Urban et al. show that RECQ5 DNA helicase promotes RAD18-dependent PCNA ubiquitination and the processing of replication intermediates upon collisions between replication and transcription complexes.
Abstract
Collisions between replication and transcription machineries represent a significant source of genomic instability. RECQ5 DNA helicase binds to RNA-polymerase (RNAP) II during transcription elongation and suppresses transcription-associated genomic instability. Here, we show that RECQ5 also associates with RNAPI and enforces the stability of ribosomal DNA arrays. We demonstrate that RECQ5 associates with transcription complexes in DNA replication foci and counteracts replication fork stalling in RNAPI- and RNAPII-transcribed genes, suggesting that RECQ5 exerts its genome-stabilizing effect by acting at sites of replication-transcription collisions. Moreover, RECQ5-deficient cells accumulate RAD18 foci and BRCA1-dependent RAD51 foci that are both formed at sites of interference between replication and transcription and likely represent unresolved replication intermediates. Finally, we provide evidence for a novel mechanism of resolution of replication-transcription collisions wherein the interaction between RECQ5 and proliferating cell nuclear antigen (PCNA) promotes RAD18-dependent PCNA ubiquitination and the helicase activity of RECQ5 promotes the processing of replication intermediates.
Introduction
DNA replication and transcription are mediated by robust machineries that compete for the same regions of the genome during S phase of the cell cycle. Studies in yeast and mammalian cells have shown that replication-transcription encounters are unavoidable and represent one of the major sources of DNA breakage and chromosomal rearrangements, particularly in cells subjected to replication stress (Azvolinsky et al., 2009; Barlow et al., 2013; Helmrich et al., 2013; Jones et al., 2013; Wilson et al., 2015). A correlation between replication stress–provoked genomic instability and active transcription is particularly apparent in case of common fragile sites (CFSs) and recently identified early replicating fragile sites (ERFSs; Helmrich et al., 2011; Barlow et al., 2013). CFSs are specific genomic regions that manifest as gaps or breaks on metaphase chromosomes, particularly when DNA replication is partially inhibited (Durkin and Glover, 2007). Interestingly, CFSs are frequently located within the coding region of very long genes whose transcription takes even more than one cell cycle, making replication-transcription collisions inevitable (Helmrich et al., 2011). In contrast to late replicating CFSs, ERFSs are located within early replicating regions that contain clusters of highly transcribed genes (Barlow et al., 2013). ERFSs break spontaneously during replication, but their fragility is significantly increased by exogenously induced replication arrest in early S phase (Barlow et al., 2013). ERFS fragility is also dependent on the level of transcription activity at these loci, suggesting that it is driven by replication-transcription encounters (Barlow et al., 2013).
Despite accumulating evidence that conflicts between replication and transcription are frequent events in proliferating cells and have detrimental effects on genome integrity, little is known about the molecular mechanisms underlying their resolution. In fission yeast, the progression of replication forks through actively transcribed genes depends on DNA helicase Pfh1, suggesting a general role for accessory helicases in the displacement of transcription complexes at sites of replication-transcription collisions (Sabouri et al., 2012). However, studies in budding yeast have shown that RNA-polymerase (RNAP) II mutants defective in transcription elongation impair replication fork progression and cause genomic instability, suggesting that RNAPII transcription complex might actively participate in the resolution of replication-transcription conflicts (Felipe-Abrio et al., 2015).
Human RECQ5 belongs to the RecQ family of DNA helicases (Croteau et al., 2014). RECQ5 is known to associate with RNAPII during transcription elongation (Izumikawa et al., 2008; Kanagaraj et al., 2010). It also localizes to DNA replication foci throughout S phase and interacts physically with the proliferating cell nuclear antigen (PCNA), a key component of the replisome (Kanagaraj et al., 2006). A recent study shows that RECQ5 controls the movement of RNAPII across genes to prevent it from pausing or arrest, a condition referred to as transcription stress (Saponaro et al., 2014). RECQ5 depletion results in transcription-dependent chromosome fragmentation during S phase and accumulation of chromosomal rearrangements with the breakpoints located in genes and CFSs (Li et al., 2011; Saponaro et al., 2014). Although the incidents of genome instability in RECQ5-depleted cells colocalize with the areas of elevated transcription stress (Saponaro et al., 2014), it is unclear whether RECQ5 operates directly at sites of interference between replication and transcription.
Here, we demonstrate that RECQ5 associates with transcription complexes in DNA replication foci and counteracts replication fork stalling in RNAPI- and RNAPII-transcribed genes. We present evidence for a novel molecular mechanism involved in the resolution of replication-transcription collisions wherein RECQ5 promotes RAD18-dependent PCNA ubiquitination by directly interacting with PCNA, and the helicase activity of RECQ5 promotes the processing of replication intermediates protected by BRCA1-dependent RAD51 filaments.
Results
RECQ5 associates with RNAPI transcription complexes
Previous studies have suggested that RECQ5 acts as an elongation factor of the RNAPII transcription machinery (Saponaro et al., 2014). To assess whether RECQ5 is also involved in RNAPI transcription, we tested by chromatin immunoprecipitation (ChIP) whether RECQ5 associates with rDNA. Chromatin prepared from asynchronously growing HEK293 cells was precipitated with antibodies against RECQ5 or the largest catalytic subunit of RNAPI, RPA194. Immunoprecipitated DNA was subjected to quantitative real-time PCR (qPCR) analysis using primer pairs covering the entire rDNA repeat unit: (a) the promoter region (amplicon H42); (b) the transcription start site (TSS); (c) the pre-rRNA coding region (H0.4-H13); and (d) the intergenic spacer (IGS; H18 and H27; Fig. 1 A). We found that RECQ5 was significantly enriched on the pre-rRNA coding region, showing a distribution pattern similar to that of RPA194 (Fig. 1 B). Moreover, inhibition of transcription by actinomycin D (ActD) resulted in the accumulation of RPA194 and RECQ5 at the pre-rRNA TSS (Fig. 1 B).
Figure 1.
RECQ5 associates with RNAPI during transcription elongation. (A) Scheme of the human rDNA repeat unit showing location of the amplicons (in red) used in this study. ETS, external transcribed spacer; IGS, intergenic spacer. (B) Distribution of RECQ5 and RNA polymerase I (RPA194 subunit) along the rDNA repeat unit in nontreated or ActD-treated (1 µg/ml, 1 h) HEK293 cells determined by ChIP assay. (C) Coimmunoprecipitation of RPA194 with RECQ5 from extracts of HEK293 cells. Where indicated, cell extract was treated with benzonase nuclease to degrade nucleic acids. (D) Interaction of RPA194 or RPB1 (the largest catalytic subunit of RNAPII) with wild-type and mutant forms of GFP-RECQ5 expressed ectopically in HEK293 cells. Cell extracts were subjected to IP with GFP-Trap_A beads. Bound proteins were analyzed by WB. The RECQ5 variants were visualized by Ponceau S staining. IIo, hyperphosphorylated form of RPB1; IIa, hypophosphorylated form of RPB1. (E) Binding of GFP-RECQ5 and GFP-RECQ5ΔCt to rDNA in HEK293 cells determined by ChIP assay using anti-GFP antibody. Expression of GFP-RECQ5 and GFP-RECQ5ΔCt was confirmed by WB (right). (F) RPA194 density along rDNA repeat unit in mock- or RECQ5-depleted HEK293 cells determined by ChIP assay. RECQ5 down-regulation (using siRECQ5 #1) was confirmed by WB (right). For B, E, and F, data are represented as mean ± SD.
To determine whether RECQ5 interacts with the components of RNAPI transcription complex, a HEK293 cell extract was subjected to immunoprecipitation (IP) with anti-RECQ5 antibody, and the immunoprecipitated material was tested for the presence of RPA194. To exclude RNA- or DNA-mediated interactions, the extract was treated with benzonase nuclease. We found that RPA194 coprecipitated with anti-RECQ5 antibody but not with control IgG, suggesting that RECQ5 forms a complex with RNAPI (Fig. 1 C). RECQ5-RNAPI interaction was confirmed by coimmunoprecipitation of RECQ5 with ectopically expressed RPA43-GFP (Fig. S1 A).
To determine which region of RECQ5 is required for its interaction with RNAPI, different C-terminally truncated variants of GFP-tagged RECQ5 were overexpressed in HEK293 cells and tested for RPA194 binding by IP (Fig. S1, B and C). The results showed that the interaction between RECQ5 and RPA194 was abolished by deletion of the last 61 C-terminal amino acids of the RECQ5 polypeptide (Fig. 1 D and Fig. S1, B and C). It should be noted that this RECQ5 variant (RECQ5ΔCt) also lacks a part of the Set2-Rpb1-interacting (SRI) domain, which binds to the hyperphosphorylated C-terminal repeat domain (CTD) of RNAPII and mediates the association of RECQ5 with RNAPII-transcribed genes (Kanagaraj et al., 2010). Accordingly, RECQ5ΔCt showed reduced binding to the hyperphosphorylated form of RNAPII (IIo), whereas its binding to the hypophosphorylated form of RNAPII (IIa), mediated by the internal RNAPII–interacting (IRI) domain, was similar to that of wild-type RECQ5 (Kanagaraj et al., 2010; Fig. 1 D). Importantly, a RECQ5 variant lacking the IRI and SRI domains (GFP-RECQ5ΔIRIΔSRI), which failed to bind RNAPII, still retained RPA194 binding activity (Fig. 1 D), suggesting that the RNAPI-interacting domain of RECQ5 is located adjacent to the SRI domain.
Finally, by ChIP assay using anti-GFP antibody, we compared the binding of ectopically expressed GFP-RECQ5 and GFP-RECQ5ΔCt to rDNA in HEK293 cells. The results showed that the C-terminal region of RECQ5 is required for the efficient binding of RECQ5 to the pre-rRNA coding region of the rDNA repeat unit (Fig. 1 E). Collectively, these results provide evidence that RECQ5 binds to RNAPI during rDNA transcription.
RECQ5 depletion increases RNAPI density in the pre-rRNA coding region
To explore the possible involvement of RECQ5 in processes associated with RNAPI-directed transcription, we analyzed the effect of RECQ5 depletion on RPA194 density along rDNA repeat units in HEK293 cells. Interestingly, we observed a significant enrichment of RNAPI in the pre-rRNA coding region, but not on the promoter, in cells lacking RECQ5 (Fig. 1 F). These cells did not show any alteration in the cell cycle profile compared with mock-depleted cells (Fig. S1 D), excluding the possibility that the observed increase in RNAPI occupancy on rDNA was caused by an increase in the proportion of S/G2 cells, where RNAPI-directed transcription reaches its highest level (Grummt, 2003). These data suggest that RECQ5 might counteract RNAPI transcription stalling.
RECQ5 depletion leads to amplification of DNA segments within the pre-rRNA coding region in cells exposed to replication stress
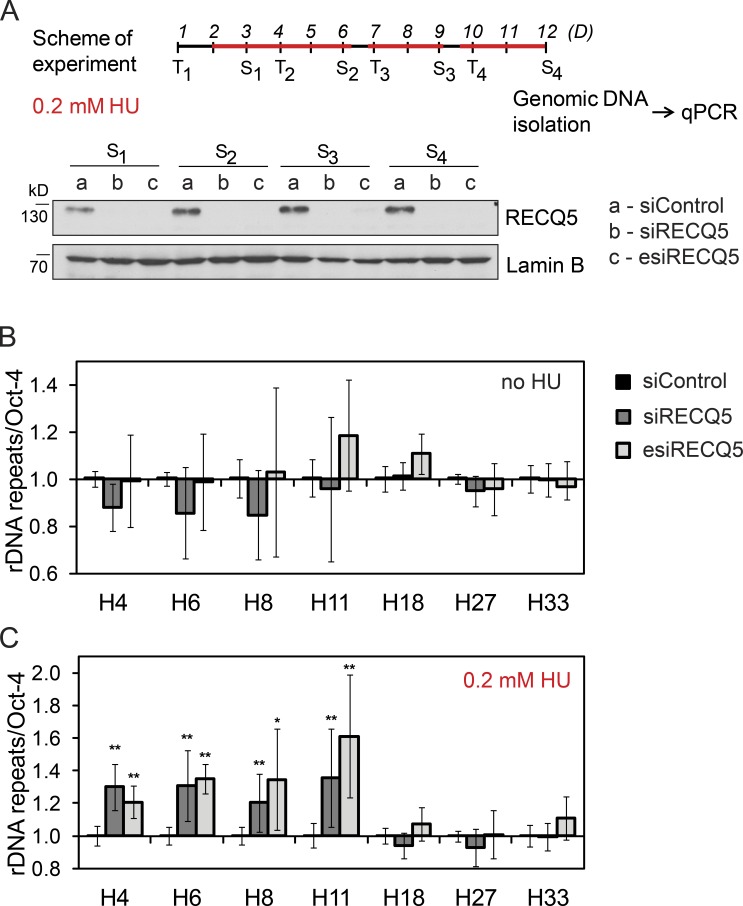
To determine whether RECQ5 is required for the stability of rDNA arrays, it was kept knocked down in HEK293 cells for 12 d by successive siRNA transfections (Fig. 2 A). Subsequently, genomic DNA was isolated, and rDNA copy numbers at selected amplicons were measured by qPCR relative to Oct-4 gene, which is not transcribed in HEK293 cells (Nejepinska et al., 2012). To assess whether RECQ5 prevents rDNA recombination arising as a consequence of interference between transcription and replication, cells were also exposed to mild replication stress generated by 0.2 mM hydroxyurea (HU; Fig. 2 A). We found that RECQ5 depletion caused DNA copy number variations, particularly within the pre-rRNA coding region of the rDNA repeat unit (Fig. 2 B). Importantly, upon HU treatment, a significant amplification of DNA sequences within the pre-rRNA coding region was observed in RECQ5-depleted cells, but not in mock-depleted cells (Fig. 2 C). Such structural changes of rDNA likely arose as the consequence of DNA double-strand breaks generated within the pre-rRNA coding region by clashes between transcription and replication complexes, suggesting an important role for RECQ5 in preventing rDNA instability arising from interference between replication and transcription.
Figure 2.
Ribosomal DNA instability in RECQ5-depleted cells. (A) Experimental scheme. RECQ5 was kept down-regulated in HEK293 cells for 12 d by successive transfections (T) of either siRECQ5#1 or esiRECQ5. Samples of cells were collected after each splitting (S) and analyzed by WB (bottom). Lamin B was used as loading control. Cells were treated with 0.2 mM HU at indicated time intervals (red lines) or left untreated. (B and C) Effect of RECQ5 depletion on rDNA stability in the absence or presence of 0.2 mM HU. The rDNA copy number at indicated amplicons (see Fig. 1 A) was measured by qPCR of genomic DNA (∼2 ng) relative to the Oct-4 gene. Data are represented as mean ± SD. *, P < 0.0005; **, P < 0.00005 (two-tailed unpaired t test).
Depletion of RECQ5 causes replisome stalling in actively transcribed genes
To explore the hypothesis that RECQ5 is involved in the resolution of collisions between replication and transcription machineries, we investigated whether RECQ5 depletion leads to replication fork stalling at rDNA arrays. To this end, we measured DNA polymerase ε (Pol ε) occupancy along the rDNA repeat unit in mock- and RECQ5-depleted HEK293 cells released synchronously from a nocodazole (NC) block. The characteristic feature of rDNA arrays is that the actively transcribed rDNA repeats are replicated in early S phase, whereas the silent repeats are replicated in late S phase (Li et al., 2005). Therefore, we first determined rDNA replication timing in our experimental setup. By IP of BrdU pulse-labeled DNA followed by PCR analysis, we found that the first wave of rDNA replication occurred 6–9 h after NC block release, whereas the late-replicating rDNA was detected 12–16 h after release (Fig. 3 A). Hence, mock- and RECQ5-depleted cells were cross-linked at 8 or 14 h after NC block release and subjected to ChIP assay using antibody against the catalytic subunit of Pol ε. We found that RECQ5 depletion was associated with a significant enrichment of Pol ε on rDNA 8 h after NC block release (Fig. 3, B–D), when the replication of actively transcribed repeats occurred (Fig. 3 A). In contrast, the Pol ε occupancy profile at the late-replicating portion of rDNA (14 h after NC block release) was not affected by RECQ5 knockdown (Fig. 3 D). This suggests that RECQ5 counteracts replication fork stalling in transcriptionally active rDNA repeats during early S phase.
Figure 3.
RECQ5 depletion leads to replication fork stalling in actively transcribed genes. (A) rDNA replication timing in HEK293 cells. Cells were released from NC block and pulse labeled with BrdU for 30 min before harvesting at the indicated time points. BrdU-labeled DNA was immunoprecipitated and analyzed by PCR using primer pair for amplicon H6. (B) Cell cycle distribution of HEK293 cells released from NC block for indicated periods of time in the presence or absence of 0.2 mM HU. (C) Western blot analysis of RECQ5 protein levels in chromatin (10 µg) used for ChIP assays. (D) DNA Pol ε occupancy on rDNA repeat unit in HEK293 cells transfected with either control siRNA (siControl) or RECQ5 siRNA (siRECQ5#1). Chromatin was isolated at indicated time points after the release from NC block and subjected to ChIP assay. (E) As in D, but cells were released from NC block into medium containing 0.2 mM HU. (F) Effect of RECQ5 knockdown on Pol ε occupancy on the indicated RNAPII-transcribed genes (RPS19, RPL22, and ACTG1). Chromatin for ChIP assay was prepared as in E. ATCG1-I represents a nontranscribed intergenic region located ∼2 kb downstream of ACTG1 stop codon. For D, E, and F, data are represented as mean ± SD.
We also examined the effect of RECQ5 depletion on replication fork progression through rDNA in cells exposed to mild replication stress. Mock- and RECQ5-depleted HEK293 cells were released from NC block in medium containing 0.2 mM HU, and the level of Pol ε binding to rDNA was analyzed 8 or 14 h after NC block release. As expected, HU treatment slowed down cell cycle progression, with cells being in early S phase at 14 h after release from the NC block (Fig. 3 B). Importantly, we observed that RECQ5 knockdown caused a gradual accumulation of Pol ε on the rDNA repeat unit over the analyzed time period (Fig. 3 E).
Finally, we investigated whether RECQ5 depletion led to replication fork stalling in RNAPII-transcribed genes. We used chromatin prepared from HEK293 cells exposed to 0.2 mM HU to analyze Pol ε occupancy on the coding regions of several constitutively transcribed genes. We chose the ACTG1 gene, which was previously shown to be occupied by RECQ5 in a RNAPII-dependent manner (Kanagaraj et al., 2010), and the genes encoding the ribosomal proteins RPS19 and RPL22. We found that RECQ5 depletion resulted in a significant enrichment of Pol ε on the coding regions of all three genes at 14 h after NC block release (Fig. 3 F). Importantly, no significant enrichment of Pol ε was observed at amplicon ACTG1-I, which represents an intergenic region located ∼2 kb downstream of the ACTG1 transcription termination site (Fig. 3 F). Collectively, these results suggest that RECQ5 counteracts replication fork stalling in actively transcribed genes, particularly under conditions of replication stress.
RECQ5 associates with transcription machinery in replication foci
RECQ5 was previously shown to accumulate in DNA replication foci (Kanagaraj et al., 2006). To identify the core complex bound by RECQ5 in these foci, we used the FRAP technique to measure the intranuclear mobility of GFP-tagged RECQ5 before and after the blockage of transcription or replication. HEK293 cells were transfected with vectors expressing GFP-tagged RECQ5 and RFP-tagged PCNA and brought to early S phase by the release from NC block. Live cell imaging revealed that GFP-RECQ5 formed discrete nuclear foci that largely colocalized with RFP-PCNA foci (Fig. 4 A, left). FRAP measurements of GFP-RECQ5 in circular regions containing one to three replication foci showed a very high mobility of RECQ5, in both the replication foci and the surrounding area (Fig. 4, right). In contrast, PCNA is characterized by a very slow fluorescence recovery, on the order of minutes, indicating a stable association with replication forks (Sporbert et al., 2002; unpublished data). Thus, the observed fast fluorescence recovery of GFP-RECQ5 in replication foci suggests that RECQ5 is not stably bound to the replication machinery. This was also supported by our observation that GFP-RECQ5 mobility in replication foci was not affected by aphidicolin (Aph), which inhibits DNA replication (Fig. 4). In contrast, inhibition of transcription by ActD dramatically impaired the mobility of GFP-RECQ5 in replication foci (Fig. 4), whereas no change in the mobility of GFP-RECQ5 outside the replication foci was observed (Fig. 4, magnified sections). These findings suggest that RECQ5 associates with transcription machinery in replication foci.
Figure 4.
RECQ5 associates with transcription complexes in replication foci. (A, left) GFP-RECQ5 foci selected for FRAP analysis colocalize with PCNA-RFP foci in mock-, aphidicolin-, and ActD-treated HEK293 cells. (right) Representative images from a FRAP sequence showing the fluorescence recovery of GFP-RECQ5 in replication foci under indicated conditions. Bleached regions are depicted in white circles and also separately magnified for ActD-treated cells. FRAP measurements of GFP-RECQ5 were performed 6–9 h after release of cells from NC block. (B) Graph showing mean ± SD (n = 8–12) of relative fluorescence recovery in the bleached regions as a function of time.
BRCA1 and RAD51 function at sites of replication-transcription interference
To gain insight into the role of RECQ5 in the resolution of conflicts between replication and transcription, we explored its functional relationship with proteins that are involved in the repair of damaged replication forks. BRCA1, an important tumor suppressor, has been implicated in the stabilization and processing of stalled replication forks, and it is known to be recruited to ERFSs both after HU-induced replication arrest and during normal replication (Schlacher et al., 2012; Barlow et al., 2013; Willis et al., 2014). BRCA1 forms discrete nuclear foci in unperturbed S-phase cells (Scully et al., 1997). Interestingly, we found that the formation of BRCA1 foci in U2OS cells was impaired by cordycepin, which causes premature termination of transcription (Fig. 5, A and B; and Fig. S2 A), thus lowering the probability of replication-transcription clashes (Jones et al., 2013). In contrast, the number of BRCA1 foci increased if cells were exposed to ActD, which is capable of arresting transcription complexes on DNA, thereby creating a barrier to replication fork movement (Fig. 5, A–C; and Fig. S2 A). In agreement with this notion, RNAPI transcription complexes arrested on DNA by ActD treatment (Fig. 1 B) caused a significant increase in Pol ε occupancy on the rDNA repeat unit (Fig. S3 A). Neither of the transcription inhibitors used altered the percentage of S-phase cells (Fig. S2 B). Importantly, inhibition of DNA replication by Aph or HU significantly decreased the number of BRCA1 foci in both mock- and ActD-treated cells (Fig. 5 C and Fig. S2, C and D), suggesting that the formation of these foci is driven by interference between replication and transcription. To further characterize the nature of BRCA1 foci, we performed FRAP experiments to measure GFP-BRCA1 mobility in these sites before and after replication or transcription arrest. We found that BRCA1 mobility in cells treated with Aph (replication arrest) was similar to that measured in mock-treated cells (Figs. 5 D and S2 E). In contrast, BRCA1 mobility was dramatically reduced in cells treated with ActD (transcription arrest; Figs. 5 D and S2 E). These data suggest that BRCA1 localized in S-phase foci associates with the transcription machinery at sites of replication-transcription interference.
Figure 5.
RECQ5 depletion leads to accumulation of unresolved replication intermediates at sites of replication-transcription interference. (A) Representative images of mock- and RECQ5-depleted (siRECQ5#2) U2OS cells stained for BRCA1 and EdU. Nuclei were stained with DAPI. EdU (20 µM) was added to cell cultures 1 h before fixation. Where indicated, cells were treated with cordycepin (50 µM) for 2 h or ActD (1 µg/ml) for 1 h before fixation. (B, left) Quantification of BRCA1 foci counts in EdU-positive nuclei of mock- or RECQ5-depleted U2OS cells represented in A. (right) Western blot analysis of U2OS cells transfected with indicated siRNAs. (C) Analysis of BRCA1 foci counts in EdU-positive nuclei of U2OS cells exposed to inhibitors of replication (5 µM Aph or 2 mM HU) for 90 min and/or transcription inhibitor (1 µg/ml ActD) for 1 h before fixation. Note that EdU was added 1 h before addition of Aph or HU. (D) Graph showing time dependency of relative fluorescence recovery for bleached nuclear regions containing GFP-BRCA1 foci in mock-, Aph-, and ActD-treated HEK293 cells. HEK293 cells were transfected with GFP-BRCA1 and PCNA-RFP (to identify cells in S phase) constructs, and FRAP measurements were performed 6–9 h after the release of cells from NC block. (E) Representative images of mock-, BRCA1-, RECQ5-, and BRCA1/RECQ5-depleted U2OS cells stained for RAD51 and EdU. Nuclei were stained with DAPI. (F) Analysis of RAD51 foci counts in EdU-positive nuclei of cells treated with 50 µM cordycepin for 2 h, or as in C. (G) Effect of RECQ5 depletion on the frequency of BRCA1-positive and BRCA1-negative RAD51 foci in U2OS cells. (H) Analysis of RAD51 foci counts in EdU-positive nuclei of RECQ5-proficient and RECQ5-deficient cells treated or not with the Rad51 inhibitor B02 (20 µM) for 6 h. (I) Representative images of mock- or RECQ5-depleted U2OS cells analyzed by alkaline BrdU comet assay. U2OS cells were pulse labeled with 20 µM BrdU for 20 min and harvested at the indicated time points after the BrdU wash-off to perform alkaline comet assay followed by immunostaining of BrdU-labeled DNA and staining with SYBR Gold. (J and K) Statistical analysis of BrdU-positive comet tail lengths measured for mock-, RECQ5-, and BRCA1-depleted U2OS cells. Whiskers indicate 10th to 90th percentile. ns, not significant; *, P < 0.01; ***, P < 0.0001 (two-tailed unpaired t test). Data in B–D and F–H are represented as mean ± SD.
BRCA1 acts to form RAD51 filaments at arrested replication forks to protect them from nucleolytic degradation (Schlacher et al., 2012). Like BRCA1, RAD51 forms discrete nuclear foci in unperturbed S-phase cells, and the formation of these foci is strictly dependent on the presence of BRCA1 (Scully et al., 1997; Fig. 5 E). Of note, RAD51 foci colocalize with BRCA1 foci only partially, suggesting that BRCA1 dissociates from these sites upon RAD51 recruitment (Scully et al., 1997; Fig. S2 F). In agreement with the immobilization of BRCA1 upon ActD treatment, the formation of RAD51 foci in S-phase cells was strongly attenuated by ActD (Fig. 5 F and Fig. S2, C and G), which further confirmed that BRCA1 activity is dependent on active transcription. RAD51 foci formation was also impaired in cells treated with the transcription terminator cordycepin or replication inhibitors HU and Aph (Fig. 5 F and Fig. S2, C and G). Combined treatment of cells with ActD and either HU or Aph further decreased the number of RAD51 foci per cell (Fig. 5 F and Fig. S2, C and G), suggesting that the formation of these foci is driven by replication-transcription interference.
Depletion of RECQ5 causes accumulation of unresolved replication intermediates
Next, we analyzed the frequency of BRCA1 foci in cells depleted of RECQ5. Interestingly, RECQ5 depletion led to a reduction in the frequency of BRCA1 foci in unperturbed cells but not in ActD-treated cells (Fig. 5, A and B; and Fig. S2 A). These results suggest that RECQ5 is not required for BRCA1 foci formation, but rather affects late steps of the process initiated by BRCA1 at sites of replication-transcription interference. Thus, we investigated the effect of RECQ5 depletion on the frequency of RAD51 foci in S-phase cells. We found that in RECQ5-deficient cells, the number of RAD51 foci that did not colocalize with a BRCA1 focus was increased compared with RECQ5-proficient cells, whereas the number of colocalizing foci was not affected (Fig. 5 G). Interestingly, the RAD51 inhibitor B02, which strongly inhibited RAD51 foci formation in mock-depleted cells, did not significantly reduce RAD51 foci in RECQ5-depleted cells (Fig. 5 H). This suggests that in the absence of RECQ5, cells accumulate replication intermediates stabilized by RAD51.
To further explore this hypothesis, we monitored replication fork elongation in RECQ5-proficient and RECQ5-deficient cells by alkaline BrdU comet assay (Mórocz et al., 2013). In this assay, the stretches of newly synthesized DNA pulse labeled with BrdU are separated from the parental strand by alkaline denaturation and electrophoresis and are visualized as a comet tail by immunostaining with anti-BrdU antibody (Fig. 5 I). If DNA replication continues after BrdU pulse labeling, the nascent DNA strands become continuous and remain in the comet head (Fig. 5 I). In our experiments, we measured comet tail lengths at time points of 0, 2, and 4 h after the BrdU pulse. We found that at 4 h after BrdU labeling, RECQ5-depleted cells showed substantially longer comet tails than mock-depleted cells, confirming that RECQ5 deficiency is associated with accumulation of unresolved replication intermediates (Fig. 5, I and J). Similarly, BRCA1-depleted cells also displayed longer comet tail lengths compared with mock-depleted cells (Fig. 5 K). Importantly, cells depleted for both RECQ5 and BRCA1 did not show a further increase in comet tail length as compared with cells depleted for either protein (Fig. 5 K). Collectively, the data described point to a coordinated action of RECQ5 and BRCA1/RAD51 at sites of replication-transcription interference to promote replication fork recovery.
RECQ5 is required for RAD18-dependent PCNA ubiquitination at sites of replication-transcription interference
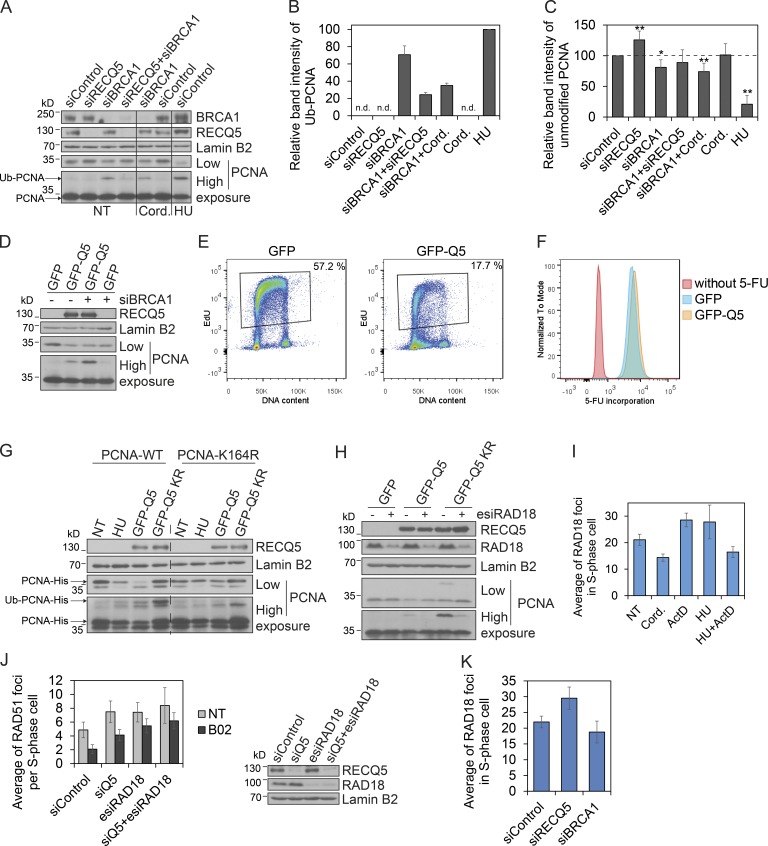
Studies in budding yeast have shown that PCNA sliding clamp is unloaded from the lagging strand arm of stalled replication forks in a manner dependent on its ubiquitination at lysine 164 (Yu et al., 2014). To explore the possible involvement of RECQ5 and BRCA1 in replisome remodeling during the resolution of conflicts between replication and transcription machineries, we examined effects of their depletion on the level of ubiquitinated PCNA (Ub-PCNA) in chromatin fraction of unperturbed HEK293 cells. Ub-PCNA was monitored by Western blotting (WB) as a form of PCNA with reduced electrophoretic mobility that is readily detected in chromatin fraction of cells exposed to HU (Fig. 6, A and B). Depletion of BRCA1, but not that of RECQ5, substantially increased the level of Ub-PCNA in chromatin (Fig. 6, A and B). This PCNA ubiquitination was largely alleviated by cordycepin, suggesting that it arises as a consequence of interference between replication and transcription (Fig. 6, A and B). Importantly, almost no Ub-PCNA was detected in chromatin of cells depleted for both BRCA1 and RECQ5, suggesting that Ub-PCNA formation in the absence of BRCA1 depends on RECQ5 (Fig. 6, A and B). Moreover, depletion of RECQ5 alone increased the level of unmodified PCNA in chromatin, suggesting that RECQ5 might promote PCNA unloading (Fig. 6, A [low exposure] and C).
Figure 6.
RECQ5 promotes RAD18-dependent PCNA ubiquitination at sites of replication-transcription interference. (A) Western blot analysis of PCNA, RECQ5, BRCA1, and Lamin B2 (loading control) levels in chromatin fractions isolated from HEK293 cells transfected with indicated siRNAs. Where indicated, cells were treated with 2 mM HU for 14 h or 50 µM cordycepin (Cord.) for the last 2 h. NT, nontreated. For unmodified PCNA, high and low exposures are shown. (B and C) Quantitative analysis of band intensity of ubiquitinated PCNA (Ub-PCNA) and unmodified PCNA, respectively, on Western blots represented in A. PCNA intensity was normalized to Lamin B2 intensity. Data are expressed as percentage of Ub-PCNA levels in chromatin of HU-treated cells or as percentage of unmodified PCNA levels in chromatin of mock-depleted cells, respectively. n.d., signal was not detected. *, P < 0.01; **, P < 0.005 (two-tailed unpaired t test). (D) Western blot analysis of PCNA levels in chromatin fractions of mock- or BRCA1-depleted HEK293 cells transiently overexpressing either GFP or GFP-RECQ5. Lamin B2 was used as loading control. (E) Flow cytometry analysis of EdU incorporation (y axis) and DNA content (propidium iodide, x axis) in HEK293 cells performed 48 h after transfection of vectors expressing either GFP or GFP-RECQ5. Only GFP-positive cells were analyzed. (F) Flow cytometry analysis of 5-FU incorporation into nascent RNA in HEK293 cells expressing either GFP or GFP-RECQ5. Mock-treated HEK293 cells (without 5-FU) served as noise-signal control. (G) Western blot analysis of PCNA levels in chromatin fractions isolated from HEK293 cells transiently expressing wild-type (WT) or mutant (K164R) forms of His-tagged PCNA. Cells were treated with 2 mM HU for 14 h or transfected with vectors expressing either wild-type (GFP-Q5) or helicase/ATPase-dead mutant (GFP-Q5 K58R) of RECQ5. (H) Western blot analysis of PCNA levels in chromatin of HEK293 cells transiently overexpressing GFP, GFP-Q5, or GFP-Q5 K58R that were either mock- or RAD18-depleted. (I) Analysis of RAD18 foci counts in EdU-positive nuclei of U2OS cells exposed to transcription inhibitors cordycepin (Cord.; 50 µM for 2 h) and ActD (1 µg/ml for 1 h) and/or replication inhibitor (2 mM HU) for 90 min before fixation. Note that EdU was added 1 h before addition of HU or fixation. (J, left) Analysis of RAD51 foci counts in EdU-positive nuclei of mock-, RECQ5-, and/or RAD18-depleted cells treated or not with the Rad51 inhibitor B02 (20 µM) for 6 h. (right) Western blot analysis of U2OS cells transfected with indicated siRNAs. (K) Analysis of RAD18 foci counts in EdU-positive nuclei of U2OS cells transfected with indicated siRNAs. Data in B, C, and I–K are represented as mean ± SD.
To explore this further, we measured the levels of Ub-PCNA and unmodified PCNA in the chromatin fraction of HEK293 cells overexpressing GFP-RECQ5. We found that GFP-RECQ5 overexpression stimulated PCNA ubiquitination and reduced the levels of unmodified PCNA on chromatin in both mock-depleted and BRCA1-depleted cells (Fig. 6 D). A similar phenotype was seen upon replication arrest induced by HU (Fig. 6 A). Consistently, DNA synthesis in GFP-RECQ5–overexpressing cells was strongly attenuated as revealed by measurement of EdU incorporation (Fig. 6 E). In contrast, overexpression of GFP-RECQ5 did not significantly affect transcription as measured by incorporation of 5-fluorouridine (Fig. 6 F). Interestingly, BRCA1-depleted cells overexpressing GFP-RECQ5 contained significantly higher levels of chromatin-bound Ub-PCNA compared with BRCA1-proficient cells overexpressing GFP-RECQ5 or BRCA1-deficient cells expressing only GFP (Fig. 6 D). This suggests that in the absence of BRCA1, Ub-PCNA generated by a RECQ5-mediated mechanism accumulates on chromatin.
To confirm that RECQ5 promotes PCNA ubiquitination and unloading, we examined the effect of GFP-RECQ5 overexpression on chromatin binding of a His-tagged version of a nonubiquitinable PCNA mutant, PCNA-K164R. We found that overexpression of GFP-RECQ5 did not affect the binding of PCNA-K164R to chromatin, whereas it significantly reduced chromatin binding of wild-type PCNA (Fig. 6 G, low exposure). As expected, binding of PCNA-K164R to chromatin was also not affected upon HU treatment (Fig. 6 G). Interestingly, overexpression of a helicase/ATPase-dead mutant of RECQ5, GFP-RECQ5-K58R (Garcia et al., 2004), dramatically increased the level of Ub-PCNA in chromatin without reducing the level of unmodified PCNA (Fig. 6 G), suggesting a role for the helicase activity of RECQ5 in PCNA unloading.
We also tested the effect of GFP-RECQ5 overexpression on the level of Ub-PCNA in chromatin of cells lacking RAD18, an E3 ubiquitin ligase, which is recruited to stalled replication forks to monoubiquitinate PCNA (Tsuji et al., 2008). We found that depletion of RAD18 impaired PCNA ubiquitination induced by the overexpression of wild-type GFP-RECQ5 or GFP-RECQ5-K58R, suggesting that RECQ5 promotes RAD18-dependent PCNA ubiquitination (Fig. 6 H).
Next, we investigated a relationship between RECQ5 and RAD18 at the interface of replication and transcription. First, we characterized the nature of RAD18 foci in cells exposed to different transcription or replication inhibitors (Fig. S4). The formation of RAD18 foci in S-phase cells was decreased upon cordycepin treatment (Fig. 6 I), which prematurely terminates transcription. Cells exposed to HU or ActD treatment, conditions leading to replication fork blockage, showed an increased number of RAD18 foci compared with untreated cells (Fig. 6 I). Importantly, combined treatment of cells with ActD and HU significantly decreased the number of RAD18 foci per cell (Fig. 6 I), suggesting that RAD18 foci formation is dependent on replication fork stalling caused by collisions with transcription complexes. As in the case of RECQ5, RAD18 depletion increased the number of RAD51 foci in S-phase cells, and these foci also persisted after exposure of cells to B02 (Fig. 6 J). Moreover, codepletion of RAD18 and RECQ5 did not cause a further increase in the number of RAD51 foci in comparison with depletion of either protein (Fig. 6 J), suggesting that RAD18 and RECQ5 act in the same pathway to resolve replication intermediates. Importantly, RECQ5 depletion caused accumulation of RAD18 foci in S-phase cells, implying that RECQ5 is not involved in the recruitment of RAD18 to stalled forks (Fig. 6 K). Moreover, BRCA1 depletion, which causes replication stalling, did not significantly change the number of RAD18 foci in S-phase cells (Fig. 6 K), suggesting that the increased frequency of RAD18 foci in RECQ5-deficient cells is a consequence of a defect in PCNA ubiquitination, leading to persistence of RAD18 at sites of stalled replication forks.
Helicase and PCNA-interacting domains of RECQ5 have distinct roles in the resolution of replication intermediates
Next, we wanted to identify the domains of RECQ5 involved in PCNA ubiquitination. For this, we prepared GFP-RECQ5 variants containing the K58R mutation in the helicase domain (its overexpression caused accumulation of Ub-PCNA on chromatin; Fig. 6, G and H) in combination with either a R943A mutation in the SRI domain (abolishment of RECQ5 binding to RNAPII CTD) or mutations in the PCNA-interacting peptide (PIP4A, abolishment of RECQ5–PCNA interaction). We tested the effect of overexpression of these RECQ5 variants on the level of chromatin-bound PCNA in HEK293 cells. We found that the K58R/PIP4A variant largely failed to induce PCNA ubiquitination, whereas K58R/R943A stimulated PCNA ubiquitination, although to a lesser extent compared with the K58R mutant (Fig. 7 A). These data suggest that RECQ5 promotes PCNA ubiquitination by directly interacting with PCNA via the PIP motif.
Figure 7.
Distinct roles of the helicase and PCNA-interacting domains of RECQ5 in resolution of replication intermediates. (A) Western blot analysis of PCNA levels in chromatin of HEK293 cells transiently overexpressing various mutants of GFP-RECQ5 with defects in the helicase activity (K58R), RNAPIIo binding (R943A), and PCNA-interacting motif (PIP4A). (B) Western blot analysis of chromatin fraction of U2OS T-REx cells with doxycycline-regulated expression of Flag-tagged (shRNA-resistant) wild-type RECQ5 (WT), RECQ5-K58R (K58R), RECQ5-R934A (R934A), or RECQ5-PIP4A (PIP4A). Endogenous RECQ5 in these cells is down-regulated by shRNA. Parental U2OS T-REx cells were transfected with siControl or siRECQ5. (C) Western blot analysis of whole cell extracts of U2OS T-REx cells in B. (D and E) Analysis of RAD51 or RAD18 foci counts in EdU-positive nuclei of U2OS T-REx cells as in B. Data in D and E are represented as mean ± SD.
To investigate the phenotypes of RECQ5 mutants if expressed at levels comparable to that of endogenous RECQ5 protein, we established a cellular system that uses a pAIO-based vector (Ghodgaonkar et al., 2014) for inducible replacement of the endogenous RECQ5 with an shRNA-resistant mutant of interest. We prepared stable U2OS T-REx cell lines inducibly expressing Flag-tagged versions of wild-type RECQ5, RECQ5-K58R, RECQ5-R943A, and RECQ5-PIP4A, along with an shRNA targeting endogenous RECQ5. Thus, cells exposed to doxycycline (0.4 ng/ml) expressed a RECQ5 variant of interest with concomitant down-regulation of endogenous RECQ5 (Fig. S5). To detect Ub-PCNA in unchallenged cells, we used an antibody specifically recognizing ubiquityl-PCNA (Lys 164). We found that the substitution of endogenous RECQ5 with RECQ5-K58R caused accumulation of both Ub-PCNA and unmodified PCNA on chromatin (Fig. 7 B). Moreover, the lack of RECQ5 helicase activity increased the number of RAD51 foci in S-phase cells (Fig. 7 D). These results suggest that the helicase activity of RECQ5 prevents the accumulation of unresolved replication intermediates. On the contrary, the substitution of endogenous RECQ5 with RECQ5-PIP4A did not cause accumulation of unmodified PCNA nor Ub-PCNA on chromatin (Fig. 7 B). Instead, it increased the level of RAD18 in chromatin fraction and the number of RAD18 foci in S-phase cells (Fig. 7, B and E), which is consistent with our earlier finding of the requirement of RECQ5–PCNA interaction for PCNA ubiquitination. Interestingly, although the expression levels of all RECQ5 variants were comparable (Fig. 7 C), the levels of RECQ5-K58R and RECQ5-PIP4A in chromatin were significantly increased compared with that of wild-type RECQ5 (Fig. 7 B), providing a further evidence for a defect in their function. Together, these results suggest that a coordinated action of the helicase and PCNA-interacting domains of RECQ5 promotes the resolution of replication-transcription encounters.
Discussion
RECQ5 DNA helicase is essential for maintenance of genomic stability, but its exact molecular functions remain unclear. Recent studies have shown that human RECQ5 binds to RNAPII during transcription elongation and maintains genomic stability at RNAPII-transcribed genes by acting as a factor that prevents transcription pausing or arrest, a condition termed transcription stress (Kanagaraj et al., 2010; Li et al., 2011; Saponaro et al., 2014). Here, we show that RECQ5 also forms a complex with RNAPI during rDNA transcription, prevents RNAPI-transcription stress, and enforces the stability of the pre-rRNA coding regions of rDNA arrays. These results are consistent with the proposal of RECQ5 acting as a general transcription elongation factor that is important for preserving genome stability during transcription (Kanagaraj et al., 2010; Li et al., 2011; Saponaro et al., 2014). However, we also demonstrate that RECQ5 depletion causes the persistence of unresolved replication intermediates with replisomes stalled in both RNAPI- and RNAPII-transcribed genes. These findings suggest that the genome stabilization effect of RECQ5 at sites of transcription might reflect a role for RECQ5 in resolving collisions between the replication and transcription machineries. In support of this hypothesis, we have found that RECQ5 associates with active transcription complexes in DNA replication foci, suggesting that it acts at sites of concomitant replication and transcription. Moreover, our data show that BRCA1, RAD51, and RAD18, proteins involved in the stabilization and restart of stalled forks, form foci at sites of replication-transcription interference, where they cooperate with RECQ5 to resume replication. Finally, we provide evidence that RECQ5 promotes RAD18-dependent ubiquitination of PCNA by directly interacting with PCNA and that the helicase activity of RECQ5 promotes the resolution of replication intermediates stabilized by RAD51 filaments upon replication-transcription encounters.
Mechanisms that cells evolved to resolve conflicts between replication and transcription remain elusive. Studies in bacteria and yeast have shown that specific helicases act in conjunction with the replisome to disrupt transcription complexes and other obstacles that impair replication fork progression (Azvolinsky et al., 2009; Boubakri et al., 2010; Sabouri et al., 2012). However, on very long genes in mammalian cells, collisions between transcription and replication complexes occur within each round of transcription, because the synthesis of the full-length transcript of these genes takes more than one cell cycle (Helmrich et al., 2013). Therefore, to ensure proper gene expression, cells must have mechanisms that permit RNA chain elongation after the collision with replication fork. Here, we provide evidence that RECQ5, BRCA1, and RAD18 are recruited independently of each other to sites of replication-transcription collisions. BRCA1-dependent loading of RAD51 stabilizes the stalled replication forks for replication resumption (Schlacher et al., 2012). RECQ5 promotes RAD18-dependent PCNA ubiquitination and unloading from replication forks upon their collision with active transcription. It is therefore likely that RECQ5-mediated replisome remodeling at stalled forks might allow the passage of oncoming transcription complexes across the fork to complete RNA synthesis (Fig. 8 A). There is accumulating evidence suggesting a role for active transcription in the resolution of replication-transcription collisions. It was shown that RNA polymerase translocation is required for the resolution of head-on collisions between the transcription machinery and bacteriophage Φ29 DNA polymerase in Bacillus subtilis (Elías-Arnanz and Salas, 1999). Similarly, studies in budding yeast have shown that RNAPII mutants with a defect in transcription elongation impair replication fork progression and cause genomic instability (Felipe-Abrio et al., 2015). In addition, we have shown in human cells that halted RNAPI transcription complexes prevented the movement of the replisome through rDNA (Fig. S3 A). Moreover, we provide evidence that replication forks stalled by halted transcription complexes could be elongated once the RNA polymerase is allowed to resume transcription (Fig. S3 B). Thus, RNA polymerase might actively participate in the resolution of replication-transcription collisions.
Figure 8.
Model for resolution of conflicts between replication and transcription. (A) Scheme of the model. RECQ5, BRCA1, and RAD18 (R18) are recruited independently of each other to sites of replication-transcription collisions. BRCA1-dependent loading of RAD51 on stalled replication forks, which depends on active transcription, leads to fork stabilization. RECQ5 promotes RAD18-dependent PCNA ubiquitination and unloading at sites of replication-transcription interference that might allow the passage of oncoming transcription complexes across the fork to complete RNA synthesis. (B) Consequences of BRCA1 and RECQ5/RAD18 deficiencies. In the absence of BRCA1, RECQ5 can mediate PCNA ubiquitination and unloading, but the replication fork fails to restart because of the defect in RAD51 loading. In the absence of RECQ5 or RAD18, BRCA1 can promote assembly of RAD51 filaments to protect stalled replication forks, but RNA polymerase cannot translocate across the replication fork, and hence replication restart is prevented. Failure of either of these activities would lead to persistence of stalled replication forks, resulting in genomic instability.
Studies in budding yeast revealed the presence of a Fob1-dependent replication fork barrier at the 3′-end of the rDNA transcription unit that prevents head on clashes of the replisome with RNAPI transcription complexes (Takeuchi et al., 2003). However, termination of DNA replication throughout the rDNA transcription units has been observed in human cells, suggesting that replication fork barriers at the 3′ end of rDNA transcription units may not be absolute and that some leftward-moving forks pass through the barriers and enter the transcribed region (Little et al., 1993). Such events would require a resolution mechanism like the one suggested by our study.
Our study provides evidence for a coordinated action of RECQ5 and BRCA1 at sites of replication-transcription collisions, which leads to their resolution. We have found that cells lacking BRCA1 show increased levels of Ub-PCNA in chromatin, which is dependent on RECQ5 and transcription. On the other hand, cells lacking RECQ5 (as well as cells lacking RAD18) accumulate RAD51 nuclear foci that are formed in a BRCA1-dependent manner at sites of concomitant replication and transcription. These RAD51 foci in RECQ5-deficient cells likely represent unresolved replication intermediates because they display a long-term stability in the presence of RAD51 inhibitor B02, which prevents the formation of RAD51 foci in normal cells. Our findings imply that in the absence of BRCA1, RECQ5 can mediate PCNA ubiquitination and unloading followed by RNA polymerase bypass, but the replication fork fails to restart because of the defect in RAD51 loading (Schlacher et al., 2012; Fig. 8 B). In the absence of RECQ5, BRCA1 can promote assembly of RAD51 filaments to protect stalled replication forks, but RNA polymerase cannot translocate across the replication fork; hence replication restart is prevented (Fig. 8 B). Thus, both the RECQ5-RAD18-PCNA and the BRCA1-RAD51 “arms” of this mechanism are required to properly resolve replication-transcription collisions. Failure of either of these activities would lead to the persistence of stalled replication forks, resulting in genomic instability (Fig. 8 B).
Studies in budding yeast have shown that strains defective in PCNA unloading (e.g., Δelg1 or PCNA-K164R mutants) exhibit uncontrolled DNA replication and accumulate Rad52 foci, an indication of genomic instability (Yu et al., 2014). Interestingly, these studies have revealed that PCNA is unloaded only from the lagging strand arm of forks stalled by HU treatment (Yu et al., 2014). In human cells, the process of replisome remodeling at stalled forks is not understood. We have shown that the PIP motif of RECQ5 induces RAD18-dependent ubiquitination of PCNA, which accumulated on chromatin of cells expressing a helicase-dead mutant of RECQ5. Mutational inactivation of the PIP motif of RECQ5 resulted in accumulation of RAD18 on chromatin in S-phase cells, indicating a failure in PCNA ubiquitination. However, PIP motif mutations did not apparently affect PCNA unloading induced by overexpression of GFP-RECQ5, which is also dependent on the modification of PCNA Lys-164 (Fig. 7 A, compare lanes 3 and 6). Of note, overexpression of GFP-RECQ5 led to the accumulation of Ub-PCNA on chromatin, whereas unmodified PCNA was largely absent in chromatin under these conditions. A similar scenario can be observed for PCNA in HU-treated cells. Therefore, it is possible that the PIP motif of RECQ5 is involved in ubiquitination of PCNA trimers that remain at stalled forks during the resolution of replication-transcription encounters. The unloaded PCNA is likely to be ubiquitinated independently of RECQ5’s PIP motif, but the unloading process depends on the helicase activity of RECQ5. This would explain our observation of increased Ub-PCNA levels in chromatin of RECQ5-depleted U2OS cells (Fig. 7 B, lane 5).
Interestingly, we have obtained evidence that BRCA1 associates with the transcription machinery at sites of replication-transcription interference. Moreover, we have found that active transcription is required for the formation of BRCA1-dependent RAD51 foci in S-phase cells. Thus, BRCA1 might act in association with the transcription machinery during the process of resolution of replication-transcription collisions. Consistently, previous studies have shown that BRCA1 binds to hyperphosphorylated RNAPII present in the transcription elongation complex (Krum et al., 2003).
Interference between replication and transcription represents a significant source of genome instability and contributes to oncogene-induced tumorigenesis (Poveda et al., 2010). Here, we provide evidence that RECQ5 exerts its genome maintenance function through its involvement in the resolution of collisions between replication and transcription complexes. Because RECQ5 deficiency is associated with cancer susceptibility in mice (Hu et al., 2007), our study provides further evidence for the role of replication-transcription interference in cancer development.
Materials and methods
Antibodies, siRNAs, and plasmids
The following antibodies were used for WB, IP, ChIP, or immunofluorescence staining (IF): rabbit polyclonal anti-RPA194 (WB, IP, and ChIP; sc-28714; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-RECQ5 (WB, IP, ChIP, and IF; made in the laboratory), mouse monoclonal anti-GFP (WB and ChIP; ab290; Abcam), mouse monoclonal anti-Pol ε (ChIP; ab3163; Abcam), bridging antibody for mouse IgG (ChIP; 53017; Active Motif), mouse monoclonal anti–lamin B (WB; NA12; EMD Millipore), mouse monoclonal anti–lamin B2 (WB; gtx628803; GeneTex), rabbit polyclonal anti-TFIIH (WB; sc-293; Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-PCNA (WB; sc-56; Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-RNAPII (clone 7C2; WB; gift from J.-M. Egly, Institute of Genetics and Molecular and Cellular Biology, Illkirch, France), mouse monoclonal anti-BrdU (IF and IP; B2531; Sigma-Aldrich), mouse monoclonal anti-BRCA1 (WB; GTX70111; GeneTex), mouse monoclonal anti-BRCA1 (IF; sc-6954; Santa Cruz Biotechnology, Inc.), rabbit monoclonal anti-RAD18 (IF and WB; #9040; Cell Signaling Technology), rabbit monoclonal anti-ubiquityl-PCNA Lys164 (WB; #13439; Cell Signaling Technology), and mouse monoclonal anti-Flag (WB; F1804; Sigma-Aldrich). The siRNA oligonucleotides used in this study were purchased from Microsynth AG. The sequences of the sense strand of siRNA duplexes are 5′-CGUACGCGGAAUACUUCGA-3′ (siControl); 5′-GGAGAGUGCGACCAUGGCU-3′ (siRECQ5#1); 5′-CAGGUUUGUCGCCCAUUGGAA-3′ (siRECQ5#2); and 5′-CAGGAAAUGGCUGAACUAGAA-3′ (siBRCA1). The esiRECQ5 (MISSION esiRNA human RECQL5; Sigma-Aldrich) and esiRAD18 (MISSION esiRNA human RAD18; Sigma-Aldrich) are an siRNA pool. Transfections of siRNAs (10–40 nM) were done using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s instructions. Cells were harvested 48–72 h after siRNA transfection. The EcoRV–KpnI fragment of the plasmid pJP136 containing human RECQ5 ORF was subcloned into the plasmid pAIO digested with EcoRV and KpnI (Schwendener et al., 2010; Ghodgaonkar et al., 2014). The resulting construct was further modified as follows: (a) a DNA oligoduplex encoding for an shRNA corresponding to siRECQ5#1 (top strand: 5′-GATCCCCGGAGAGTGCGACCATGGCTTTCAAGAGAAGCCATGGTCGCACTCTCCTTTTTGGAAA-3′; bottom strand: 5′-AGCTTTTCCAAAAAGGAGAGTGCGACCATGGCTTCTCTTGAAAGCCATGGTCGCACTCTCCGGG-3′) was introduced between the HindIII and BglII sites; (b) silent mutations were introduced into the RECQ5 cDNA between nucleotides 102 and 120 to render the resulting RECQ5 transcript resistant to the shRNA; (c) the STOP codon in the RECQ5 cDNA was removed by site-directed mutagenesis to put the RECQ5 ORF in frame with the 3xFLAG tag present in the pAIO vector; (d) mutations were introduced into the RECQ5 ORF to generate K58R, R943A, PIP4A substitutions, respectively. Mutagenesis was performed using a QuickChange Site-Directed Mutagenesis kit (Agilent Technologies). The pEGFP-C1 derivatives expressing GFP-tagged wild-type RECQ5, RECQ5ΔIRIΔSRI, or BRCA1 were described previously (Mailand et al., 2007; Kanagaraj et al., 2010). The previously mentioned point mutations were subcloned into GFP-RECQ5 construct. The truncated forms of GFP-RECQ5 coding for amino acids 1–930 (RECQ5ΔCt), 1–651, and 1–502 were cloned by PCR using Pfu DNA polymerase (Fermentas) and reverse primers 5′-TCCTCGAGGCCCTCCTTGTAGAAAGGGGTG-3′, 5′-TCCTCGAGTTTGAGCGAGTACACATGGG-3′, and 5′-GCAGGATCCGCTTGTGGGCCTCATCTCTG-3′, respectively. The resulting amplicons were digested with ScaI–XhoI (for 1–930 and 1–651) or ScaI–BamHI (for 1–502) and inserted between the ScaI–SalI or ScaI–BamHI sites in the GFP-RECQ5 plasmid. The PCNA-RFP and RPA43-GFP expression constructs were gifts from D. Stanek (Institute of Molecular Genetics, Prague, Czech Republic) and M. Dundr (Rosalind Franklin University of Medicine and Science, Chicago, IL), respectively. pcDNA3.1 derivatives encoding for His-tagged wild-type PCNA or PCNA-K164R were described previously (Niimi et al., 2008). Plasmid transfections were performed using TransIT-2020 Reagent (Mirus Bio) according to the manufacturer’s instructions.
Cell culture
HEK293 and U2OS cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Thermo Fisher Scientific) and streptomycin/penicillin (100 U/ml). DMEM for cultivation of U2OS T-REx cells (Invitrogen) was supplemented with 10% Tet-system approved FBS (Biochrom). U2OS T-REx cells stably transfected with pAIO derivatives were selected in the presence of 1 µg/ml puromycin (InvivoGen). Doxycyclin (0.4 ng/ml) was added for 48 h to induce expression of recombinant RECQ5 proteins and down-regulate endogenous RECQ5. HEK293 cells were synchronized in mitosis by treatment with NC (0.1 µg/ml) for 14 h. Mitotic cells were shaken off, collected, washed twice with complete DMEM, and replated with fresh media. Where required, EdU was added to cell culture medium to a final concentration of 20 µM.
Flow cytometry
For analysis of cell cycle distribution by flow cytometry, cells were fixed with ethanol and stained with propidium iodide (PI; 10 µg/ml). EdU (20 µM, 1 h) or 5-FU (1 mM, 30 min) was added to medium before fixation with 4% paraformaldehyde in PBS, followed by permeabilization with 0.25% Triton X-100 in PBS for 5 min, EdU click reaction, or immunostaining with anti-BrdU antibody, respectively, and PI staining. Flow cytometry was performed using LSRII (BD) and FlowJo software (Tree Star).
Immunoprecipitation
For IP experiments, harvested cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, and 0.5% Nonidet P-40) supplemented with protease inhibitor cocktail (Roche) and incubated for 5 min on ice. After sonication and subsequent centrifugation (16,000 g for 10 min at 4°C), the supernatants (500 µg protein) were incubated at 4°C either for 1 h with 10 µl GFP-Trap_A beads (for GFP-tagged proteins; ChromoTek) or overnight with 2 µg primary antibody and ChIP-IT Protein G Magnetic Beads (Active Motif). Where indicated, mixtures were supplemented with 25 U Benzonase nuclease (EMD Millipore) and 5 mM MgCl2. Beads were washed three times in lysis buffer, and bound proteins were eluted by boiling with 2× Laemmli sample buffer.
Analysis of rDNA copy number
HEK293 cells were cultured on 6-cm dishes. The final concentration of siRNAs/esiRNA used for RECQ5 down-regulation was 40 nM at 0.4 ng/µl for the first transfection and 20 nM at 0.2 ng/µl for subsequent transfections. HU (0.2 mM) was added after cell passage or at indicated time points (Fig. 3 A). Samples for WB were collected during each passage. Genomic DNA was isolated using GenElute Mammalian Genomic DNA Miniprep kit (Sigma-Aldrich) according to the manufacturer’s instructions. Extracted DNA was diluted to a concentration of 2 ng/µl and subjected to qPCR performed on a LightCycler 480 PCR System using LightCycler 480 SYBR Green I Master Mix (Roche). The changes in rDNA copy number were calculated by the comparative ΔCp method (2-ΔΔCp) using Oct-4 gene for DNA amount normalization. The sequences of primers used for qPCR are shown in Table S1.
ChIP assays
ChIP assays were performed using the ChIP-IT Express kit (Active Motif) according to the manufacturer’s instructions. HEK293 cells were fixed with 1% formaldehyde for 10 min. Mock- or RECQ5-knockdown cells (asynchronous) were fixed 40 h after siRNA transfection. Cells released from NC block were fixed at indicated time points. NC was added 24 h after siRNA transfection to synchronize cells in mitosis, and treatment with 0.2 mM HU was started immediately after NC block release. HEK293 cells transfected with GFP-RECQ5 plasmids were fixed 24 h after transfection. Cross-linked chromatin was sheared by sonication (Diagenode Bioruptor, water-bath sonicator) to achieve DNA fragments of ∼300 bp. Fragmented chromatin containing ∼15 µg DNA was immunoprecipitated with 2 µg of specific antibody or control IgG (or bridging antibody) overnight at 4°C. In the case of ChIP with anti-GFP antibody, reactions contained only ∼7 µg chromatin. After elution, cross-link reversal, and proteinase K digestion of the immunoprecipitated chromatin, DNA was recovered using QIAquick PCR Purification kit (QIAGEN) and analyzed by qPCR. The sequences of primers used are shown in Table S1. The input chromatin was processed in the same way. DNA amounts were determined from a standard curve generated using input chromatin. Fold enrichment was calculated as a ratio of DNA amount precipitated by specific antibody versus DNA amount obtained with control IgG (or bridging antibody). Relative enrichment was calculated as the amount of precipitated DNA relative to the amount of DNA present in input chromatin (% of input), and normalized to the amplicon H27 of control sample.
Replication timing analysis
HEK293 cells were released from NC-induced mitotic arrest at indicated time points and incubated with 30 µM BrdU for 30 min before harvesting. Genomic DNA was isolated by phenol/chloroform extraction. 1 µg heat-denatured DNA was mixed with 5 µg anti-BrdU antibody and 15 µl ChIP-IT Protein G Magnetic Beads in 100 µl DNA binding buffer (50 mM Tris-HCl, pH 8, 120 mM NaCl, and 0.05% Triton X-100). After 1-h incubation, beads were washed with DNA binding buffer, and immunoprecipitated DNA was eluted by proteinase K treatment in the presence of 0.5% SDS. DNA was isolated using a QIAquick PCR Purification kit and subjected to PCR with primers amplifying the amplicon H6 (Fig. 1 B and Table S1). PCR products were analyzed by agarose electrophoresis.
Immunofluorescence microscopy and ScanR analysis
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 0.25% Triton X-100 in PBS for 5 min. Coverslips were blocked with 1% BSA in PBS before incubation with primary antibodies for 90 min. After washing in PBS, coverslips were incubated with appropriate Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific) and mounted using Vectashield reagent (Vector Laboratories). Cell images were captured on a DM 6000 fluorescence microscope (Leica Biosystems) using an oil-immersion objective, 63×/1.4 NA. For RAD18 immunostaining, cells were preextracted with 25 mM Hepes-NaOH. pH 7.5, buffer containing 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 0.3 M sucrose, and 0.5% Triton X-100 for 4 min at 4°C before fixation. In the case of BRCA1 and RAD51 immunostaining, methanol was used for permeabilization of cells (20 min at −20°C). EdU click reactions were performed in buffer containing 100 mM Tris-HCl, pH 8.5, 2 mM CuSO4, 100 mM ascorbate, and 5 µM Alexa Fluor 647 azide (Thermo Fisher Scientific) for 30 min in the dark followed by two washes with 1% BSA in PBS and immunostaining. DNA was counterstained with DAPI. For BRCA1, RAD51, and RAD18 foci counting and analysis, automated image acquisition was performed on an IX70 microscope (Olympus) equipped with ScanR imaging platform and 40×/1.3 NA oil-immersion objective. Nuclei were identified based on DAPI signal, and foci counts for each nuclear object were analyzed using a spot detection module of the Analysis ScanR software. At least 144 images were acquired and analyzed per sample (2,000 nuclei on average).
FRAP assays
FRAP measurements of GFP-RECQ5 and GFP-BRCA1 mobility in HEK293 cells were performed 6–9 h after the release from NC block. NC was added simultaneously with plasmid transfection (GFP-RECQ5/GFP-BRCA1 and PCNA-RFP), and cells were plated on glass-bottomed Petri dishes after NC removal. ActD (1 µg/ml) or Aph (10 µM) was added 30 min before the start of imaging. The cells were imaged with a SP5 confocal microscope equipped with an oil immersion objective (HCX Plan-Apochromat 63×/1.4 NA) and an environmental chamber controlling temperature (37°C) and CO2 level (5%). For GFP-RECQ5, data acquisition was performed using a 512 × 512-pixel format at a 1,000-Hz scan speed and 1.6-Airy pinhole in 8-bit resolution. After acquisition of eight prebleach images, a region of interest (circular spot 1.7 µm in diameter, covering one to three foci) was bleached with 100% laser power (488-nm line from an argon laser), and fluorescence recovery was monitored at low laser intensity in 110 iterations at 0.29-s intervals. For GFP-BRCA1, data acquisition was performed with modification in pixel format (1,024 × 1,024), and fluorescence recovery was monitored in 40 iterations at 4-s intervals. Fluorescence intensities of the bleached region and a proportionally equal distal unbleached area of the nucleus were measured at each time point using Fiji software. Values were corrected for extracellular background intensity and normalized to prebleach intensity to obtain relative fluorescence intensity.
Alkaline BrdU comet assay
Alkaline BrdU comet assay was performed as described previously (Mórocz et al., 2013). In brief, U2OS cells were pulse labeled with 20 µM BrdU for 20 min and chased in fresh medium for indicated periods of time. After washing in ice-cold PBS, 2 × 104 cells were resuspended in 40 µl of 0.95% low-melting agarose (37°C; SeaPlaque GTG agarose; Lonza) dissolved in PBS, and the suspension was spread onto an agarose-coated slide by covering with a coverslip. Agarose-embedded cells were lysed in ice-cold solution containing 10 mM Tris-HCl, pH 10, 2.5 M NaCl, 100 mM EDTA, 1% Triton X-100, and 0.5% N-lauroylsarcosine sodium salt for 90 min, with detergents added freshly before use, and washed three times for 5 min in PBS. The slides were placed in ice-cold alkaline electrophoresis solution (0.3 M NaOH and 1 mM EDTA, pH >13) for 40 min. Electrophoresis was subsequently conducted at 1 V/cm for 20 min in the same buffer. The slides were washed three times for 5 min in neutralization buffer (0.4 mM Tris-HCl, pH 7.4) and twice for 5 min in PBS and blocked with PBS-BT (PBS containing 0.1% Tween 20 and 0.1% BSA) for 30 min before immunostaining. The slides were incubated with mouse monoclonal anti-BrdU antibody (B2531; Sigma-Aldrich) for 1 h and washed three times with PBS and once with PBS-BT. Then the slides were incubated with secondary antibody Alexa Fluor 647 and SYBR Gold (both Thermo Fisher Scientific) for 45 min. After three washes in PBS, slides were covered with coverslips and kept in a humidified box until microscopy. Comet images were captured on a DM 6000 fluorescence microscope using a 20× objective, and the comet tail length was evaluated using OpenComet software (http://www.cometbio.org). At least 50 cells were analyzed for each of at least three independent experiments. Statistical analysis was done using GraphPad Prism5.
Cell fractionation
To obtain chromatin fractions, cells were resuspended in buffer A (10 mM Hepes-NaOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, and 1 mM DTT) supplemented with 1 mg/ml digitonin and incubated on ice for 5 min. Nuclei were collected by centrifugation at 1,500 g for 4 min, washed once with buffer A, resuspended in buffer B (3 mM EDTA, 0.2 mM EGTA, and 1 mM DTT), and incubated on ice for 15 min. Chromatin was separated from nucleoplasmic fraction by centrifugation at 2,000 g for 5 min and sheared by sonication in buffer B.
Online supplemental material
Fig. S1 shows interaction of ectopically expressed RECQ5 variants with RPA194. Fig. S2 shows frequencies of BRCA1 and RAD51 foci in nuclei of cells exposed to different replication and transcription inhibitors, and images from a FRAP sequence showing fluorescence recovery of GFP-BRCA1 foci in mock-, Aph-, and ActD-treated HEK293 cells. Fig. S3 shows occupancy of Pol ε on rDNA repeat units and analysis of BrdU comet tail lengths in nontreated and ActD-treated cells. Fig. S4 shows representative images of RAD18 foci in nuclei of cells exposed to different replication and transcription inhibitors. Fig. S5 shows relative mRNA abundance of endogenous and total RECQ5 in U2OS T-REx cells expressing RECQ5 variants. Table S1 lists the sequences of primers used for qPCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507099/DC1.
Supplementary Material
Acknowledgments
We thank Radek Malik and Jana Urbanova for comments on the manuscript and help with real-time PCR, and Miroslav Dundr, David Stanek, and Alan R. Lehmann for providing plasmid constructs.
This work was supported by the Czech Science Foundation (14-05743S), the Swiss National Science Foundation (31003A-146206), the Danish Council for Independent Research (DFF-1331-00262), the Swedish Research Council, the Ministry of Education, Youth and Sports of the Czech Republic (KONTAKT II LH14037), and the Neuron Fund for Support of Science.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ActD
- actinomycin D
- CFS
- common fragile site
- ChIP
- chromatin immunoprecipitation
- CTD
- C-terminal repeat domain
- ERFS
- early replicating fragile site
- HU
- hydroxyurea
- IF
- immunofluorescence staining
- IP
- immunoprecipitation
- IRI
- internal RNAPII–interacting
- NC
- nocodazole
- PCNA
- proliferating cell nuclear antigen
- PI
- propidium iodide
- Pol ε
- polymerase ε
- qPCR
- quantitative real-time PCR
- RNAP
- RNA-polymerase
- SRI
- Set2-Rpb1-interacting
- TSS
- transcription start site
- WB
- Western blotting
References
- Azvolinsky A., Giresi P.G., Lieb J.D., and Zakian V.A.. 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell. 34:722–734. 10.1016/j.molcel.2009.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.H., Faryabi R.B., Callén E., Wong N., Malhowski A., Chen H.T., Gutierrez-Cruz G., Sun H.W., McKinnon P., Wright G., et al. 2013. Identification of early replicating fragile sites that contribute to genome instability. Cell. 152:620–632. 10.1016/j.cell.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakri H., de Septenville A.L., Viguera E., and Michel B.. 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 29:145–157. 10.1038/emboj.2009.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D.L., Popuri V., Opresko P.L., and Bohr V.A.. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83:519–552. 10.1146/annurev-biochem-060713-035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin S.G., and Glover T.W.. 2007. Chromosome fragile sites. Annu. Rev. Genet. 41:169–192. 10.1146/annurev.genet.41.042007.165900 [DOI] [PubMed] [Google Scholar]
- Elías-Arnanz M., and Salas M.. 1999. Resolution of head-on collisions between the transcription machinery and bacteriophage phi29 DNA polymerase is dependent on RNA polymerase translocation. EMBO J. 18:5675–5682. 10.1093/emboj/18.20.5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe-Abrio I., Lafuente-Barquero J., García-Rubio M.L., and Aguilera A.. 2015. RNA polymerase II contributes to preventing transcription-mediated replication fork stalls. EMBO J. 34:236–250. 10.15252/embj.201488544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P.L., Liu Y., Jiricny J., West S.C., and Janscak P.. 2004. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 23:2882–2891. 10.1038/sj.emboj.7600301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodgaonkar M.M., Kehl P., Ventura I., Hu L., Bignami M., and Jiricny J.. 2014. Phenotypic characterization of missense polymerase-δ mutations using an inducible protein-replacement system. Nat. Commun. 5:4990 10.1038/ncomms5990 [DOI] [PubMed] [Google Scholar]
- Grummt I. 2003. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17:1691–1702. 10.1101/gad.1098503R [DOI] [PubMed] [Google Scholar]
- Helmrich A., Ballarino M., and Tora L.. 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 44:966–977. 10.1016/j.molcel.2011.10.013 [DOI] [PubMed] [Google Scholar]
- Helmrich A., Ballarino M., Nudler E., and Tora L.. 2013. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 20:412–418. 10.1038/nsmb.2543 [DOI] [PubMed] [Google Scholar]
- Hu Y., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21:3073–3084. 10.1101/gad.1609107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa K., Yanagida M., Hayano T., Tachikawa H., Komatsu W., Shimamoto A., Futami K., Furuichi Y., Shinkawa T., Yamauchi Y., et al. 2008. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem. J. 413:505–516. 10.1042/BJ20071392 [DOI] [PubMed] [Google Scholar]
- Jones R.M., Mortusewicz O., Afzal I., Lorvellec M., García P., Helleday T., and Petermann E.. 2013. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 32:3744–3753. 10.1038/onc.2012.387 [DOI] [PubMed] [Google Scholar]
- Kanagaraj R., Saydam N., Garcia P.L., Zheng L., and Janscak P.. 2006. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34:5217–5231. 10.1093/nar/gkl677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagaraj R., Huehn D., MacKellar A., Menigatti M., Zheng L., Urban V., Shevelev I., Greenleaf A.L., and Janscak P.. 2010. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 38:8131–8140. 10.1093/nar/gkq697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krum S.A., Miranda G.A., Lin C., and Lane T.F.. 2003. BRCA1 associates with processive RNA polymerase II. J. Biol. Chem. 278:52012–52020. 10.1074/jbc.M308418200 [DOI] [PubMed] [Google Scholar]
- Li J., Santoro R., Koberna K., and Grummt I.. 2005. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 24:120–127. 10.1038/sj.emboj.7600492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Xu X., and Liu Y.. 2011. The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol. Cell. Biol. 31:2090–2099. 10.1128/MCB.01137-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R.D., Platt T.H., and Schildkraut C.L.. 1993. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 13:6600–6613. 10.1128/MCB.13.10.6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., and Lukas J.. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900. 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Mórocz M., Gali H., Raskó I., Downes C.S., and Haracska L.. 2013. Single cell analysis of human RAD18-dependent DNA post-replication repair by alkaline bromodeoxyuridine comet assay. PLoS One. 8:e70391 10.1371/journal.pone.0070391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejepinska J., Malik R., Filkowski J., Flemr M., Filipowicz W., and Svoboda P.. 2012. dsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells. Nucleic Acids Res. 40:399–413. 10.1093/nar/gkr702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi A., Brown S., Sabbioneda S., Kannouche P.L., Scott A., Yasui A., Green C.M., and Lehmann A.R.. 2008. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc. Natl. Acad. Sci. USA. 105:16125–16130. 10.1073/pnas.0802727105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda A.M., Le Clech M., and Pasero P.. 2010. Transcription and replication: breaking the rules of the road causes genomic instability. Transcription. 1:99–102. 10.4161/trns.1.2.12665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N., McDonald K.R., Webb C.J., Cristea I.M., and Zakian V.A.. 2012. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev. 26:581–593. 10.1101/gad.184697.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro M., Kantidakis T., Mitter R., Kelly G.P., Heron M., Williams H., Söding J., Stewart A., and Svejstrup J.Q.. 2014. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 157:1037–1049. 10.1016/j.cell.2014.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendener S., Raynard S., Paliwal S., Cheng A., Kanagaraj R., Shevelev I., Stark J.M., Sung P., and Janscak P.. 2010. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 285:15739–15745. 10.1074/jbc.M110.110478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., and Livingston D.M.. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 88:265–275. 10.1016/S0092-8674(00)81847-4 [DOI] [PubMed] [Google Scholar]
- Sporbert A., Gahl A., Ankerhold R., Leonhardt H., and Cardoso M.C.. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 10:1355–1365. 10.1016/S1097-2765(02)00729-3 [DOI] [PubMed] [Google Scholar]
- Takeuchi Y., Horiuchi T., and Kobayashi T.. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17:1497–1506. 10.1101/gad.1085403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y., Watanabe K., Araki K., Shinohara M., Yamagata Y., Tsurimoto T., Hanaoka F., Yamamura K., Yamaizumi M., and Tateishi S.. 2008. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 13:343–354. 10.1111/j.1365-2443.2008.01176.x [DOI] [PubMed] [Google Scholar]
- Willis N.A., Chandramouly G., Huang B., Kwok A., Follonier C., Deng C., and Scully R.. 2014. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 510:556–559. 10.1038/nature13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.E., Arlt M.F., Park S.H., Rajendran S., Paulsen M., Ljungman M., and Glover T.W.. 2015. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 25:189–200. 10.1101/gr.177121.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Gan H., Han J., Zhou Z.X., Jia S., Chabes A., Farrugia G., Ordog T., and Zhang Z.. 2014. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell. 56:551–563. 10.1016/j.molcel.2014.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.