Tanimura et al. demonstrate that SH3P2 binds to and functions as a cytosolic anchor for myosin 1E (Myo1E). ERK signaling–dependent phosphorylation of SH3P2 induces the dissociation of bound Myo1E and its consequent localization to the tips of lamellipodia, where it promotes cell motility.
Abstract
Signaling by extracellular signal–regulated kinase (ERK) plays an essential role in the induction of cell motility, but the precise mechanism underlying such regulation has remained elusive. We recently identified SH3P2 as a negative regulator of cell motility whose function is inhibited by p90 ribosomal S6 kinase (RSK)–mediated phosphorylation downstream of ERK. We here show that myosin 1E (Myo1E) is a binding partner of SH3P2 and that the interaction of the two proteins in the cytosol prevents the localization of Myo1E to the plasma membrane. Serum-induced phosphorylation of SH3P2 at Ser202 by RSK results in dissociation of Myo1E from SH3P2 in the cytosol and the subsequent localization of Myo1E to the tips of lamellipodia mediated by binding of its TH2 domain to F-actin. This translocation of Myo1E is essential for lamellipodium extension and consequent cell migration. The ERK signaling pathway thus promotes cell motility through regulation of the subcellular localization of Myo1E.
Introduction
Cell motility plays a central role in various biological processes, including embryogenesis, immune surveillance, and wound healing, with spatiotemporal regulation of such motility being essential for homeostasis in multicellular organisms (Lauffenburger and Horwitz, 1996). Cell motility is induced by multiple extracellular cues, including gradients of chemokines, growth factors, and extracellular matrix components. These molecules engage cell surface receptors and thereby initiate a cascade of events such as activation of the phosphatidylinositol 3-kinase (PI3K) and extracellular signal–regulated kinase (ERK) signaling pathways that function downstream of the small GTP-binding protein Ras (Guo and Giancotti, 2004). Activated PI3K catalyzes the production of phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3), which triggers the formation of lamellipodia at the leading edge of a migrating cell via activation of the small GTPase Rac1 and the protein kinase Akt and thereby promotes cell motility (Raftopoulou and Hall, 2004; Vanhaesebroeck et al., 2012; Xue and Hemmings, 2013). Activated ERK also modulates cell motility through direct phosphorylation of several molecules, including myosin light chain kinase (Klemke et al., 1997), cortactin (Martinez-Quiles et al., 2004), WAVE2 (Danson et al., 2007; Nakanishi et al., 2007; Mendoza et al., 2011), and FAK (Hunger-Glaser et al., 2003). We recently showed that the Src homology 3 (SH3) domain–containing protein SH3P2 is a negative regulator of cell motility whose function is abrogated by p90 ribosomal S6 kinase (RSK)–mediated phosphorylation at Ser202 downstream of ERK (Tanimura et al., 2011). However, the mechanism by which SH3P2 regulates cell motility has remained elusive.
Myosin 1E (Myo1E) is an actin-dependent molecular motor that is widely expressed in vertebrate tissues (McConnell and Tyska, 2010). Myo1E is a class 1 myosin, a defining feature of which is the ability to interact with both cell membranes and actin filaments via a C-terminal tail homology 1 (TH1) domain and an N-terminal motor domain, respectively. This spatial segregation of membrane and actin-binding sites suggests that class 1 myosins have the potential to serve as divalent cross-linking proteins that physically connect and generate force between actin filaments and membranes and thereby to regulate plasma membrane tension. Whereas most class 1 myosins are “short tailed” in that they possess only the TH1 domain in the tail region, Myo1E also contains a proline-rich membrane binding (TH2) domain and a protein–protein interaction (SH3) domain and is therefore classified as “long tailed.” Myo1E has been proposed to function in a manner dependent on interactions mediated by its SH3 domain as a transporter or recruiter of effector proteins involved in myosin-based as well as actin nucleation–based force generation at the plasma membrane. It thus contributes to the accumulation of effector molecules such as dynamin, synaptojanin-1, and the N-WASP–WIP complex at the membrane–cytoskeleton interface to support endocytosis as well as cell motility (Krendel et al., 2007; Cheng et al., 2012). However, the molecular mechanisms by which the function of Myo1E, and in particular its intracellular localization, are regulated have remained unknown. We have now identified Myo1E as a binding partner of SH3P2. We found that RSK-mediated phosphorylation of SH3P2 induces the dissociation of Myo1E from SH3P2 in the cytosol, which results in the localization of Myo1E to the tips of lamellipodia and thereby promotes cell motility.
Results
Identification of Myo1E as a binding partner of SH3P2
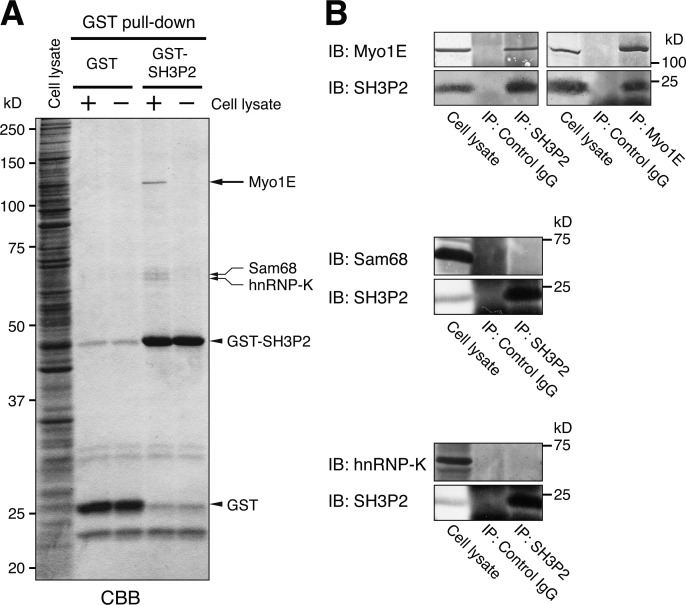
To identify proteins that interact with SH3P2, we performed a pull-down assay with MKN1 cell lysates and a GST-SH3P2 fusion protein as the bait. An ∼120-kD protein was found to bind specifically to SH3P2 (Fig. 1 A) and was identified by mass spectrometry (MS) as Myo1E. Specific interaction between endogenous SH3P2 and Myo1E was confirmed by reciprocal coimmunoprecipitation assays (Fig. 1 B). The pull-down assay also revealed the binding of ∼66- and ∼68-kD proteins to SH3P2, and these two proteins were identified by MS as hnRNP-K and Sam68, respectively. However, given that coimmunoprecipitation assays did not detect a specific interaction between endogenous SH3P2 and either of these two RNA-binding proteins (Fig. 1 B), we did not analyze them further.
Figure 1.
SH3P2 specifically interacts with Myo1E. (A) Glutathione-Sepharose beads coupled with GST or GST-SH3P2 were incubated with MKN1 cell lysates (+) or with the lysis buffer alone (−), after which bead-bound proteins were subjected to SDS-PAGE followed by staining with Coomassie brilliant blue (CBB). Bands corresponding to proteins of ∼120, ∼68, and ∼66 kD (arrows) pulled down by GST-SH3P2 were identified by MS analysis as Myo1E, Sam68, and hnRNP-K, respectively. (B) MKN1 cell lysates were subjected to immunoprecipitation (IP) with antibodies to SH3P2 or to Myo1E (or with control IgG), and the resulting precipitates as well as the whole-cell lysates (10% of the input for immunoprecipitation) were subjected to immunoblot analysis (IB) with antibodies to the indicated proteins. All data are representative of at least three separate experiments.
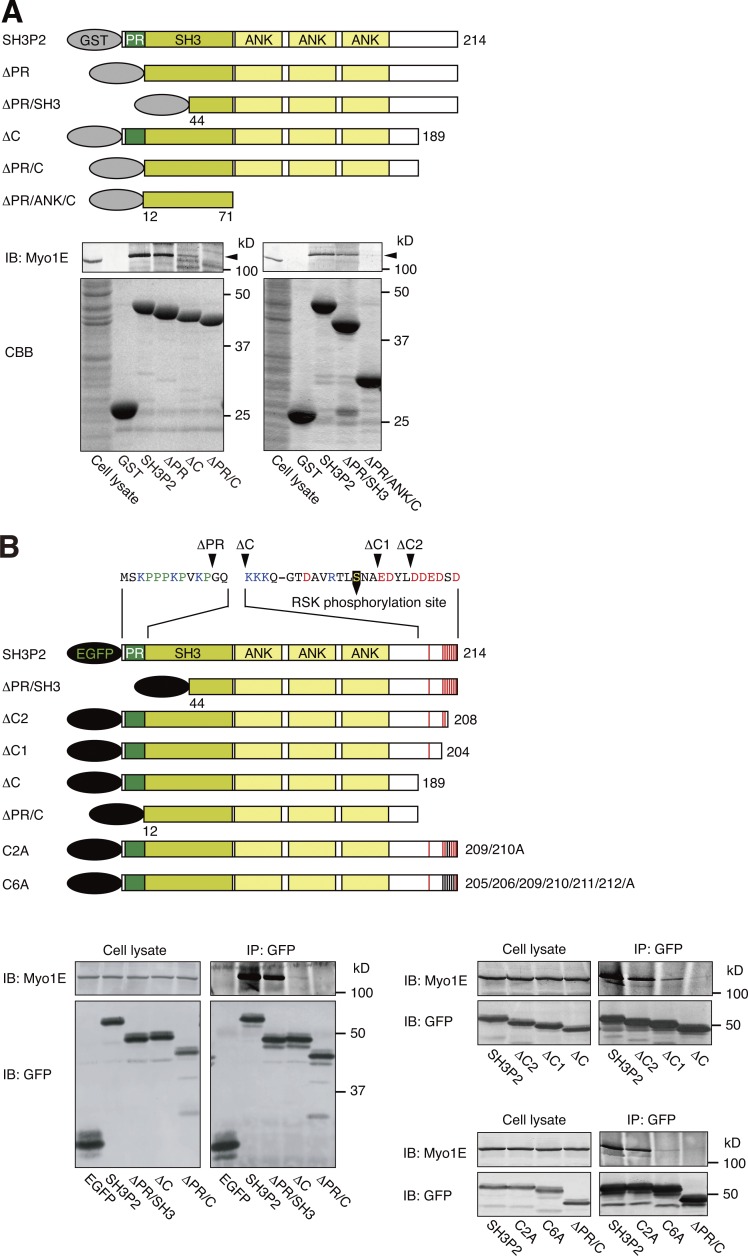
SH3P2 interacts with Myo1E via its N-terminal proline-rich region and C-terminal acidic amino acid cluster
SH3P2 consists of an N-terminal proline-rich (PR) region, an SH3 domain, three ankyrin (ANK) repeats, and a C-terminal (C) region (Fig. 2 A). To determine the region of SH3P2 responsible for interaction with Myo1E, we first performed pull-down assays with MKN1 cell lysates and GST-fused SH3P2 deletion mutants as baits. We found that SH3P2(ΔPR) and SH3P2(ΔPR/SH3) as well as SH3P2(ΔC) interacted with endogenous Myo1E, albeit to a slightly or substantially reduced extent compared with wild-type SH3P2, respectively, whereas neither SH3P2(ΔPR/C) nor SH3P2(ΔPR/ANK/C) showed such an interaction (Fig. 2 A). Coimmunoprecipitation analysis with lysates of MKN1 cells expressing EGFP-tagged SH3P2 deletion mutants showed that deletion of the PR/SH3 or C regions attenuated the interaction of SH3P2 with Myo1E slightly or substantially, respectively, and that deletion of both PR and C regions abolished the interaction (Fig. 2 B).
Figure 2.
SH3P2 interacts with Myo1E via its N-terminal proline-rich region and C-terminal acidic amino acid cluster. (A) Glutathione-Sepharose beads coupled with GST or GST-tagged wild-type or indicated mutant forms of human SH3P2 were incubated with MKN1 cell lysates, after which bead-bound proteins as well as the whole-cell lysates (10% of the input for the pull-down assays) were subjected to SDS-PAGE followed by staining with Coomassie brilliant blue (CBB) or immunoblot analysis (IB) with antibodies to Myo1E. Arrowheads indicate the Myo1E band. (B) MKN1 cells were transfected for 24 h with vectors encoding EGFP-tagged wild-type or mutant forms of human SH3P2, lysed, and subjected to immunoprecipitation (IP) with antibodies to GFP. The resulting precipitates as well as the whole-cell lysates (10% of the input for immunoprecipitation) were subjected to immunoblot analysis with antibodies to Myo1E and to GFP. Amino acid sequences of the PR region and the C-terminal acidic amino acid cluster are shown. The RSK phosphorylation site (Ser202), acidic amino acids, basic amino acids, and proline are indicated in yellow, red, blue, and green, respectively. All data are representative of at least three separate experiments.
The SH3P2 proteins of various mammalian species contain a conserved acidic amino acid cluster at the C terminus (Fig. S1 A), whose potential role in the interaction with Myo1E we next examined. Coimmunoprecipitation analysis with lysates of MKN1 cells expressing EGFP-tagged SH3P2(ΔC2), SH3P2(ΔC1), or SH3P2(ΔC) revealed that the extent of interaction of these mutants with endogenous Myo1E decreased as the number of deleted acidic amino acid residues increased (Fig. 2 B). Furthermore, replacement of two (C2A mutant: Asp209/Asp210) or six (C6A mutant: Glu205/Asp206/Asp209/Asp210/Glu211/Asp212) acidic amino acid residues of SH3P2 with alanine reduced the extent of association with Myo1E slightly or markedly, respectively (Fig. 2 B). Together, these results suggested that both the PR region and the C-terminal acidic amino acid cluster, but not the SH3 domain or ANK repeats, of SH3P2 are required for efficient interaction with Myo1E.
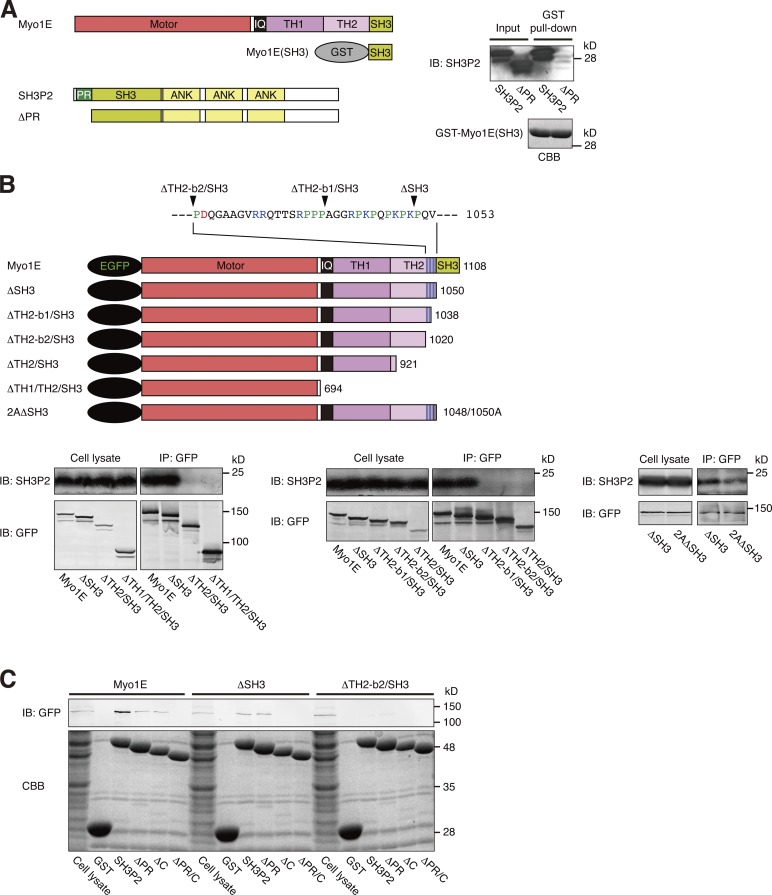
Myo1E interacts with SH3P2 via its SH3 domain and the C-terminal positively charged region of its TH2 domain
Myo1E comprises an actin-activated motor head domain, a light-chain binding neck (IQ) domain, as well as TH1, TH2, and SH3 domains (Fig. 3 A). To determine the region of Myo1E responsible for interaction with SH3P2, we first examined the potential interaction between the SH3 domain of Myo1E and the PR region of SH3P2, given that SH3 domains recognize PR sequences within specific peptide sequence contexts (Ren et al., 1993; Feng et al., 1994). A pull-down assay with the GST-fused SH3 domain of Myo1E as bait showed that Myo1E(SH3) bound to recombinant SH3P2, but not to SH3P2(ΔPR) (Fig. 3 A), suggesting that the SH3 domain of Myo1E indeed contributes to the interaction with SH3P2 by binding to its PR region.
Figure 3.
Myo1E interacts with SH3P2 via its SH3 domain and the C-terminal positively charged region of its TH2 domain. (A) Glutathione-Sepharose beads coupled with GST-fused human Myo1E(SH3) were incubated with recombinant human SH3P2 or SH3P2(ΔPR), after which bead-bound proteins as well as recombinant SH3P2 or SH3P2(ΔPR) (10% of the input for the pull-down assay) were subjected to SDS-PAGE followed by staining with Coomassie brilliant blue (CBB) or immunoblot analysis (IB) with antibodies to SH3P2. (B) MKN1 cells were transfected for 24 h with vectors encoding EGFP-tagged wild-type or indicated mutant forms of human Myo1E, lysed, and subjected to immunoprecipitation (IP) with antibodies to GFP. The resulting precipitates as well as the whole-cell lysates (10% of the input for immunoprecipitation) were subjected to immunoblot analysis with antibodies to SH3P2 and to GFP. The amino acid sequence of the C-terminal positively charged region of the TH2 domain of Myo1E is shown, with acidic amino acids, basic amino acids, and proline being indicated in red, blue, and green, respectively. (C) MKN1 cells were transfected for 24 h with vectors encoding EGFP-tagged wild-type or mutant forms of human Myo1E, lysed, and subjected to pull-down assays with GST or GST-fused wild-type or mutant forms of human SH3P2. Bead-bound proteins as well as the whole-cell lysates (10% of the input for the pull-down assays) were subjected to SDS-PAGE followed by staining with Coomassie brilliant blue or by immunoblot analysis with antibodies to GFP. All data are representative of at least three separate experiments.
Coimmunoprecipitation analysis with lysates of MKN1 cells expressing EGFP-tagged Myo1E deletion mutants revealed that although deletion of the SH3 domain alone did not substantially affect the binding of Myo1E to endogenous SH3P2, deletion of TH1–TH2-SH3 or TH2-SH3 domains abolished such binding (Fig. 3 B), suggesting the presence of another binding site (or sites) within the TH2 domain. Seven basic amino acids are clustered in the C-terminal region of the TH2 domain, which is conserved among mammalian Myo1E proteins (Fig. S1 B). We next focused on the potential role of these basic amino acid residues in the interaction of Myo1E with SH3P2. Coimmunoprecipitation analysis with lysates of MKN1 cells expressing EGFP-tagged Myo1E mutants revealed that deletion of sequences containing all seven (ΔTH2-b2/SH3 mutant: Arg1029/Arg1030/Arg1035/Arg1042/Lys1044/Lys1048/Lys1050) or the final four (ΔTH2-b1/SH3 mutant: Arg1042/Lys1044/Lys1048/Lys1050) of these basic amino acid residues abrogated the interaction of Myo1E(ΔSH3) with SH3P2 (Fig. 3 B). Furthermore, replacement of Lys1048 and Lys1050 with alanine (2AΔSH3 mutant) reduced the extent of the interaction between Myo1E(ΔSH3) and SH3P2 (Fig. 3 B). These results suggested that the positively charged region located at the C terminus of the TH2 domain also contributes to the interaction of Myo1E with SH3P2, most likely through binding to the acidic amino acid cluster of the latter protein.
To confirm the presence of these two binding interfaces for the interaction between SH3P2 and Myo1E, we performed pull-down assays with MKN1 cell lysates expressing EGFP-tagged Myo1E deletion mutants and with GST-fused SH3P2 deletion mutants as baits. As expected, SH3P2 bound to Myo1E and to Myo1E(ΔSH3), but not to Myo1E(ΔTH2-b2/SH3). SH3P2(ΔPR), which possesses the C-terminal acidic amino acid cluster, bound to Myo1E and to Myo1E(ΔSH3), but not to Myo1E(ΔTH2-b2/SH3). SH3P2(ΔC), which possesses the PR region, bound to Myo1E, but not to Myo1E(ΔSH3) or Myo1E(ΔTH2-b2/SH3). Finally, SH3P2(ΔPR/C) failed to bind to Myo1E, Myo1E(ΔSH3), or Myo1E(ΔTH2-b2/SH3) (Fig. 3 C and Fig. S2).
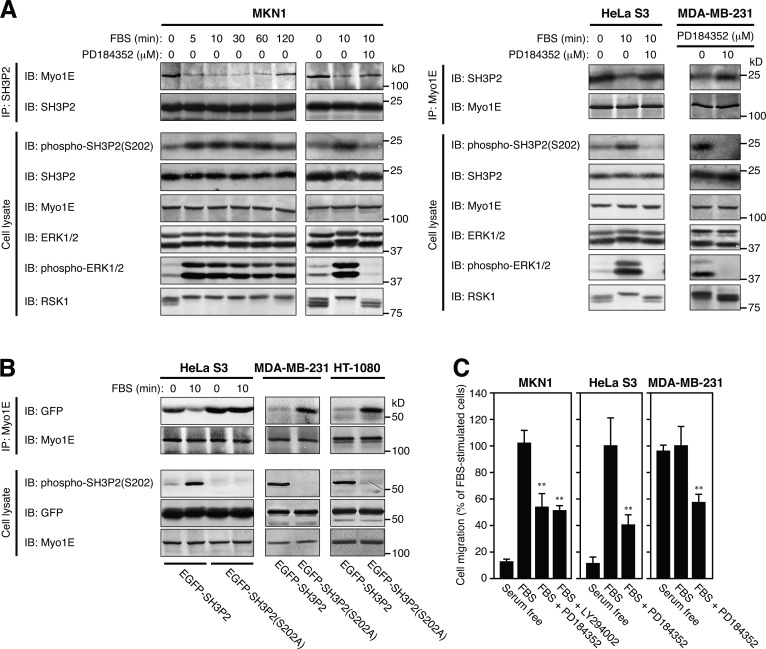
RSK-mediated phosphorylation of SH3P2 at Ser202 induces dissociation of Myo1E
We previously showed that RSK1, acting downstream of the ERK pathway, phosphorylates SH3P2 at Ser202 and thereby inhibits its ability to suppress cell motility (Tanimura et al., 2011). This serine residue is located in the C-terminal region of SH3P2, which is required for the interaction with Myo1E (Fig. 2 B). We therefore examined whether RSK-mediated phosphorylation of SH3P2 affects its interaction with Myo1E.
Coimmunoprecipitation assays with antibodies to SH3P2 and to Myo1E revealed an inverse relation between the extent of SH3P2 phosphorylation (resulting from ERK pathway activation) and the extent of SH3P2 interaction with Myo1E in several tumor cell lines (Fig. 4 A). Marked association of these two proteins was thus observed in HeLa S3 (78.7 ± 11.6% of Myo1E in cell lysates was associated with SH3P2) and MKN1 (78.4 ± 6.8%) cells, in which the activity of the ERK pathway is limited (Hoshino et al., 1999). Stimulation of these cells with serum, however, induced SH3P2 phosphorylation and reduced the level of SH3P2–Myo1E interaction within 10 min (22.4 ± 3.9% in MKN1 cells); both of these effects were partially reversed at 120 min after the onset of stimulation, and they were prevented by prior exposure of the cells to PD184352, an inhibitor of the ERK kinase MEK. In contrast, only a low level of SH3P2–Myo1E interaction (6.1 ± 2.1%) was apparent in MDA-MB-231 cells, in which the ERK pathway is constitutively activated (Hoshino et al., 1999), and treatment of these cells with PD184352 induced dephosphorylation of SH3P2 and increased the association between these two proteins (17.1 ± 4.0%).
Figure 4.
RSK-mediated phosphorylation of SH3P2 at Ser202 induces the dissociation of Myo1E from SH3P2. (A) MKN1 or HeLa S3 cells were deprived of serum for 12 h, incubated with or without 10 µM PD184352 for 30 min, and then stimulated with 10% FBS for the indicated times. Exponentially growing MDA-MB-231 cells were incubated with or without 10 µM PD184352 for 4 h. Cell lysates were then subjected to immunoprecipitation (IP) with antibodies to SH3P2 or to Myo1E, and the resulting precipitates as well as the whole-cell lysates (10% of the input for immunoprecipitation) were subjected to immunoblot analysis (IB) with antibodies to the indicated proteins. (B) HeLa S3, MDA-MB-231, or HT-1080 cells were transfected for 24 h with vectors for EGFP-SH3P2 or EGFP-SH3P2(S202A), and the transfected HeLa S3 cells were then deprived of serum for 12 h before stimulation with 10% FBS for 10 min. Cell lysates were subjected to immunoprecipitation with antibodies to Myo1E, and the resulting precipitates as well as the whole-cell lysates (10% of the input for immunoprecipitation) were subjected to immunoblot analysis with antibodies to the indicated proteins. (C) The motility of MKN1, HeLa S3, or MDA-MB-231 cells was measured with a transwell migration assay for 6 h in the absence or presence of 10% FBS, 10 µM PD184352, or 10 µM LY294002 as indicated. The numbers of cells that had migrated to reach the lower surface of the filter were 212 ± 20, 63 ± 14, and 177 ± 26 for FBS-stimulated MKN1, HeLa S3, and MDA-MB-231 cells, respectively. Data are representative of at least three separate experiments (A and B) or are means ± SD for three separate experiments (C). **, P < 0.01 versus FBS-stimulated cells.
The unphosphorylatable mutant SH3P2(S202A) showed marked association with Myo1E in HeLa S3 cells, and serum stimulation of these cells did not induce the dissociation of Myo1E from SH3P2(S202A) (Fig. 4 B). Furthermore, SH3P2(S202A), but not wild-type SH3P2, also showed a high level of association with Myo1E in MDA-MB-231 and HT-1080 cells, in the latter of which the ERK pathway is also constitutively activated (Hoshino et al., 1999). Finally, serum-induced cell motility in MKN1 and HeLa S3 cells and the enhanced basal level of motility in MDA-MB-231 cells were markedly attenuated by PD184352 (Fig. 4 C). Together, these results indicated that RSK-mediated phosphorylation of SH3P2 at Ser202 induces the dissociation of Myo1E from SH3P2, with this dissociation being required for the induction of cell motility.
Myo1E localizes to lamellipodial tips after dissociation from SH3P2
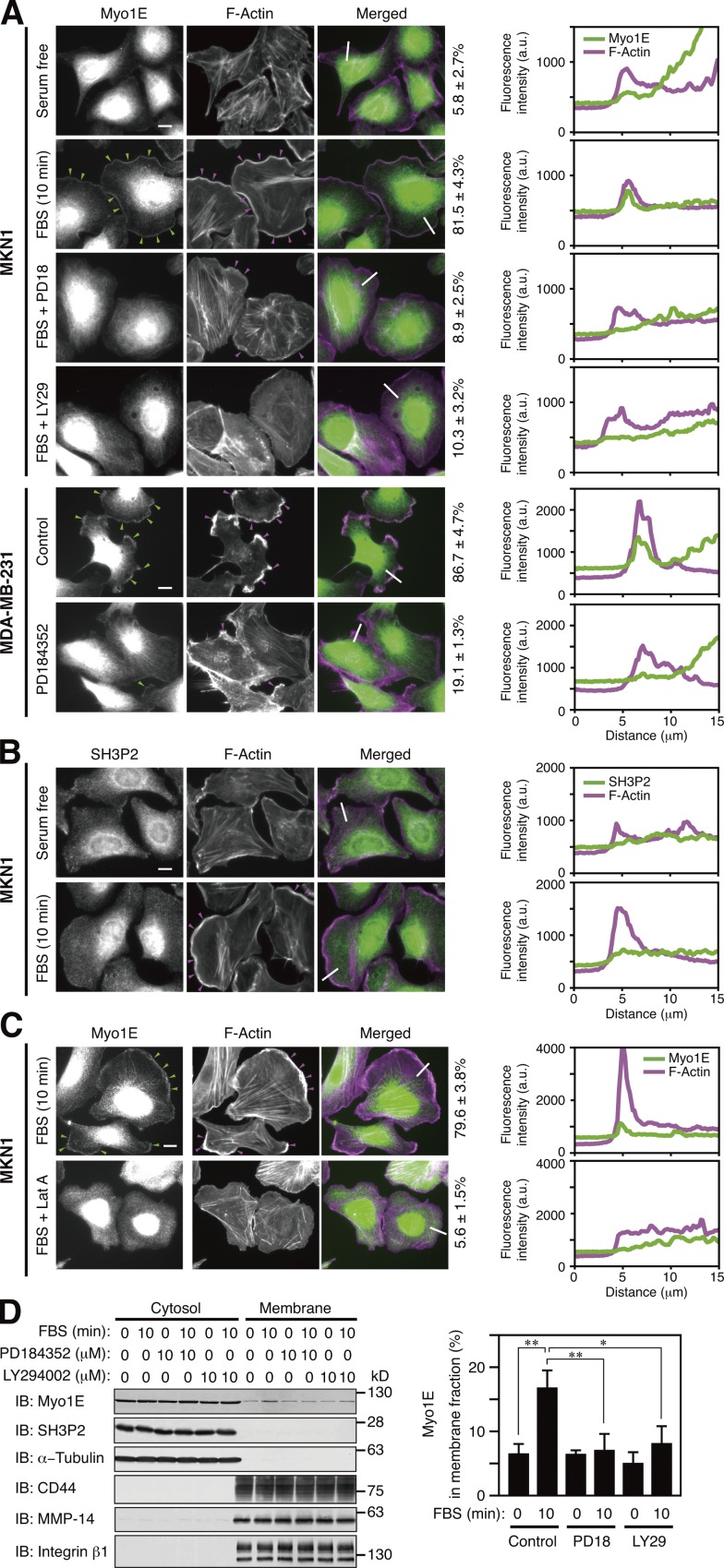
Both Myo1E and SH3P2 were localized to the cytosol of serum-deprived MKN1 cells (Fig. 5, A and B). Serum stimulation of the cells induced rapid translocation of Myo1E to sites of actin polymerization in lamellipodia, which define the tips of these structures (Fig. 5 A), whereas it had no substantial effect on the subcellular localization of SH3P2 (Fig. 5 B).
Figure 5.
Myo1E localizes to lamellipodial tips after dissociation from SH3P2. (A–C) MKN1 cells deprived of serum for 12 h were incubated with or without 10 µM PD184352 (PD18), 10 µM LY294002 (LY29), or 50 nM latrunculin A (Lat A) for 30 min and then stimulated with 10% FBS for 10 min. Exponentially growing MDA-MB-231 cells were incubated with or without 10 µM PD184352 for 4 h. All cells were then fixed and stained with antibodies to Myo1E (A and C, green) or to SH3P2 (B, green) as well as with phalloidin (A–C, magenta). Green arrowheads indicate Myo1E colocalized with F-actin, whereas magenta arrowheads indicate lamellipodia. Bars, 10 µm. The percentages of cells in which Myo1E and F-actin were colocalized are shown at the right of the merged images as means ± SD for three separate experiments, with n ≥ 80 (A) or 40 (C) cells in each experiment. Fluorescence intensity profiles along the white lines in the merged images are also shown on the right (a.u., arbitrary units). Data are representative of at least three separate experiments. (D) MKN1 cells deprived of serum for 12 h were incubated with or without PD184352 or LY294002 for 30 min and then stimulated with 10% FBS for 10 min. The cells were then subjected to subcellular fractionation, and 20% of the cytosolic fraction and 50% of the membrane fraction were subjected to immunoblot analysis (IB) with antibodies to the indicated proteins (left). α-Tubulin was analyzed as a cytosolic marker and CD44, MMP-14, and integrin β1 as membrane markers. The proportion of Myo1E in the membrane fraction was determined by measurement of immunoblot signals as mean ± SD values for three separate experiments (right). *, P < 0.05; **, P < 0.01.
Activation of both the ERK and PI3K signaling pathways is required for the induction of actin polymerization and subsequent lamellipodium formation (Wennström et al., 1994; Weiger et al., 2009; Mendoza et al., 2011; Vanhaesebroeck et al., 2012). Prior treatment of MKN1 cells with PD184352 thus inhibited the serum-induced formation of lamellipodia and suppressed the translocation of Myo1E to the plasma membrane (Fig. 5 A). Furthermore, the PI3K inhibitor LY294002 suppressed the serum-induced production of PI(3,4,5)P3 (Fig. S3 A), lamellipodium formation, translocation of Myo1E to the cell periphery (Fig. 5 A), and cell motility (Fig. 4 C). In addition, exponentially growing MDA-MB-231 cells exhibited lamellipodia at their periphery in which F-actin and Myo1E were colocalized, and PD184352 treatment of the cells induced a marked reduction in the extent of lamellipodium formation and abolished the association of Myo1E with lamellipodial tips (Fig. 5 A). Finally, a low concentration of latrunculin A, which inhibits actin assembly through sequestration of monomeric actin, also suppressed translocation of Myo1E to the cell periphery in MKN1 cells (Fig. 5 C), suggesting that serum-induced rearrangement of F-actin, rather than the production of PI(3,4,5)P3, is critical for localization of Myo1E to the plasma membrane.
Subcellular fractionation confirmed that most Myo1E and almost all SH3P2 resided in the cytosolic fraction of resting MKN1 cells, that serum stimulation induced the translocation of a substantial proportion (∼10%) of Myo1E to the membrane fraction without affecting the cytosolic localization of SH3P2, and that the serum-induced association of Myo1E with the membrane fraction was suppressed by PD184352 and by LY294002 (Figs. 5 D and S3 B). These results suggested that after its dissociation from SH3P2, Myo1E translocates from the cytosol to the tips of lamellipodia via binding to F-actin.
Role of the TH2 domain in the localization of Myo1E to lamellipodial tips
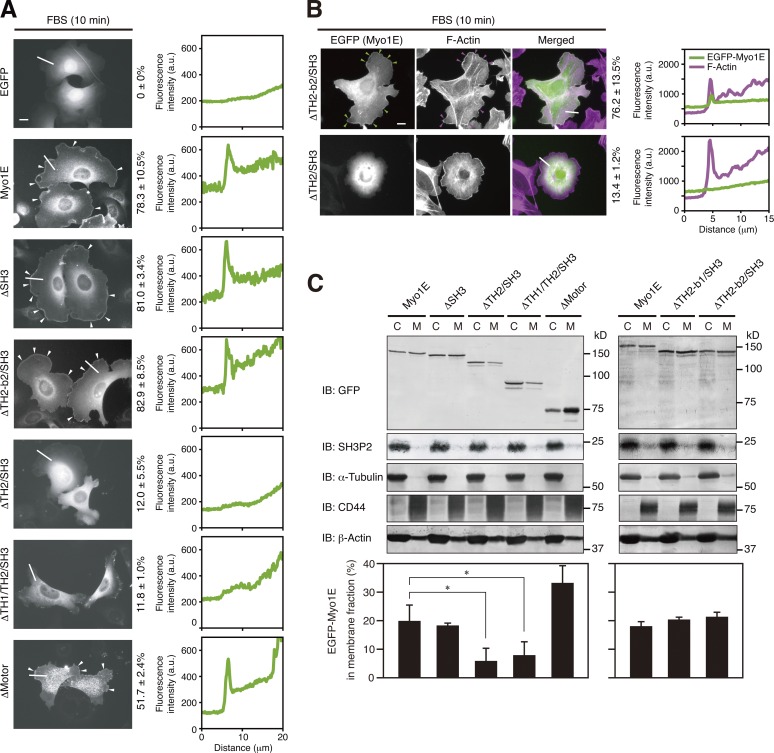
To examine how Myo1E interacts with the plasma membrane, we determined the localization of EGFP-tagged Myo1E and its deletion mutants in transfected MKN1 cells. Serum stimulation of the cells induced the formation of lamellipodia (Fig. 5 A), to which wild-type Myo1E, Myo1E(ΔSH3), and Myo1E(ΔTH2-b2/SH3), but not Myo1E(ΔTH2/SH3) or Myo1E(ΔTH1/TH2/SH3), were found to be localized (Fig. 6 A). Myo1E(ΔTH2-b2/SH3) thus colocalized with F-actin at extended lamellipodial tips, whereas Myo1E(ΔTH2/SH3) did not (Fig. 6 B). Although lamellipodia were less well defined in cells expressing a mutant of Myo1E lacking the motor head domain (Myo1E(ΔMotor)) than in those expressing the wild-type protein, localization of this mutant to the periphery of serum-stimulated cells was observed (Fig. 6 A).
Figure 6.
The TH2 domain mediates the localization of Myo1E to lamellipodial tips. (A and B) MKN1 cells were transfected for 24 h with vectors for the indicated EGFP-tagged Myo1E constructs, deprived of serum for 12 h, and then stimulated with 10% FBS for 10 min. The cells were fixed and then examined for EGFP fluorescence (B, green) with or without staining with phalloidin (B, magenta). White arrowheads (A) indicate Myo1E proteins localized to lamellipodial tips, whereas green and magenta arrowheads (B) indicate colocalization of Myo1E and F-actin, respectively, at lamellipodial tips. Bars, 10 µm. The percentages of cells in which EGFP-Myo1E proteins were localized to lamellipodial tips (A) or colocalized with F-actin (B) are shown at the right of the images as means ± SD for three separate experiments, with n ≥ 80 (A) or 30 (B) cells in each experiment. Fluorescence intensity profiles along the white lines are also shown on the right. Data are representative of at least three separate experiments. (C) MKN1 cells transfected for 24 h with vectors for the indicated EGFP-Myo1E constructs were subjected to subcellular fractionation, and 20% of the cytosolic (C) fraction and 50% of the membrane (M) fraction were subjected to immunoblot analysis (IB) with antibodies to the indicated proteins (top). The proportion of each EGFP-Myo1E construct in the membrane fraction was determined by measurement of immunoblot signals as mean ± SD values for three separate experiments (bottom). *, P < 0.05.
Subcellular fractionation revealed that EGFP-tagged Myo1E and its deletion mutants as well as β-actin were detected in both cytosolic and membrane fractions of transfected MKN1 cells under exponentially growing conditions, although the proportion of each Myo1E protein in the membrane fraction varied (Fig. 6 C). The proportion of Myo1E mutants lacking all or most of the TH2 domain (Myo1E(ΔTH2/SH3) and Myo1E(ΔTH1/TH2/SH3)) in the membrane fraction was thus reduced compared with that for wild-type Myo1E or deletion mutants containing all or most of the TH2 domain (Myo1E(ΔSH3), Myo1E(ΔMotor), Myo1E(ΔTH2-b1/SH3), and Myo1E(ΔTH2-b2/SH3)). Together with the previous demonstration that the TH2 domain of Myo1E preferentially associates with dynamic F-actin (Yu and Bement, 2007), these results suggested that after its dissociation from SH3P2, Myo1E localizes to lamellipodial tips in a manner dependent on interaction between the TH2 domain and F-actin. However, the C-terminal positively charged region of the TH2 domain, which mediates the interaction of Myo1E with SH3P2 (Fig. 3), does not appear to be required for the association of Myo1E with F-actin.
SH3P2 suppresses the localization of Myo1E to lamellipodial tips
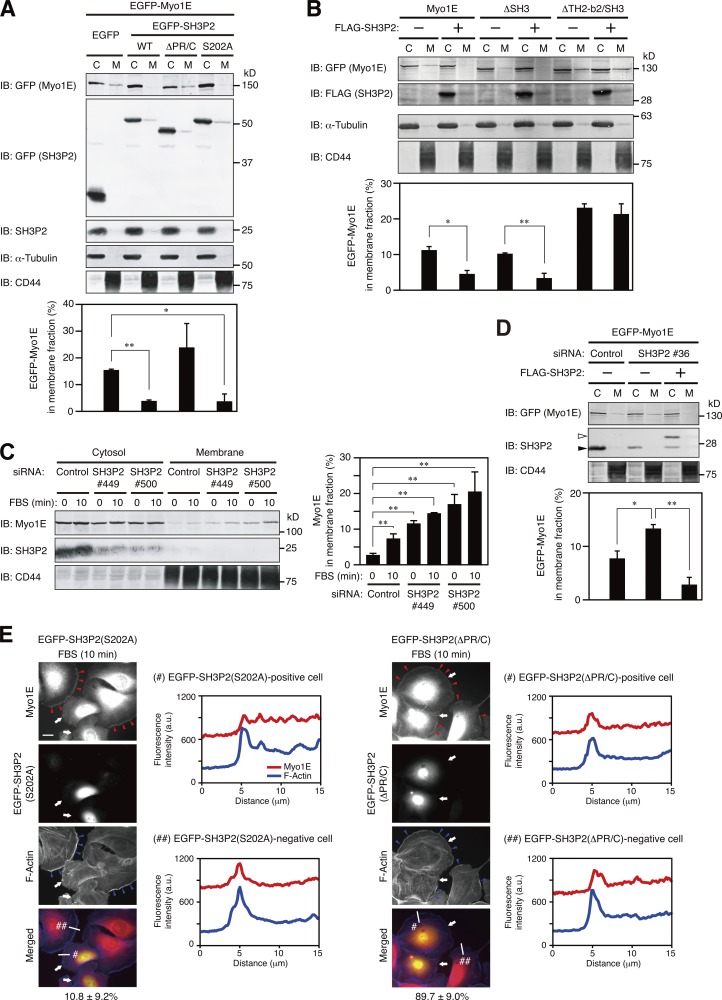
We further examined the physiological relevance of the SH3P2–Myo1E interaction. Forced expression of wild-type SH3P2 or SH3P2(S202A), but not that of an SH3P2 mutant lacking the Myo1E binding regions (SH3P2(ΔPR/C)), prevented the membrane association of Myo1E in MKN1 cells (Fig. 7 A). Incubation of a membrane fraction of MKN1 cells with recombinant Myo1E in vitro confirmed that the membrane association of Myo1E was suppressed by prior incubation with GST-SH3P2 in a concentration-dependent manner, but not by that with GST-SH3P2(ΔPR/C) (Fig. S4).
Figure 7.
SH3P2 suppresses the localization of Myo1E to lamellipodial tips. (A and B) MKN1 cells transfected for 24 h with vectors for EGFP-Myo1E and the indicated EGFP-SH3P2 constructs (WT, wild type; A) or for FLAG-tagged SH3P2 and the indicated EGFP-Myo1E constructs (B) were subjected to subcellular fractionation, and 20% of the cytosolic (C) fraction and 50% of the membrane (M) fraction were subjected to immunoblot analysis (IB) with antibodies to the indicated proteins (top). The proportion of EGFP-Myo1E constructs in the membrane fraction was determined by measurement of immunoblot intensities as mean ± SD values for three separate experiments (bottom). *, P < 0.05; **, P < 0.01. (C) MKN1 cells transfected with an SH3P2 siRNA (#449 or #500) or a control siRNA for 48 h were deprived of serum for 12 h and then stimulated with 10% FBS for 10 min. The cells were then subjected to subcellular fractionation followed by quantitative immunoblot analysis, and the proportion of Myo1E in the membrane fraction was determined, as in A and B. (D) MKN1 cells were transfected first with an siRNA targeting the 5′-UTR sequence of SH3P2 (#36) or a control siRNA for 48 h and then with vectors for EGFP-Myo1E and FLAG-SH3P2 for 24 h. The cells were subjected to subcellular fractionation followed by quantitative immunoblot analysis, and the proportion of EGFP-Myo1E in the membrane fraction was determined as in A and B. Open and closed arrowheads indicate FLAG-SH3P2 and endogenous SH3P2 bands, respectively. (E) MKN1 cells transfected with vectors for EGFP-SH3P2(S202A) or EGFP-SH3P2(ΔPR/C) for 24 h were deprived of serum for 12 h, stimulated with 10% FBS for 10 min, fixed, and stained with antibodies to Myo1E (red) and with phalloidin (blue). EGFP fluorescence was monitored directly. Red arrowheads indicate Myo1E localized to lamellipodial tips, blue arrowheads indicate lamellipodia, and white arrows indicate cells expressing EGFP-SH3P2(S202A) or EGFP-SH3P2(ΔPR/C). Bar, 20 µm. The percentages of EGFP-positive cells in which Myo1E and F-actin were colocalized are shown below the merged images as means ± SD for three separate experiments, with n ≥ 40 cells in each experiment. Fluorescence intensity profiles along the white lines in the merged images are also shown. Data are representative of at least three separate experiments.
Forced expression of SH3P2 also suppressed the membrane association of Myo1E(ΔSH3), which interacts with SH3P2, but not that of Myo1E(ΔTH2-b2/SH3), which does not interact with SH3P2 (Fig. 7 B). RNAi-mediated depletion of SH3P2 increased the association of Myo1E with the membrane fraction even under resting conditions, and the extent of this association was not increased further by serum stimulation of the cells (Fig. 7 C). Knockdown of SH3P2 with an siRNA targeting the 5′-UTR sequence also increased the association of Myo1E with the membrane fraction, and this effect was suppressed by forced expression of SH3P2 (Fig. 7 D). Finally, whereas expression of SH3P2(S202A) attenuated the serum-induced localization of Myo1E to lamellipodial tips, expression of SH3P2(ΔPR/C) did not (Fig. 7 E). Together, these results suggested that SH3P2 functions as a cytoplasmic anchor for Myo1E to suppress its localization to lamellipodial tips.
Myo1E promotes active lamellipodium formation and thereby increases cell motility
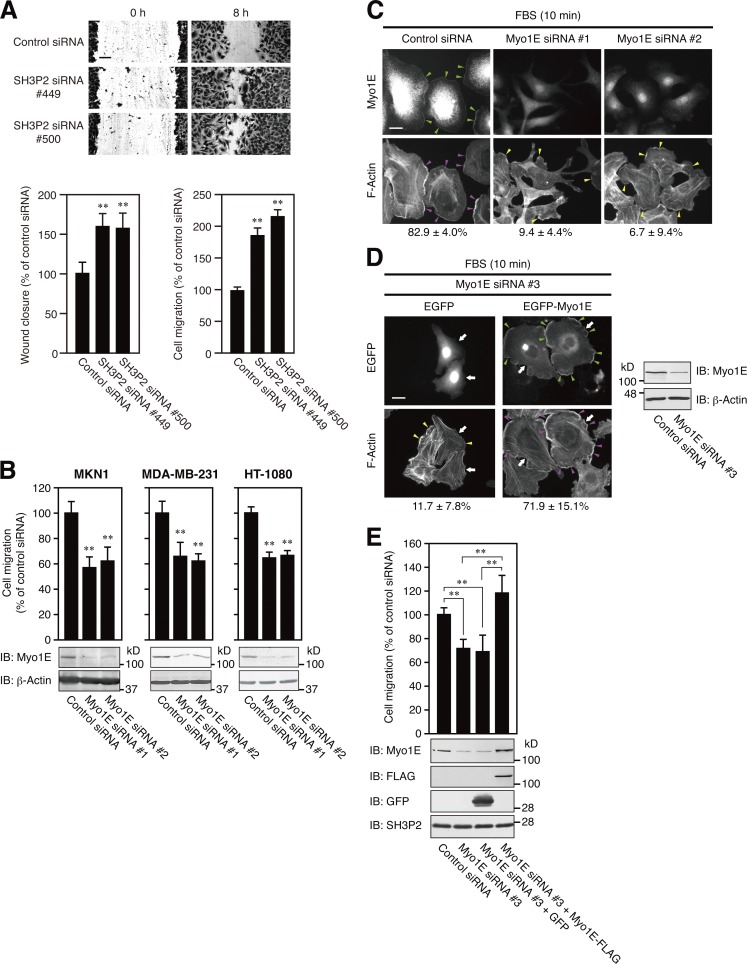
RNAi-mediated depletion of SH3P2 increased the motility of MKN1 cells as revealed by both wound-healing and transwell migration assays (Fig. 8 A). In contrast, RNAi-mediated knockdown of Myo1E suppressed motility in MKN1, MDA-MB-231, and HT-1080 cells (Fig. 8 B). Knockdown of Myo1E by itself with an siRNA targeting the ORF (Fig. 8 C) or the 5′-UTR sequence (Fig. 8 D) did not abolish the serum-induced formation of lamellipodia in MKN1 cells. However, lamellipodia formed under such conditions were poorly developed compared with those formed in cells transfected with a control siRNA. Importantly, forced expression of EGFP-Myo1E restored the formation of extended lamellipodia in cells depleted of endogenous Myo1E with the siRNA targeting the 5′-UTR sequence (Fig. 8 D). Furthermore, rescue of Myo1E expression by infection with a lentivirus encoding FLAG epitope–tagged Myo1E restored the motility of Myo1E-depleted MKN1 cells (Fig. 8 E). Together, these results suggest that Myo1E is necessary for the formation of active lamellipodia required for cell motility.
Figure 8.
Myo1E facilitates lamellipodium formation and thereby promotes cell motility. (A) MKN1 cells transfected with an SH3P2 siRNA (#449 or #500) or a control siRNA were subjected to a wound-healing assay (top and bottom left panels) or a transwell migration assay (bottom right panel) in the presence of 10% FBS for 8 or 6 h, respectively. Bar, 100 µm. The number of cells that had migrated to reach the lower surface of the filter in the transwell migration assay was 98 ± 7 for cells transfected with the control siRNA (bottom right panel). Quantitative data are means ± SD for three separate experiments. **, P < 0.01 versus cells transfected with the control siRNA. (B) MKN1, MDA-MB-231, and HT-1080 cells transfected with a Myo1E siRNA (#1 or #2) or a control siRNA were subjected to a transwell migration assay in the presence of 10% FBS for 6 h. Cell lysates were also subjected to immunoblot analysis (IB) with antibodies to Myo1E and to β-actin (loading control). The numbers of cells that had migrated to reach the lower surface of the filter were 159 ± 17, 166 ± 15, and 288 ± 12 for control siRNA–transfected MKN1, MDA-MB-231, and HT-1080 cells, respectively. Quantitative data are means ± SD for three separate experiments. **, P < 0.01 versus cells transfected with the control siRNA. (C) MKN1 cells transfected with a Myo1E siRNA (#1 or #2) or a control siRNA for 48 h were deprived of serum for 12 h, stimulated with 10% FBS for 10 min, fixed, and stained with antibodies to Myo1E and with phalloidin. Green arrowheads indicate Myo1E localized to lamellipodial tips, magenta arrowheads indicate extended lamellipodia (total length of >50 µm per cell), and yellow arrowheads indicate immature lamellipodial structures (total length of <50 µm per cell). Bar, 20 µm. The percentages of cells with extended lamellipodia are shown below the images as means ± SD for three separate experiments, with n ≥ 60 cells in each experiment. (D) MKN1 cells were transfected first with an siRNA targeting the 5′-UTR sequence of Myo1E (#3) for 48 h and then with a vector for EGFP or EGFP-Myo1E for 24 h. The cells were then deprived of serum for 12 h, stimulated with 10% FBS for 10 min, fixed, and examined for EGFP fluorescence and stained with phalloidin. Green arrowheads indicate Myo1E localized to lamellipodial tips, magenta arrowheads indicate extended lamellipodia (total length of >50 µm per cell), yellow arrowheads indicate immature lamellipodial structures (total length of <50 µm per cell), and white arrows indicate cells positive for EGFP or EGFP-Myo1E. Bar, 20 µm. The percentages of EGFP-positive cells with extended lamellipodia are shown below the images as means ± SD for three separate experiments, with n ≥ 40 cells in each experiment. Cell lysates were also subjected to immunoblot analysis with antibodies to Myo1E and to β-actin. Data are representative of at least three separate experiments. (E) MKN1 cells were transfected with an siRNA targeting the 5′-UTR sequence of Myo1E (#3) for 24 h, infected with lentiviruses encoding GFP or Myo1E-FLAG for 24 h, and then cultured in fresh medium for 24 h. The cells were subjected to a transwell migration assay in the presence of 10% FBS for 6 h. Cell lysates were also subjected to immunoblot analysis with antibodies to the indicated proteins. The number of cells that had migrated to reach the lower surface of the filter was 205 ± 15 for control siRNA–transfected MKN1 cells. Quantitative data are means ± SD for three separate experiments. **, P < 0.01.
Discussion
The molecular mechanisms underlying cell motility, especially with regard to the integration of signaling pathways and cytoskeletal systems, remain poorly understood. Activation of both the PI3K and ERK pathways downstream of Ras is essential for the induction of cell motility. We recently identified SH3P2 as a negative regulator of cell motility whose function is inhibited by RSK-mediated phosphorylation (Tanimura et al., 2011). We have now identified Myo1E as a binding partner of SH3P2 and uncovered the molecular mechanism by which SH3P2 inhibits cell motility.
We first examined how SH3P2 interacts with Myo1E with the use of pull-down assays with GST-fused SH3P2 or Myo1E deletion mutants as baits as well as coimmunoprecipitation assays with MKN1 cells expressing such deletion mutants tagged with EGFP. Our results suggest that SH3P2 interacts with Myo1E in the cytosol through two binding interfaces. The PR region of SH3P2 thus interacts with the SH3 domain of Myo1E, and the C-terminal acidic amino acid cluster of SH3P2 interacts with the positively charged region located at the C terminus of the TH2 domain of Myo1E (Fig. 9, 1a). We confirmed that the SH3 domain of Myo1E interacts directly with SH3P2 in a manner dependent on the PR region of SH3P2. However, we failed to demonstrate direct interaction between the TH2 domain of Myo1E and SH3P2 or between the C-terminal acidic amino acid cluster of SH3P2 and Myo1E, possibly because such small domains of SH3P2 and Myo1E alone are not able to maintain their native conformations when expressed as EGFP-tagged or GST-fused constructs. Structural analysis of the SH3P2–Myo1E complex will be required to provide a full understanding of the mode of interaction between these two proteins.
Figure 9.
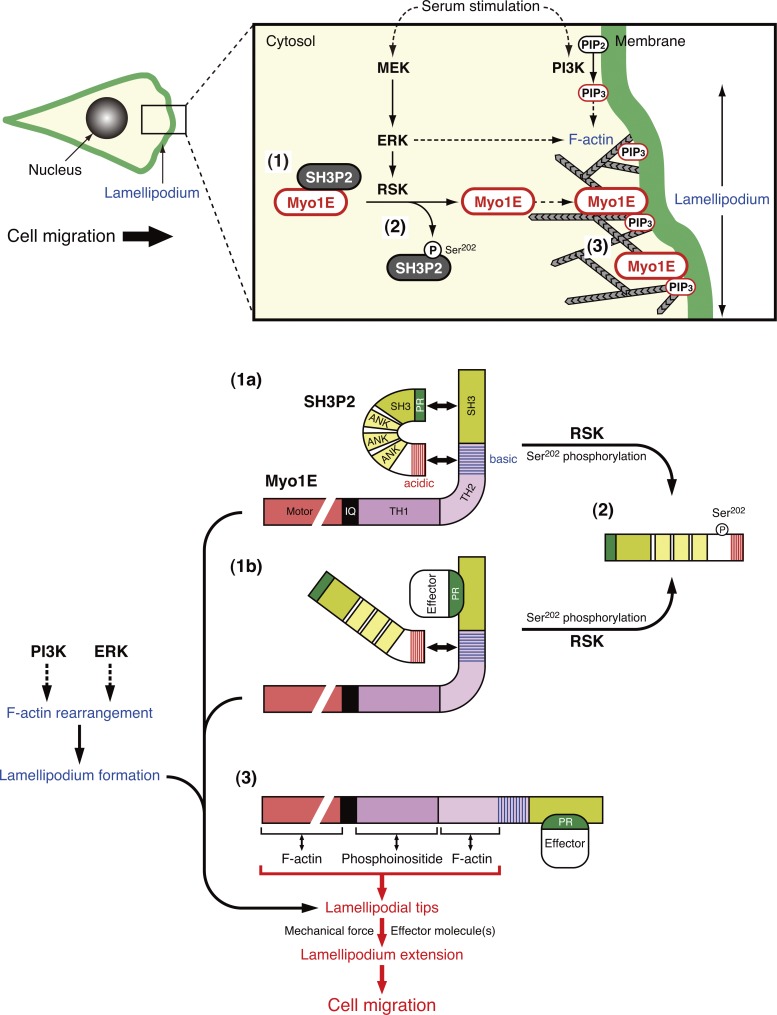
Model for the regulation of cell motility by the RSK-SH3P2-Myo1E axis. SH3P2 binds to and functions as a cytosolic anchor for Myo1E (1). SH3P2 interacts with Myo1E through two binding interfaces (the PR region of SH3P2 with the SH3 domain of Myo1E, and the C-terminal acidic amino acid cluster of SH3P2 with the positively charged region located at the C terminus of the TH2 domain of Myo1E; 1a), with the latter interaction appearing to be responsible for the association of the two proteins in most SH3P2-Myo1E complexes in cells (1b). Serum stimulation of cells induces activation of the ERK signaling pathway and the consequent phosphorylation of SH3P2 at Ser202 by RSK, resulting in the dissociation of Myo1E from SH3P2 (2). Serum stimulation also induces activation of the PI3K signaling pathway, resulting in the production of PI(3,4,5)P3 and consequent rearrangement of F-actin at the leading edge of the cell. Myo1E released from SH3P2 moves along the rearranged actin filaments and localizes at the tips of lamellipodia via binding of its TH2 domain to F-actin (3). The mechanical force generated by Myo1E–F-actin interaction at the plasma membrane together with the function of effector molecules transported or recruited by Myo1E through binding to its SH3 domain coordinately induce active lamellipodium formation and thereby promote cell migration. PIP2, PI(4,5)P2; PIP3, PI(3,4,5)P3.
Serum stimulation of cells induces activation of the ERK signaling pathway and the consequent phosphorylation of SH3P2 at Ser202 by RSK, resulting in the dissociation of Myo1E from SH3P2 (Fig. 9, step 2). Free Myo1E binds directly to membranes via association of its TH1 domain with anionic phospholipids such as PI(4,5)P2 and PI(3,4,5)P3, with such interactions being relatively nonspecific and electrostatic in nature rather than stereospecific (Feeser et al., 2010). We have now shown that the TH2 domain is required for the preferential localization of Myo1E to lamellipodial tips via binding to F-actin. This domain was previously shown to be required for the localization of Myo1E to invadosomes (Ouderkirk and Krendel, 2014). Long-tailed class 1 myosins bind to F-actin via the TH2 domain as well as the motor domain (Doberstein and Pollard, 1992; Jung and Hammer, 1994; Rosenfeld and Rener, 1994). Furthermore, the TH2 domain was shown to mediate the targeting of Xenopus laevis Myo1E to newly assembled actin in vivo (Yu and Bement, 2007) and to be required for the targeting of Myo1B to dynamic actin waves in Dictyostelium discoideum (Brzeska et al., 2014). The binding of the TH2 domain to F-actin thus provides a mechanism by which this class of myosin can be targeted specifically to sites of active actin assembly. Together, these previous and our present observations suggest that the TH2 domain plays a key role in the localization of Myo1E to lamellipodial tips through its binding to F-actin, whereas the TH1 domain may facilitate the association of Myo1E with the plasma membrane by binding to anionic phospholipids.
Myo1E localized at the plasma membrane interacts with F-actin to generate mechanical force and thereby regulates plasma membrane tension (McConnell and Tyska, 2010). Myo1E has also been shown to transport or recruit several effector molecules such as dynamin, synaptojanin-1, and the N-WASP–WIP complex to the membrane–cytoskeleton interface in a process dependent on the binding of the PR region of the respective cargo to the SH3 domain of Myo1E (Krendel et al., 2007; Cheng et al., 2012; Gupta et al., 2013). The TH2 domain of Myo1E also contains three conserved PR regions (Fig. S1 B), through which Myo1E might bind to SH3 domain–containing proteins and transport or recruit them to the plasma membrane. These observations suggest that the mechanical force generated by Myo1E–F-actin interaction at the plasma membrane together with the functions of effector molecules transported or recruited by Myo1E through binding to its SH3 domain and possibly to its TH2 domain coordinately induce active lamellipodium formation and thereby promote cell migration (Fig. 9, step 3).
Given that SH3P2 also binds to the SH3 domain of Myo1E via its PR region, SH3P2 may regulate the interaction of PR region–containing effector molecules with Myo1E in a competitive manner. In this regard, deletion of its SH3 domain did not affect the binding of Myo1E to SH3P2 in MKN1 cells. The SH3 domain of most Myo1E molecules in these cells may be occupied by effector proteins rather than by SH3P2. Thus, although SH3P2 is able to bind to Myo1E through the two binding interfaces identified in the present study, the interaction between the C-terminal acidic amino acid cluster of SH3P2 and the positively charged region at the C terminus of the TH2 domain of Myo1E may be responsible for the association of the two proteins in most SH3P2–Myo1E complexes in cells (Fig. 9, 1b).
The major finding of the present study is that SH3P2 regulates the intracellular localization of Myo1E in an ERK pathway–dependent manner. The cytosolic protein SH3P2 thus forms a complex with most Myo1E to prevent its localization to the plasma membrane in cells in which basal ERK activity is limited. On cell stimulation, however, activated RSK1 phosphorylates SH3P2 at Ser202 and thereby induces the dissociation of Myo1E from SH3P2 and the subsequent translocation of Myo1E to the tips of lamellipodia, a process that is essential for the induction of cell motility. Binding of SH3P2 likely renders Myo1E inaccessible to F-actin and possibly to effector molecules, thereby preventing Myo1E localization to lamellipodial tips. SH3P2 therefore functions as a cytoplasmic anchor for Myo1E and thereby negatively regulates cell motility.
Activation of the PI3K and ERK signaling pathways by serum stimulation results in the production of PI(3,4,5)P3 and consequent F-actin rearrangement at the leading edge of the cell as well as in phosphorylation of SH3P2 and consequent dissociation of Myo1E, respectively. The ERK pathway also contributes to the rearrangement of F-actin via ERK-mediated phosphorylation of WAVE2 and Abi1, components of the WAVE2 regulatory complex (Mendoza et al., 2011). Myo1E thus appears to be positioned at an integration point for the ERK and PI3K pathways in the coordinated regulation of cell motility, with Myo1E released from its cytoplasmic anchor SH3P2 moving along the rearranged actin filaments toward the leading edge of the cell. Myo1E thereby localized at lamellipodial tips, and the effector molecules recruited by binding to its SH3 domain then cooperatively promote lamellipodium extension and consequent cell migration. Further studies are required to determine the precise molecular mechanisms by which the downstream targets of Myo1E facilitate cell motility.
Materials and methods
Antibodies and reagents
Rabbit polyclonal antibodies to SH3P2 and to Ser202-phosphorylated SH3P2 were prepared as described previously (Tanimura et al., 2011). Rabbit polyclonal antibodies to Myo1E were generated in response to a GST fusion protein containing the SH3 domain of human Myo1E (amino acids 1,039–1,108) and were purified with an affinity column containing the His6-tagged antigen. Antibodies to ERK1/2, RSK1, Sam68, and hnRNP-K were obtained from Santa Cruz Biotechnology, Inc.; those to GFP (mouse/rat) were obtained from Nacalai Tesque; those to diphosphorylated ERK1/2, α-tubulin, FLAG, and β-actin were obtained from Sigma-Aldrich; those to Akt and Ser473-phosphorylated Akt were obtained from Cell Signaling Technology; those to integrin β1 (CD29) were obtained from BD; those to MMP-14 (MT1-MMP) were obtained from EMD Millipore; and those to CD44 were obtained from R&D Systems. PD184352 was synthesized as described previously (Tanimura et al., 2003), LY294002 was obtained from Wako Pure Chemical Industries, and latrunculin A was obtained from Cayman Chemical.
Cell culture and migration assays
The human tumor cell lines MKN1, HT-1080 (Health Science Research Resources Bank), HeLa S3, and MDA-MB-231 (ATCC) were cultured in DMEM supplemented with 10% FBS. Cell motility was analyzed with a transwell migration assay as described previously (Tanimura et al., 2011) as well as with a wound-healing assay. In the latter, a scratch wound was introduced in a monolayer of MKN1 cells with a plastic micropipette tip. After incubation for 8 h to allow wound healing, cells were fixed with 100% methanol and stained with Coomassie brilliant blue. Images were acquired with an Axiovert 200M microscope (ZEISS) equipped with an AxioCam MRm camera (ZEISS), and the width of the wound area was measured with the use of AxioVision software version 4.8.2 (ZEISS).
Subcellular fractionation
Subcellular fractionation of MKN1 cells was performed as described previously (Higuchi et al., 2008), with minor modifications. In brief, cells grown in each well of a 12-well plate were scraped into 200 µl hypotonic lysis buffer (25 mM Tris-HCl, pH 7.4, 25 mM NaCl, 1 mM sodium orthovanadate, 10 mM NaF, 10 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 25 mM p-nitrophenyl phosphate, 20 nM okadaic acid, 0.2 mM sodium molybdate, 0.5 mM EGTA, 1 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin; Hoshino et al., 1999) and disrupted with the use of a Dounce homogenizer. The homogenate was centrifuged at 700 g for 5 min at 4°C to remove nuclei and intact cells, and the resulting supernatant was centrifuged at 20,000 g for 30 min at 4°C to obtain a cytosolic fraction (supernatant) and pellet. After washing with the hypotonic lysis buffer, the pellet was suspended in 100 µl of the hypotonic lysis buffer containing 1% Triton X-100 and centrifuged at 20,000 g for 30 min at 4°C to obtain a membrane fraction (supernatant). Portions corresponding to 20% or 50% of the cytosolic or membrane fractions, respectively, prepared from cells grown in each well of a 12-well plate were analyzed for the distribution of Myo1E.
Cell lysis and immunoblot analysis
Cell lysates were prepared with a solution containing 25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM sodium orthovanadate, 10 mM NaF, 10 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 25 mM p-nitrophenyl phosphate, 20 nM okadaic acid, 0.2 mM sodium molybdate, 0.5 mM EGTA, 1 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin. Cell lysates were fractionated by SDS-PAGE, and the separated proteins were transferred electrophoretically to a polyvinylidene difluoride membrane and probed with primary antibodies and HRP-conjugated secondary antibodies (Promega). Immune complexes were detected with the use of enhanced chemiluminescence reagents (GE Healthcare) and an ImageQuant LAS4000 instrument (GE Healthcare). Image analysis was performed with National Institutes of Health ImageJ version 1.46r software. For quantitation of immunoblot signals, linearity of band intensities was examined with the use of serial dilutions of each sample and the intensity of each band was measured within the linear range of detection (Janes, 2015). Representative results of such quantitation for the distribution of Myo1E, CD44, integrin β1, and MMP-14 in cytosolic and membrane fractions of MKN1 cells are shown in Fig. S5.
Coimmunoprecipitation analysis
Cell lysates were incubated with antibodies for 6 h at 4°C with gentle shaking. Immune complexes were precipitated with Protein A/G PLUSAgarose (Santa Cruz Biotechnology, Inc.), washed three times with cell lysis buffer, and then subjected to SDS-PAGE followed by either staining with Coomassie brilliant blue or immunoblot analysis. The proportion of Myo1E associated with SH3P2 was calculated by comparison of the intensity of the SH3P2 and Myo1E bands in immunoprecipitates prepared with antibodies to these proteins with that of those in the corresponding cell lysates.
GST pull-down assays
Bacterially expressed GST, GST-fused wild-type or mutant forms of SH3P2, or GST-Myo1E(SH3) (10 µg for each) bound to glutathione-Sepharose beads (GE Healthcare) was incubated with gentle shaking for 6 h at 4°C with MKN1 cell lysate or recombinant SH3P2 or SH3P2(ΔPR). The bead-bound proteins were then subjected to SDS-PAGE followed by either staining with Coomassie brilliant blue or immunoblot analysis.
Plasmids
The mammalian expression vectors pEGFP-C1-SH3P2 and pEGFP-C1-SH3P2(S202A) were generated as described previously (Tanimura et al., 2011). For generation of expression plasmids encoding human SH3P2 amino acids 1 to 189 (ΔC), 1 to 204 (ΔC1), 1 to 208 (ΔC2), 44 to 214 (ΔPR/SH3), or 12 to 189 (ΔPR/C), the corresponding cDNA fragments were amplified from wild-type SH3P2 cDNA by PCR with the primer pairs (forward with the underlined EcoRI site and reverse with the underlined XhoI site, respectively) 5′-CGGAATTCGATGTCGAAGCCGCCACCC-3′ and 5′-GCCCTCGAGTTACAGCAGAGATGCACAGGC-3′, 5′-CGGAATTCGATGTCGAAGCCGCCACCC-3′ and 5′-GCCCTCGAGTTAGGCATTGCTTAATGTTC-3′, 5′-CGGAATTCGATGTCGAAGCCGCCACCC-3′ and 5′-GCCCTCGAGTTAGAGATAGTCCTCGGCAT-3′, 5′-CCGAATTCCATGAGCGATACCAATTGG-3′ and 5′-GCCCTCGAGTTAATCTGAGTCTTCATC-3′, and 5′-CCGAATTCAGGGCAAGTTAAAGTCTTC-3′ and 5′-GCCCTCGAGTTACAGCAGAGATGCACAGGC-3′, respectively. The PCR products were digested with EcoRI and XhoI, and the resulting fragments were cloned into EcoRI- and SalI-digested pEGFP-C1 (Takara Bio Inc.). For generation of an expression plasmid encoding FLAG-tagged SH3P2, a cDNA fragment amplified from wild-type SH3P2 cDNA by PCR with the primers (forward with the underlined EcoRI site and reverse with the underlined XhoI site, respectively) 5′-GCCGAATTCGCGATGTCGAAGCCGCCACCC-3′ and 5′-GCCCTCGAGTTAATCTGAGTCTTCATCATC-3′ was digested with EcoRI and XhoI and then cloned into EcoRI- and XhoI-digested pcDNA3 (Invitrogen) together with the DNA sequence for an N-terminal FLAG tag. For construction of an expression plasmid encoding human Myo1E, a cDNA fragment was amplified from HeLa S3 cell cDNA by PCR with the forward primer 5′-GCCGGATTCATGGGAAGCAAAGGTGTC-3′ (underline indicates the BamHI site) and reverse primer 5′-GCCCTCGAGTCAGATCTTGGTCACATAG-3′ (underline indicates the XhoI site). The PCR product was digested either with BamHI and EcoRI to yield a cDNA encoding Myo1E amino acids 1 to 851 (N terminal) or with EcoRI and XhoI to yield a cDNA encoding Myo1E amino acids 852 to 1,108 (C terminal). The cDNA encoding N terminus was cloned into BglII- and EcoRI-digested pEGFP-C1, and that encoding the C terminus was then cloned into the EcoRI- and SalI-digested pEGFP-C1 N terminus. For generation of expression plasmids encoding human Myo1E amino acids 1 to 694 (ΔTH1/TH2/SH3) or 695 to 1,108 (ΔMotor), the corresponding cDNA fragments were amplified from wild-type Myo1E cDNA by PCR with the primer pairs (forward with the underlined BamHI site and reverse with underlined XhoI site) 5′-GCCGGATTCATGGGAAGCAAAGGTGTC-3′ and 5′-GCCCTCGAGTCACTTTCTCTCTCTCATCTC-3′ for Myo1E(ΔTH1/TH2/SH3) and 5′-GCCGGATTCTATGATGGGTATGCACGAGTGATACAG-3′ and 5′-GCCCTCGAGTCAGATCTTGGTCACATAG-3′ for Myo1E(ΔMotor). The PCR products were digested with BamHI and XhoI, and the resulting fragments were cloned into BglII- and SalI-digested pEGFP-C1. For generation of expression plasmids encoding human Myo1E amino acids 1 to 1,050 (ΔSH3), 1 to 1,038 (ΔTH2-b1/SH3), 1 to 1,020 (ΔTH2-b2/SH3), or 1 to 921 (ΔTH2/SH3), cDNA fragments corresponding to Myo1E amino acids 695 to 1,050, 695 to 1,038, 695 to 1,020, or 695 to 921 were amplified from Myo1E(ΔMotor) cDNA by PCR with the forward primer 5′-GCCGGATTCTATGATGGGTATGCACGAGTGATACAG-3′ (underline indicates the BamHI site) and reverse primers 5′-GCCCTCGAGTCACTTGGGCTTGGGCTGGGG-3′, 5′-GCCCTCGAGTCATGGGGGAGGCCGACTGG-3′, 5′-GCCCTCGAGTCAGACCTTGAGGAAATCC-3′, or 5′-GCCCTCGAGTCATCCGATGCTGACCTGCAG-3′ (underline indicates XhoI sites), respectively. The PCR products were digested with EcoRI and XhoI, and the resulting cDNA fragments corresponding to amino acids 852 to 1,050, 852 to 1,038, 852 to 1,020, or 852 to 921 were cloned into the EcoRI- and SalI-digested pEGFP-C1 N terminus. For generation of an expression plasmid encoding human GRP1(PH), the corresponding cDNA fragment was amplified from HeLa S3 cell cDNA by PCR with the forward primer 5′-GCCGAATTCGGACGACGGGAACGACCTG-3′ (underline indicates the EcoRI site) and reverse primer 5′-GCCCTCGAGCCTCGTTGCCAACATGTC-3′ (underline indicates the XhoI site). The PCR product was digested with EcoRI and XhoI and then cloned into EcoRI- and SalI-digested pEGFP-C1.
The bacterial expression vectors pGEX6P3-SH3P2 and pGEX6P3-SH3P2(ΔC) were generated as described previously (Tanimura et al., 2011). For generation of bacterial expression plasmids encoding human SH3P2 amino acids 12 to 214 (ΔPR), 44 to 214 (ΔPR/SH3), 12 to 189 (ΔPR/C), or 12 to 71 (ΔPR/ANK/C), the corresponding cDNA fragments were amplified from wild-type SH3P2 cDNA by PCR with the primer pairs (forward with the underlined EcoRI site and reverse with the underlined XhoI site, respectively) 5′-CCGAATTCAGGGCAAGTTAAAGTCTTC-3′ and 5′-GCCCTCGAGTTAATCTGAGTCTTCATC-3′, 5′-CCGAATTCCATGAGCGATACCAATTGG-3′ and 5′-GCCCTCGAGTTAATCTGAGTCTTCATC-3′, 5′-CCGAATTCAGGGCAAGTTAAAGTCTTC-3′ and 5′-GCCCTCGAGTTACAGCAGAGATGCACAGGC-3′, and 5′-CCGAATTCAGGGCAAGTTAAAGTCTTC-3′ and 5′-GCCCTCGAGTTATTCTGCCTGCTCAGCCAC-3′, respectively. The PCR products were digested with EcoRI and XhoI, and the resulting fragments were cloned into EcoRI- and XhoI-digested pGEX6P3 (GE Healthcare). For generation of a bacterial expression plasmid encoding human Myo1E amino acids 1,039 to 1,108 (SH3), the corresponding cDNA was amplified from Myo1E(ΔMotor) cDNA by PCR with the forward primer 5′-GCGAATTCAGCAGGGGGCAGACCCAAG-3′ (underline indicates the EcoRI site) and the reverse primer 5′-GCCCTCGAGTCAGATCTTGGTCACATAG-3′ or 5′-GCCCTCGAGGATCTTGGTCACATAGTTG-3′ (underline indicates XhoI sites). The PCR products were digested with EcoRI and XhoI, and the resulting fragments were cloned into EcoRI- and XhoI-digested pGEX6P3 for GST-Myo1E(SH3) or into EcoRI- and XhoI-digested pET24b (Novagen) for His6-tagged Myo1E(SH3), respectively.
Mutagenesis of Asp209 and Asp210 of SH3P2 to Ala (C2A) was performed with the use of a PrimeSTAR Mutagenesis Basal kit (Takara Bio Inc.) and with pEGFP-C1-SH3P2 and the mutagenic primers (forward and reverse, respectively) 5′-TATCTCGCTGCTGAAGACTCAGATTAACTC-3′ and 5′-GTCTTCAGCAGCGAGATAGTCCTCGGCATT-3′. For mutagenesis of Glu205, Asp206, Asp209, Asp210, Glu211, and Asp212 of SH3P2 to Ala (C6A), Glu205 and Asp206 were changed to Ala (205/206A) with pEGFP-C1-SH3P2 and the mutagenic primers 5′-AATGCCGCGGCCTATCTCGATGATGAAGAC-3′ and 5′-GAGATAGGCCGCGGCATTGCTTAATGTTCG-3′; Asp209 and Asp210 were then changed to Ala (C4A) with pEGFP-C1-205/206A and the mutagenic primers 5′-TATCTCGCTGCTGAAGACTCAGATTAACTC-3′ and 5′-GTCTTCAGCAGCGAGATAGGCCGCGGCATT-3′; and Glu211 and Asp212 were then changed to Ala (C6A) with pEGFP-C1-C4A and the mutagenic primers 5′-GCTGCTGCAGCCTCAGATTAACTCGACGGT-3′ and 5′-ATCTGAGGCTGCAGCAGCGAGATAGGCCGC-3′. Mutagensesis of Lys1048 and Lys1050 of Myo1E(ΔSH3) to Ala (2AΔSH3) was performed with pEGFP-C1-Myo1E(ΔSH3) and the mutagenic primers 5′-CAGCCCGCGCCCGCGTGACTCGACGGTACCGCG-3′ and 5′-GAGTCACGCGGGCGCGGGCTGGGGCTTGGGTCT-3′.
The GST- or His6-tagged protein fragments were expressed in Escherichia coli and purified as described previously (Tanimura et al., 1998, 2007). For recombinant SH3P2 or SH3P2(ΔPR), the GST tag was cleaved with PreScission protease (GE Healthcare). Transfection of tumor cells with pEGFP-C1–based plasmids was performed with the use of Lipofectamine 2000 (Invitrogen).
Production of and infection with recombinant lentiviruses
An human immunodeficiency virus–1–based lentiviral vector system was obtained from D. Trono (École Polytechnique Fédérale de Lausanne, Switzerland; Zufferey et al., 1997). A cDNA for FLAG-tagged human Myo1E (Myo1E-FLAG) was introduced into pRRL. Recombinant lentiviruses were produced by transfection of HEK293T cells in a 10-cm dish for 72 h with 1.5 µg pMD2.G, 3.0 µg pMDLg/pRRE, 1.5 µg pRSV-Rev, and 6.0 µg pRRL-CMV-GFP-Sin-18 or pRRL-CMV-Myo1E-FLAG. Culture supernatants containing lentiviruses were centrifuged at 1,200 g for 10 min, and the resulting soluble fraction was passed through a 0.45-µm filter (EMD Millipore). Lentiviruses in the filtrate were then collected by precipitation with polyethylene glycol and suspended in 500 µl Opti-MEM (Gibco). MKN1 cells that had been transfected with Myo1E siRNA for 24 h in a 24-well plate were infected with recombinant lentiviruses encoding GFP or Myo1E-FLAG (10 µl of the respective virus suspension) for 24 h and then incubated in fresh culture medium for 24 h.
RNAi
siRNAs for human Myo1E (#1, Stealth select RNAi MYO1EHSS106891; #2, Stealth select RNAi MYO1EHSS106893) and for human SH3P2 (#449 and #500; Tanimura et al., 2011) were obtained from Invitrogen. The sequences 5′-GAGTACGGATTAAGCGGCCGAAGAA-3′ (Myo1E #3) and 5′-GAGGCGACGGTTGTAAGCCAGACAA-3′ (SH3P2 #36) were designed to generate siRNA duplexes specific for the 5′-UTR sequence of human Myo1E or SH3P2 mRNAs (Invitrogen). Stealth RNAi Negative Control Medium GC Duplex was used as a control (Invitrogen). Subconfluent cultures of cells were transfected with siRNAs (20 nM) for 48 or 72 h with the use of Lipofectamine RNAiMAX (Invitrogen).
In-gel digestion and protein identification
Bands of interest were excised from Coomassie brilliant blue–stained gels, cut into 1-mm pieces, dehydrated with 50% acetonitrile containing 25 mM NH4HCO3, and dried with the use of a vacuum centrifuge. The gel pieces were rehydrated with digestion solution (50 mM NH4HCO3 containing sequencing-grade modified trypsin [Promega] at 10 µg/ml) and incubated overnight at 37°C, and the resulting peptides were extracted with 5% trifluoroacetic acid in 50% acetonitrile. The peptide mixture was subjected to liquid chromatography, electrospray ionization, and tandem MS analysis (LTQ-XL; Thermo Fisher Scientific). MS/MS data were extracted with Proteome Discoverer v.3.3 (Thermo Fisher Scientific), and spectra were searched against a human subdatabase obtained from the public nonredundant protein database (International Protein Index version 3.84 presented by The European Bioinformatics Institute). Each protein was identified on the basis of more than two unique peptides.
Immunofluorescence microscopy
Cells grown on glass coverslips and exposed to various agents were fixed with 3.7% paraformaldehyde and permeabilized with 0.25% Triton X-100. After blocking of nonspecific sites with 2.5% BSA in PBS, the cells were immunostained with antibodies to Myo1E (1/200 dilution) or to SH3P2 (1/200), and immune complexes were detected with Alexa Fluor 488– or 546–conjugated goat antibodies to rabbit IgG (1/200 dilution; Molecular Probes). F-actin was visualized with the use of Alexa Fluor 350–labeled phalloidin (Molecular Probes). Images were acquired with an Axiovert 200M microscope equipped with a FluoArc mercury lamp, a Plan-NEOFLUAR 40×/0.75 objective lens, and an AxioCam MRm camera (ZEISS). For comparison of the localization of Myo1E or of EGFP-Myo1E and its mutants, images were acquired with the same exposure time in respective experiments (typically 2 s for Myo1E and 5 s for EGFP). Images were analyzed with AxioVision software version 4.8.2 (ZEISS).
Statistical analysis
Data are presented as means ± SD and were analyzed with the two-tailed Student’s t test. A p-value of <0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the sequence alignment for the N-terminal PR region and C-terminal acidic amino acid cluster of mammalian SH3P2 proteins as well as for the TH2 domain of mammalian Myo1E proteins. Fig. S2 shows the putative binding interfaces for the interaction between SH3P2 and Myo1E. Fig. S3 shows that LY294002 suppresses the serum-induced production of PI(3,4,5)P3 and localization of Myo1E to lamellipodial tips. Fig. S4 shows the membrane association of recombinant Myo1E is suppressed by prior incubation with GST-SH3P2. Fig. S5 shows quantitative immunoblot analysis of Myo1E in the cytosolic and membrane fractions of MKN1 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503123/DC1.
Supplementary Material
Acknowledgments
This work was supported by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Takeda Science Foundation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ERK
- extracellular signal–regulated kinase
- MS
- mass spectrometry
- Myo1E
- myosin 1E
- PI3K
- phosphatidylinositol 3-kinase
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PR
- proline-rich
- RSK
- p90 ribosomal S6 kinase
- SH3
- Src homology 3
- TH
- tail homology
References
- Brzeska H., Pridham K., Chery G., Titus M.A., and Korn E.D.. 2014. The association of myosin IB with actin waves in dictyostelium requires both the plasma membrane-binding site and actin-binding region in the myosin tail. PLoS One. 9:e94306 10.1371/journal.pone.0094306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Grassart A., and Drubin D.G.. 2012. Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis. Mol. Biol. Cell. 23:2891–2904. 10.1091/mbc.E11-04-0383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson C.M., Pocha S.M., Bloomberg G.B., and Cory G.O.. 2007. Phosphorylation of WAVE2 by MAP kinases regulates persistent cell migration and polarity. J. Cell Sci. 120:4144–4154. 10.1242/jcs.013714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein S.K., and Pollard T.D.. 1992. Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J. Cell Biol. 117:1241–1249. 10.1083/jcb.117.6.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeser E.A., Ignacio C.M., Krendel M., and Ostap E.M.. 2010. Myo1e binds anionic phospholipids with high affinity. Biochemistry. 49:9353–9360. 10.1021/bi1012657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Chen J.K., Yu H., Simon J.A., and Schreiber S.L.. 1994. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 266:1241–1247. 10.1126/science.7526465 [DOI] [PubMed] [Google Scholar]
- Guo W., and Giancotti F.G.. 2004. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 5:816–826. 10.1038/nrm1490 [DOI] [PubMed] [Google Scholar]
- Gupta P., Gauthier N.C., Cheng-Han Y., Zuanning Y., Pontes B., Ohmstede M., Martin R., Knölker H.J., Döbereiner H.G., Krendel M., and Sheetz M.. 2013. Myosin 1E localizes to actin polymerization sites in lamellipodia, affecting actin dynamics and adhesion formation. Biol. Open. 2:1288–1299. 10.1242/bio.20135827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Onishi K., Kikuchi C., and Gotoh Y.. 2008. Scaffolding function of PAK in the PDK1-Akt pathway. Nat. Cell Biol. 10:1356–1364. 10.1038/ncb1795 [DOI] [PubMed] [Google Scholar]
- Hoshino R., Chatani Y., Yamori T., Tsuruo T., Oka H., Yoshida O., Shimada Y., Ari-i S., Wada H., Fujimoto J., and Kohno M.. 1999. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 18:813–822. 10.1038/sj.onc.1202367 [DOI] [PubMed] [Google Scholar]
- Hunger-Glaser I., Salazar E.P., Sinnett-Smith J., and Rozengurt E.. 2003. Bombesin, lysophosphatidic acid, and epidermal growth factor rapidly stimulate focal adhesion kinase phosphorylation at Ser-910: requirement for ERK activation. J. Biol. Chem. 278:22631–22643. 10.1074/jbc.M210876200 [DOI] [PubMed] [Google Scholar]
- Janes K.A. 2015. An analysis of critical factors for quantitative immunoblotting. Sci. Signal. 8:rs2 10.1126/scisignal.2005966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., and Hammer J.A. III. 1994. The actin binding site in the tail domain of Dictyostelium myosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology region 2). FEBS Lett. 342:197–202. 10.1016/0014-5793(94)80500-8 [DOI] [PubMed] [Google Scholar]
- Klemke R.L., Cai S., Giannini A.L., Gallagher P.J., de Lanerolle P., and Cheresh D.A.. 1997. Regulation of cell motility by mitogen-activated protein kinase. J. Cell Biol. 137:481–492. 10.1083/jcb.137.2.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M., Osterweil E.K., and Mooseker M.S.. 2007. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 581:644–650. 10.1016/j.febslet.2007.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D.A., and Horwitz A.F.. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369. 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Martinez-Quiles N., Ho H.Y., Kirschner M.W., Ramesh N., and Geha R.S.. 2004. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol. 24:5269–5280. 10.1128/MCB.24.12.5269-5280.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell R.E., and Tyska M.J.. 2010. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 20:418–426. 10.1016/j.tcb.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M.C., Er E.E., Zhang W., Ballif B.A., Elliott H.L., Danuser G., and Blenis J.. 2011. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell. 41:661–671. 10.1016/j.molcel.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi O., Suetsugu S., Yamazaki D., and Takenawa T.. 2007. Effect of WAVE2 phosphorylation on activation of the Arp2/3 complex. J. Biochem. 141:319–325. 10.1093/jb/mvm034 [DOI] [PubMed] [Google Scholar]
- Ouderkirk J.L., and Krendel M.. 2014. Myosin 1e is a component of the invadosome core that contributes to regulation of invadosome dynamics. Exp. Cell Res. 322:265–276. 10.1016/j.yexcr.2014.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M., and Hall A.. 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 265:23–32. 10.1016/j.ydbio.2003.06.003 [DOI] [PubMed] [Google Scholar]
- Ren R., Mayer B.J., Cicchetti P., and Baltimore D.. 1993. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 259:1157–1161. 10.1126/science.8438166 [DOI] [PubMed] [Google Scholar]
- Rosenfeld S.S., and Rener B.. 1994. The GPQ-rich segment of Dictyostelium myosin IB contains an actin binding site. Biochemistry. 33:2322–2328. 10.1021/bi00174a045 [DOI] [PubMed] [Google Scholar]
- Tanimura S., Chatani Y., Hoshino R., Sato M., Watanabe S., Kataoka T., Nakamura T., and Kohno M.. 1998. Activation of the 41/43 kDa mitogen-activated protein kinase signaling pathway is required for hepatocyte growth factor-induced cell scattering. Oncogene. 17:57–65. 10.1038/sj.onc.1201905 [DOI] [PubMed] [Google Scholar]
- Tanimura S., Asato K., Fujishiro S.H., and Kohno M.. 2003. Specific blockade of the ERK pathway inhibits the invasiveness of tumor cells: down-regulation of matrix metalloproteinase-3/-9/-14 and CD44. Biochem. Biophys. Res. Commun. 304:801–806. 10.1016/S0006-291X(03)00670-3 [DOI] [PubMed] [Google Scholar]
- Tanimura S., Hirano A.I., Hashizume J., Yasunaga M., Kawabata T., Ozaki K., and Kohno M.. 2007. Anticancer drugs up-regulate HspBP1 and thereby antagonize the prosurvival function of Hsp70 in tumor cells. J. Biol. Chem. 282:35430–35439. 10.1074/jbc.M707547200 [DOI] [PubMed] [Google Scholar]
- Tanimura S., Hashizume J., Kurosaki Y., Sei K., Gotoh A., Ohtake R., Kawano M., Watanabe K., and Kohno M.. 2011. SH3P2 is a negative regulator of cell motility whose function is inhibited by ribosomal S6 kinase-mediated phosphorylation. Genes Cells. 16:514–526. 10.1111/j.1365-2443.2011.01503.x [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Stephens L., and Hawkins P.. 2012. PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13:195–203. 10.1038/nrm3290 [DOI] [PubMed] [Google Scholar]
- Weiger M.C., Wang C.C., Krajcovic M., Melvin A.T., Rhoden J.J., and Haugh J.M.. 2009. Spontaneous phosphoinositide 3-kinase signaling dynamics drive spreading and random migration of fibroblasts. J. Cell Sci. 122:313–323. 10.1242/jcs.037564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennström S., Hawkins P., Cooke F., Hara K., Yonezawa K., Kasuga M., Jackson T., Claesson-Welsh L., and Stephens L.. 1994. Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr. Biol. 4:385–393. 10.1016/S0960-9822(00)00087-7 [DOI] [PubMed] [Google Scholar]
- Xue G., and Hemmings B.A.. 2013. PKB/Akt-dependent regulation of cell motility. J. Natl. Cancer Inst. 105:393–404. 10.1093/jnci/djs648 [DOI] [PubMed] [Google Scholar]
- Yu H.Y., and Bement W.M.. 2007. Multiple myosins are required to coordinate actin assembly with coat compression during compensatory endocytosis. Mol. Biol. Cell. 18:4096–4105. 10.1091/mbc.E06-11-0993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R.J., Naldini L., and Trono D.. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875. 10.1038/nbt0997-871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.