. doi: 10.1117/1.JBO.19.3.036004. [DOI] [PubMed] [Google Scholar]
- 27.Kesavan SV, Momey F, Cioni O, David-Watine B, Dubrulle N, Shorte S, Sulpice E, Freida D, Chalmond B, Dinten JM, Gidrol X, Allier C. Scientific reports. 2014;4:5942. doi: 10.1038/srep05942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pushkarsky I, Lyb Y, Weaver W, Su TW, Mudanyali O, Ozcan A, Di Carlo D. Scientific reports. 2014;4:4717. doi: 10.1038/srep04717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenbaum A, Zhang Y, Feizi A, Chung PL, Luo W, Kandukuri SR, Ozcan A. Science Translational Medicine. 2014;6:267ra175. doi: 10.1126/scitranslmed.3009850. [DOI] [PubMed] [Google Scholar]
- 30.Greenbaum A, Luo W, Su T-W, Göröcs Z, Xue L, Isikman SO, Coskun AF, Mudanyali O, Ozcan A. Nature Methods. 2012;9:889–895. doi: 10.1038/nmeth.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhen G, Lee SA, Yang S, Yang C. Lab on a Chip. 2010;10:3125–3129. doi: 10.1039/c0lc00213e. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Zheng G, Lee LM. Lab on a Chip. 2012;12:3566–3575. doi: 10.1039/c2lc40517b. [DOI] [PubMed] [Google Scholar]
- 33.Bianco V, Paturzo M, Marchesano V, Gallotta I, Di Schiavi E, Ferraro P. Lab on a Chip. 2015;15:2117–2124. doi: 10.1039/c5lc00143a. [DOI] [PubMed] [Google Scholar]
- 34.Isikman SO, Bishara W, Zhu H, Ozcan A. Proceedings of Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2011;2011:8463–8466. doi: 10.1109/IEMBS.2011.6092088. [DOI] [PubMed] [Google Scholar]
- 35.Bishara W, Zhu H, Ozcan A. Optics Express. 2010;18:27499–27510. doi: 10.1364/OE.18.027499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momey F, Berdeu A, Bordy T, Dinten J-M, Marcel FK, Picollet-D'hahan N, Gidrol X, Allier C. Biomedical Optics Express. 2016;7:949–962. doi: 10.1364/BOE.7.000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenbaum A, Akbari N, Feizi A, Luo W, Ozcan A. PLoS One. 2013;8:e76475. doi: 10.1371/journal.pone.0076475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isikman SO, Greenbaum A, Luo W, Coskun AF, Ozcan A. PLoS One. 2012;7:e45044. doi: 10.1371/journal.pone.0045044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doyle AD, Wang FW, Matsumoto K, Yamada KM. The Journal of cell biology. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul CD, Shea DJ, Mahoney MR, Chai A, Laney V, Hung WC, Konstantopoulos K. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2016;30:2161–2170. doi: 10.1096/fj.201500199R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung WC, Chen SH, Paul CD, Stroka KM, Lo YC, Yang JT, Konstantopoulos K. The Journal of Cell Biology. 2013;202:807–824. doi: 10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dikeman DA, Rivera Rosado LA, Horn TA, Alves CS, Konstantopoulos K, Yang JT. American journal of physiology. Cell physiology. 2008;295:C151–C159. doi: 10.1152/ajpcell.00169.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grazioli A, Alves CS, Konstantopoulos K, Yang JT. Developmental biology. 2006;293:165–177. doi: 10.1016/j.ydbio.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 45.Wang P, Chen SH, Hung WC, Paul C, Zhu F, Guan PP, Huso DL, Kontrogianni-Konstantopoulos A, Konstantopoulos K. Oncogene. 2015;34:4558–4569. doi: 10.1038/onc.2014.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu PH, Giri A, Sun SX, Wirtz D. Proceedings of the National Acadamy of Sciences USA. 2014;111:3949–54. doi: 10.1073/pnas.1318967111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu PH, Giri A, Wirtz D. Nature Protocols. 2015;10:517–27. doi: 10.1038/nprot.2015.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stroka KM, Konstantopoulos K. American journal of physiology. Cell physiology. 2014;306:C98–c109. doi: 10.1152/ajpcell.00289.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adler J, Pagakis SN. Journal of microscopy. 2003;210:131–137. doi: 10.1046/j.1365-2818.2003.01160.x. [DOI] [PubMed] [Google Scholar]
- 50.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Seminars in cell & developmental biology. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weigelin B, Bakker G-J, Friedl P. IntraVital. 2012;1:32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boneschansker L, Yan J, Wong E, Briscoe DM, Irimia D. Nature communications. 2014;5:4787. doi: 10.1038/ncomms5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamza B, Irimia D. Lab on a chip. 2015;15:2625–2633. doi: 10.1039/c5lc00245a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sencan I, Coskun AF, Sikora U, Ozcan A. Scientific Reports. 2014;4:3760. doi: 10.1038/srep03760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coskun AF, Sencan I, Su TW, Ozcan A. PLoS One. 2011;6:e15955. doi: 10.1371/journal.pone.0015955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meijering E, Dzyubachyk O, Smal I. Methods in enzymology. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 57.Do TV, Xiao F, Bickel LE, Klein-Szanto AJ, Pathak HB, Hua X, Howe C, O'Brien SW, Maglaty M, Ecsedy JA, Litwin S, Golemis EA, Schilder RJ, Godwin AK, Connolly DC. Oncogene. 2014;33:539–549. doi: 10.1038/onc.2012.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.