Abstract
Curving biological membranes establishes the complex architecture of the cell and mediates membrane traffic to control flux through subcellular compartments. Common molecular mechanisms for bending membranes are evident in different cell biological contexts across eukaryotic phyla. These mechanisms can be intrinsic to the membrane bilayer (either the lipid or protein components) or can be brought about by extrinsic factors, including the cytoskeleton. Here, we review examples of membrane curvature generation in animals, fungi, and plants. We showcase the molecular mechanisms involved and how they collaborate and go on to highlight contexts of curvature that are exciting areas of future research. Lessons from how membranes are bent in yeast and mammals give hints as to the molecular mechanisms we expect to see used by plants and protists.
Introduction
Membranes are the frontier between the inside and outside of cells and separate diverse intracellular compartments while being hubs of signaling activity. The exchange of molecules and signals across this barrier requires many processes to bend, invaginate, protrude, fuse, and break membranes. Furthermore, expanding the surface area of membranes for reactions is achieved by sculpting folds and tubules into intracellular organelles. Pure lipid bilayers remain flat, and stimulating them to curve requires energy (Helfrich, 1973; Helfrich and Jakobsson, 1990). The energy needed is provided by modification of the lipid composition or from the membrane-associated proteins that make up ∼50% of the membrane surface. Protein mechanisms range from different types of binding interaction to oligomerization processes and mechanochemical ATP and GTPases. Membranes can be positively curved (toward the cytoplasm) or negatively curved (away from the cytoplasm), and their deformability varies depending on the tension in the membrane (McMahon and Gallop, 2005). The local restriction of curvature to specific areas implies lateral compartmentalization within the fluid mosaic membrane (Singer, 1972; Kusumi et al., 2011).
In the last few years, rapid progress has been made in diversifying membrane curvature research beyond membrane trafficking to organelle architecture. The molecular mechanisms of membrane curvature rarely act alone, but instead cooperate in diverse ways to achieve the highly complex and well-regulated membrane architectures needed by cells. In this review, we describe how diverse membranes are shaped in plant, animal, and fungal cells (Fig. 1). First, we give a brief description of the common molecular mechanisms that are used to bend membranes. We go on to discuss how the mechanisms are used together to generate the curvatures present at the plasma membrane (PM) and intracellular organelles (Table 1). Lastly, we look ahead at some other contexts for membrane curvature that are important topics for further investigation.
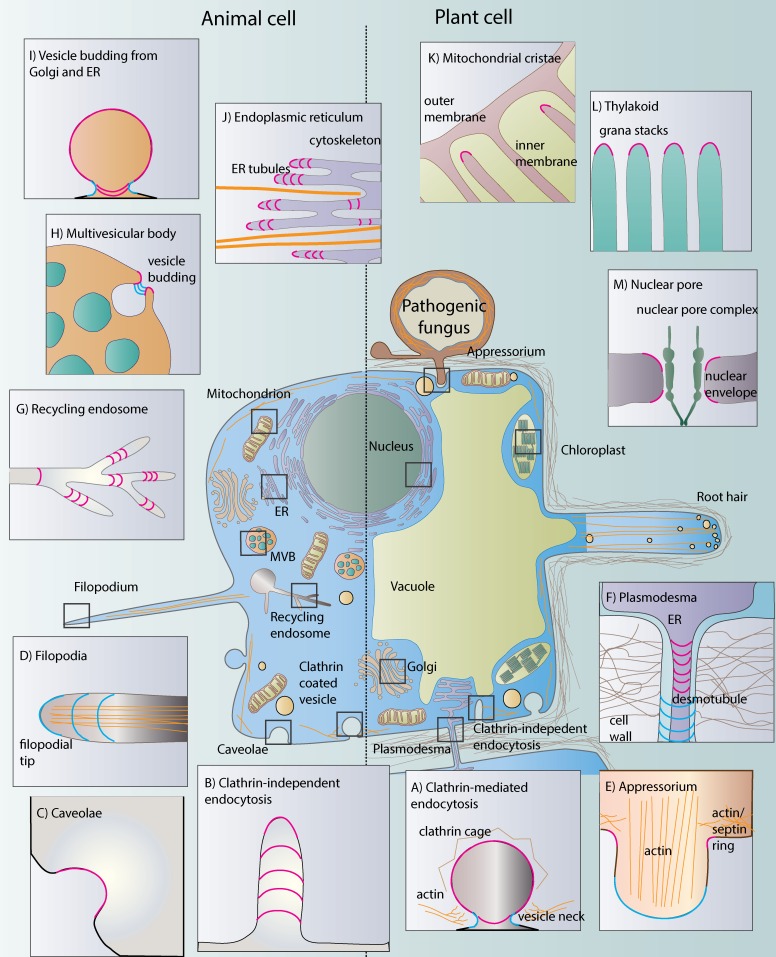
Figure 1.
Cell shaping through membrane curvature. We highlight prominent examples of membrane curvature in cells where recent progress has revealed insights into the underlying molecular mechanisms. (A) Multiple bending mechanisms in CME; (B) clathrin-independent endocytosis; (C) caveolae formation; (D) induction of negative curvature at the PM for protrusions like filopodia; (E) bulging of the fungal appressorium; (F) constriction of the PM sleeve and the ER-derived desmotubule at plasmodesmata; (G) dynamic membrane curvature at endosomal tubules; (H) establishment of a protrusion into the MVB to induce vesicle budding; (I) vesicle budding from the ER and Golgi; (J) shaping of morphologically distinct and highly dynamic ER sheets and tubules; (K) sculpting and maintenance of mitochondrial cristae membranes; (L) shaping the surface-maximized grana in plant thylakoids; and (M) assembly and persistence of the nuclear pore complex in the highly bent nuclear envelope. Positive curvature is shown in pink lines, and negative curvature is shown in blue.
Table 1. An overview of processes and proteins in membrane bending.
| Process | Protein name | Curvature | Molecular mechanism | Figure | References |
|---|---|---|---|---|---|
| Appressorium | Rvs167 | Positive | BAR domain, scaffolding | 3 A | Dagdas et al., 2012 |
| Appressorium | Septins | Positive | Oligomerization, scaffolding | 3 C | Bezanilla et al., 2015; Bridges et al., 2016 |
| Caveolae | Caveolin | Positive | Oligomerization, hairpin insertion | 2 C | Monier et al., 1995 |
| Caveolae | Cavins | Positive | Oligomerization, clustering | 2 C | Hansen et al., 2009 |
| Caveolae | EHD2 | Positive | Oligomerization, GTPase | 3 C | Daumke et al., 2007; Morén et al., 2012 |
| Caveolae | PACSIN2 | Positive | BAR domain, scaffolding | 3 A | Senju et al., 2011 |
| Endocytosis, clathrin mediated | Actin | Positive, negative | Substrate for motor proteins, membrane pulling, polymerization pushing against the membrane, ATPase | 3 G & H | Collins et al., 2011; Lewellyn et al., 2015 |
| Endocytosis, clathrin mediated | Amphiphysin | Positive | Amphipathic helix, bilayer asymmetry, BAR domain scaffolding and oligomerization | 2 E | Peter et al., 2004; Isas et al., 2015 |
| Endocytosis, clathrin mediated | CALM | Positive | Amphipathic helix, bilayer asymmetry | 2 D | Miller et al., 2015 |
| Endocytosis, clathrin mediated | CIP4 | Positive | F-BAR domain, oligomerization, scaffolding | 3 A & C | Frost et al., 2008 |
| Endocytosis, clathrin mediated | Clathrin | Positive | Scaffolding via oligomerization, crowding | 3 C & E | Fotin et al., 2004; Avinoam et al., 2015 |
| Endocytosis, clathrin mediated | Dynamin | Positive | Oligomerization, scaffolding, GTPase | 3 C | Chappie et al., 2011; Morlot et al., 2012 |
| Endocytosis, clathrin mediated | Epsin | Positive | Amphipathic helix, bilayer asymmetry, crowding | 2 D, 3 E | Ford et al., 2002; Stachowiak et al., 2012, 2013; Busch et al., 2015 |
| Endocytosis, clathrin mediated | FCHo1/2 | Positive | F-BAR domain, scaffolding | 3 A | Henne et al., 2007, 2010 |
| Endocytosis, Shiga toxin uptake | Gb3 | Negative | Lipid receptor, clustering, crowding | 2 C | Römer et al., 2007 |
| Endocytosis, Shiga toxin uptake | Shiga toxin | Negative | Clustering, crowding | 2 C | Römer et al., 2007 |
| Endosomal tubules | Dynein | Positive | Microtubule motor, pulling of membrane compartments, ATPase | 3 H | Day et al., 2015 |
| Endosomal tubules | Kinesin | Positive | Microtubule motor, pulling of membrane compartments, ATPase | 3 H | Roux et al., 2002; Delevoye et al., 2014 |
| Endosomal tubules | SNX1 | Positive | BAR domain, scaffolding, oligomerization | 3 A | Carlton et al., 2004 |
| ER shaping | Actin | Positive | Substrate for motor proteins, membrane pulling, ATPase | 3 H | Quader et al., 1989; Liebe and Menzel, 1995; Estrada et al., 2003; Sparkes et al., 2009 |
| ER shaping | Atlastins | Positive | Hydrophobic domain insertion, clustering, GTPase | 2 C | Hu et al., 2009 |
| ER shaping | DP1/YOP1 | Positive | Insertion of wedge-shaped membrane-binding domain | 2 C | Voeltz et al., 2006 |
| ER shaping | Microtubules | Positive | Substrate for motor proteins, membrane pulling | 3 H | Waterman-Storer and Salmon, 1998 |
| ER shaping | Myosin | Positive | Actin motor protein, ER tubule pulling, ATPase | 3 H | Estrada et al., 2003 |
| ER shaping | RTNs | Positive | Insertion of hairpin membrane-binding domain | 2 C | Voeltz et al., 2006 |
| ER shaping | Sey1 | Positive | Hydrophobic domain insertion, clustering | 2 C | Hu et al., 2009 |
| ER, anterograde trafficking | COPII | Positive | Scaffolding via oligomerization | 3 C | Lee et al., 2005; Manneville et al., 2008 |
| ER, anterograde trafficking | Sar1 | Positive | Amphipathic helix, dimerization, scaffolding | 2 D & F; 3 C | Lee et al., 2005; Beck et al., 2008; Krauss et al., 2008 |
| ER, anterograde trafficking | Sec12/31 | Positive | Oligomerization, scaffolding, crowding | 3 C & E | Copic et al., 2012 |
| ER, anterograde trafficking | Sec23/24 | Positive | Curved membrane-binding domain, scaffolding | 3 A | Bi et al., 2002 |
| Filopodia | Actin | Negative | Polymerization within membrane compartment, membrane pushing | 3 F | Liu et al., 2008 |
| Filopodia | Dopamine transporter | Negative | Shaped transmembrane domain | 2 B | Caltagarone et al., 2015 |
| Filopodia | IRSp53 | Negative | I-BAR domain, scaffolding | 3 B | Mattila et al., 2007 |
| Filopodia | MIM | Negative | I-BAR domain, scaffolding | 3 B | Mattila et al., 2007; Saarikangas et al., 2009, 2015 |
| Filopodia | srGAP2 | Negative | F-BAR domain, scaffolding | 3 B | Guerrier et al., 2009 |
| Golgi, retrograde trafficking | Arf1 | Positive | Amphipathic helix, dimerization, scaffolding via oligomerization | 2 D & F; 3 C | Lee et al., 2005; Beck et al., 2008; Krauss et al., 2008 |
| Golgi, retrograde trafficking | ArfGAP1 | Positive | Amphipathic helix, bilayer asymmetry | 2 D | Bigay et al., 2003; Antonny et al., 2005 |
| Golgi, retrograde trafficking | COPI | Positive | Scaffolding via oligomerization | 3 C | Lee et al., 2005; Manneville et al., 2008 |
| Mitochondrial cristae | F1Fo-ATPase | Positive | Dimerization, shaping via transmembrane domain | 2 B & C | Jiko et al., 2015 |
| Mitochondrial cristae | Mic10 | Positive | Insertion of hairpin transmembrane domain, clustering | 2 C | Harner et al., 2011; Barbot et al., 2015 |
| MVBs | ESCRT-III | Negative | Scaffolding, helical oligomerization | 3 D | Hanson et al., 2008; Henne et al., 2012; Cashikar et al., 2014; McCullough et al., 2015 |
| MVBs | Lysobisphosphatidic acid | Negative | Lipid shape | 2 A | Matsuo et al., 2004 |
| Nuclear pore | Nup53 | Positive | Amphipathic helix, bilayer asymmetry | 2 F | Vollmer et al., 2012 |
| Nuclear pore | Nup133 | Positive | Amphipathic helix, bilayer asymmetry | 2 F | Doucet and Hetzer, 2010 |
| Nuclear pore | Nup153 | Positive | Amphipathic helix, bilayer asymmetry | 2 F | Mészáros et al., 2015; Vollmer et al., 2015 |
| Thylakoid shaping | CURT1 | Positive | Amphipathic helix, bilayer asymmetry | 2 D | Armbruster et al., 2013 |
The molecular mechanisms of membrane bending
It is helpful to consider membrane-intrinsic forces that act by introducing local asymmetry to the bilayer distinct from membrane-extrinsic forces that are contributed by peripheral interacting proteins acting outside the lipid bilayer itself. One way of altering the curvature of the membrane is by modifying the local lipid composition, whether it be the lipid headgroup, tail, or cholesterol enrichment. The shape of the individual lipids gives a spontaneous curvature to the membrane (Fig. 2 A). In model membranes, lipids of the same kind tend to cluster together, and protein transmembrane domains prefer to accumulate specific lipids as a lipid coat. Asymmetry within the bilayer is also introduced by the intrinsic shape of protein membrane-spanning segments (Fig. 2 B) or the asymmetric insertion of hydrophobic protein domains, e.g., hairpins (Fig. 2 C) or amphipathic helices (Fig. 2, D–F). The contributions made by protein monomers can be greatly enhanced by oligomerization, which also stabilizes membrane curvature (Fig. 2, C and F).
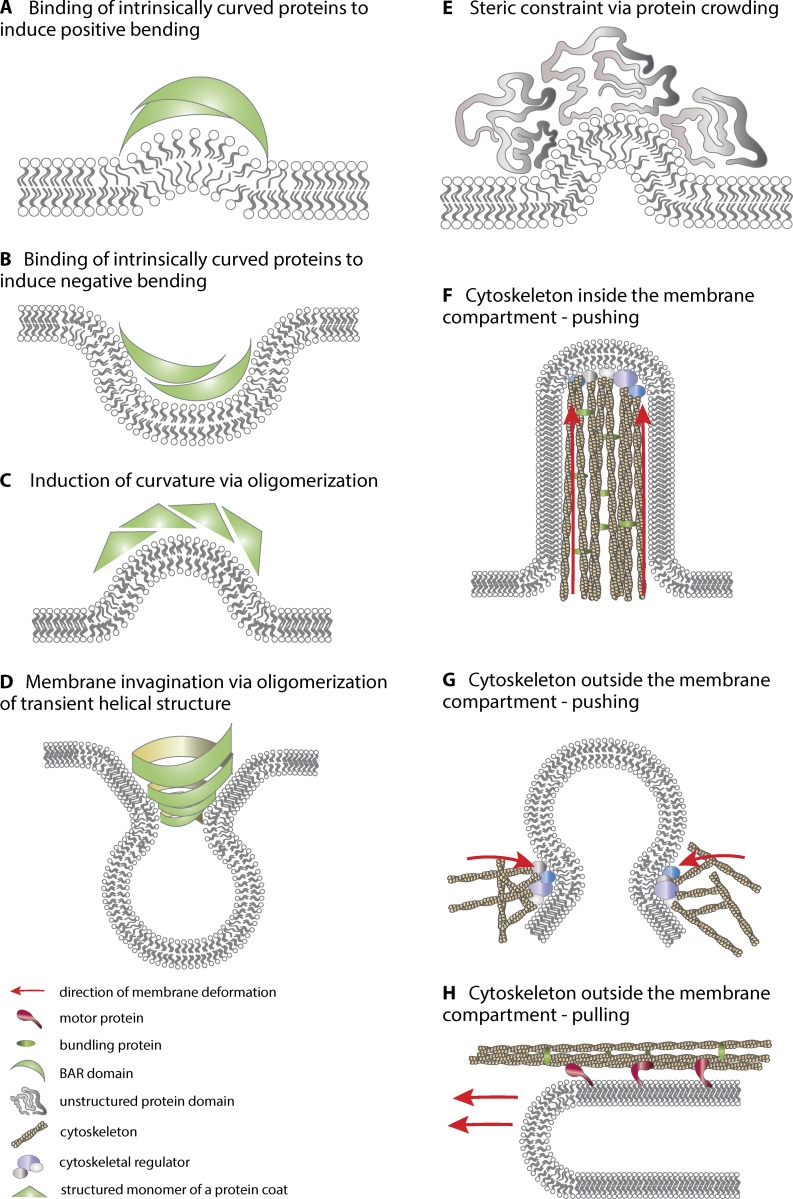
Figure 2.
Mechanisms of direct membrane bending within the lipid bilayer. (A) The shape of the lipid molecules gives rise to spontaneous curvature of the membrane, and individual lipids can be curvature promoting. (B) Transmembrane proteins can introduce curvature into the bilayer by the shape of their transmembrane domain. (C) Individual transmembrane protein without curved shapes can bend the membrane via oligomerization and clustering. Interactions with the membrane can be (a) through the formation of single leaflet hairpins; (b) via transmembrane regions; or (c) via protein interactions with lipid receptors. (D) Many curvature-inducing proteins have peptide sequences that are disordered in solution and that fold into α-helices upon interacting with the cell membrane, creating one hydrophobic and one polar face, termed amphipathic helices. The hydrophobic side of the helix penetrates like a wedge into the outer leaflet of the membrane, inducing curvature. (E) Amphipathic helices can cooperate with other proteins, such as the curved scaffold proteins that contain a BAR domain. (F) Amphipathic helices can also be present in the component proteins of large membrane-associated complexes (also called protein coats) and thus anchor them tightly onto the bilayer.
In addition to membrane-intrinsic mechanisms, various cytoplasmic membrane-extrinsic protein machineries modify membrane shape. Inherently curved peripheral binding proteins act as monomeric scaffolds or as homo- or heterodimers. For example, the Bin-amphiphysin-Rvs (BAR) domain, which characterizes the protein superfamily with the same name, can be curved to various degrees, generating either positive or negative curvature. By individual proteins, curvatures can therefore be manipulated at the nanometer scale (Fig. 3, A and B). Both curved and flat monomers can also further oligomerize to larger curved scaffolds that manipulate shape at up to a micrometer scale (Fig. 3, C and D; for more details, see McMahon and Boucrot, 2015). In contrast to true scaffolding, protein crowding involves large, unstructured protein regions introducing asymmetry in the membrane by sheer volume increase and steric constraint (Fig. 3 E). Cytoskeletal elements can push or pull membranes by polymerization or with the help of motor proteins (Fig. 3, F–H).
Figure 3.
Indirect membrane bending through peripheral exertion of force onto the bilayer. (A and B) Inherently curved proteins, such as the BAR domain protein superfamily, act as scaffolds to introduce curvature. The BAR domain shape is caused by the dimer interface and kinks in the α-helices. This can produce positive curvature, such as that formed by the BAR and FCH-BAR (F-BAR) domain proteins (A), or negative curvature, as formed by inverse BAR (I-BAR) domains (B). (C and D) Scaffold proteins do not need to be curved themselves to deform membranes; curvature can arise from the interactions between monomers or dimers to give either positive (C) or negative curvature (D). (E) Large, unstructured protein regions introduce an asymmetry in the membrane by protein crowding. (F–H) Cytoskeletal proteins can push on membranes to enforce a protrusion (F), constrict the neck of a membrane invagination (G), or pull at tubular/circular membrane compartments with the help of motor proteins (H).
In the following sections of this review, we examine a range of cellular contexts to illustrate how the core molecular mechanisms of membrane curvature are combined by cells to sculpt membrane architecture.
Membrane invaginations at the PM
Endocytosis is the main process by which eukaryotic cells internalize extracellular material. As clathrin-mediated endocytosis (CME) has been an important model for elucidating how membranes deform to make vesicles, we consider this pathway in detail (Table 1 and Fig. 1 A). The FCH-BAR (F-BAR) domain proteins FCHo1/2, which have a shallowly curved shape, are early arrivals to incipient sites of CME, yet depart from the budding intermediate (Fig. 3 A; Henne et al., 2010). In vitro, these proteins form variable 20–130-nm-wide tubules from PI(4,5)P2 liposomes by orienting the F-BAR domain differently on the membrane, which is twisted relative to similar domains in other proteins (Henne et al., 2007). Some F-BAR domain proteins that are recruited later to sites of CME, including Cdc42-interacting protein 4 (CIP4), take advantage of both curvature scaffolding and higher oligomerization mechanisms. Their membrane binding leads to activation of actin polymerization, probably at regions where membrane tension is high (Fig. 3, A and C; Frost et al., 2008; Boulant et al., 2011; Collins et al., 2011).
As the major PM phosphoinositide, PI(4,5)P2 is used during CME for recruiting proteins and is also metabolized in response to curvature, which tunes this function (Chang-Ileto et al., 2011; Schmid and Mettlen, 2013). Three key PI(4,5)P2 binding proteins implicated in membrane curvature are epsin, CALM (clathrin assembly lymphoid myeloid leukemia), and the N-BAR domain containing protein amphiphysin. They have amphipathic helices at their N termini, which fold and protrude hydrophobic residues upon membrane binding (Fig. 2 D; Ford et al., 2002; Peter et al., 2004; Miller et al., 2015). All three proteins likely help to convert an early, shallowly curved intermediate into a deeply invaginated pit (Fig. 2 E). CALM, epsin, and amphiphysin also bind to clathrin and adapters via long, natively unstructured regions that contain short motifs for protein–protein interactions and are expected to cause protein crowding (Fig. 3 E). In vitro studies show that the crowding of epsin makes a greater contribution to curvature than its amphipathic helix (Stachowiak et al., 2012; Busch et al., 2015). However, endocytic cargo is also expected to be glycosylated on the outer surface (which should promote crowding in the opposite direction), so the contribution of crowding in vivo is not yet clear (Stachowiak et al., 2013).
The clathrin lattice itself initially assembles at fairly flat membrane areas, recruited there by adapter proteins. Dynamic rearrangements of the lattice occur as the membrane bends to stabilize the curvature of the nascent vesicle (Fig. 3 C; Fotin et al., 2004; Avinoam et al., 2015). As the vesicle curves more deeply, the force generated by actin polymerization is implicated in further constriction in mammalian cells (Fig. 3 G; Collins et al., 2011). In yeast, actin–myosin interactions and myosin motor activity are essential for endocytic membrane deformation (Fig. 3 H; Lewellyn et al., 2015). In the last phase of CME, the deeply invaginated pit has a highly curved neck region, exhibiting high positive curvature in one direction and negative curvature in the other (a saddle-shaped intermediate; McMahon and Gallop, 2005). Dynamin oligomers assemble in helices around the neck of endocytic vesicles (Fig. 3 C), constricting and increasing pitch to break the membrane using energy from GTP hydrolysis (Chappie et al., 2011; Morlot et al., 2012).
Whereas CME generally leads to the formation of uniform vesicles, clathrin-independent endocytic mechanisms invaginate membranes with varying morphologies, from small tubular or vesicular structures to large macropinosomes (Fig. 1 B). Clathrin-independent pathways are diverse and are typically named either after the ligand that is taken up or after a dominant protein used to invaginate the endocytic intermediate. In the clathrin-independent carriers/glycosylphosphatidylinositol-enriched early endosomal compartment pathway, which is activated through small G proteins, the BAR domain containing protein GRAF1 localizes to PI(4,5)P2-enriched tubular membranes, stabilizing their curvature (Table 1 and Fig. 3 A; Lundmark et al., 2008). Likewise, BAR domains are used by fast endophilin-mediated endocytosis (Boucrot et al., 2015) for endocytic membrane bending and scission in neurons and for Shiga and cholera toxin uptake (Llobet et al., 2011; Renard et al., 2015). In this case, the key protein, endophilin, combines a curved BAR domain with insertion of two amphipathic helices to bend the membrane (Fig. 2 E). Endophilin uses its src-homology 3 (SH3) domain to recruit dynamin for scission (Gallop et al., 2006; Masuda et al., 2006; Jao et al., 2010). Lipid reorganization mechanisms also participate in shiga toxin uptake. The internalization of the toxin requires its binding to the glycosphingolipid receptor globotriaosyl ceramide (Gb3). The clustering of Shiga toxin and Gb3 promotes invagination by inducing asymmetric stress in the external leaflet of the membranes, both in cells and in artificial membranes (Fig. 2 C; Römer et al., 2007). Clustering is not sufficient to induce membrane bending during the uptake of cholera toxin B subunit. Here, tubulation of the PM is driven by dynein and dynactin (Fig. 3 G; Day et al., 2015). In flotillin-dependent endocytosis, flotillins associate with membranes via hydrophobic hairpin insertions into the inner leaflet (Morrow and Parton, 2005) and form homo-/heterooligomers that colocalize with PM-invaginated microdomains (Fig. 2 C; Frick et al., 2007). These microdomains are cholesterol enriched (Fig. 2 A) and have a similar morphology to caveolae.
Caveolae are ∼70-nm-wide pit-like membrane invaginations, likely used as a buffering mechanism in response to variations in PM tension (Fig. 1 C; Cheng et al., 2015). Caveolin oligomerization is proposed to induce curvature via insertion of a cholesterol-interacting hairpin loop into the inner leaflet. Both its high density (∼150 copies per pit) and membrane penetration are predicted to deform the membrane through surface asymmetry (Fig. 2 C; Monier et al., 1995). Caveolins work cooperatively with cytoplasmic proteins, the cavins, and the BAR domain protein PACSIN2 (protein kinase C and casein kinase substrate in neuron 2; Fig. 3 A; Hansen et al., 2011; Senju et al., 2011). Overexpression of cavin-2 in vivo induces caveolae-associated tubules to form, and cavin oligomerization creates a structure that can intrinsically generate membrane curvatures (Fig. 2 C; Hansen et al., 2009). Finally, the dynamin-related ATPase EHD2 (Eps15 homology domain-containing protein) localizes to and binds caveolae via interactions with both cavin-1 and PACSIN2. EHD2 oligomerizes into rings on highly curved membranes and undergoes curvature-stimulated ATP hydrolysis to mediate scission (Fig. 3 C; Daumke et al., 2007; Morén et al., 2012).
To summarize, in CME, the mechanisms of membrane curvature have been intensively elucidated, and it is evident that a range of molecular mechanisms are used cooperatively. To understand how clathrin-independent endocytic pathways are distinct from the clathrin pathway, it has been crucial to define their alternative mechanisms of membrane deformation. In caveolae too, invagination of the PM is regulated and manipulated in multiple stages using both curvature generation and sensing via an array of molecular mechanisms in concert. We hypothesize that such mechanisms allow checks at each stage to verify the suitability of a given invagination (cargo checks for example) and allow reversibility in the process.
The endosomal pathway
Once cargo is internalized and trafficked through the endosomal pathway, it is sorted by sorting and recycling endosomes (Fig. 1 G) to reach the appropriate destination. During this process, tubular membrane compartments concentrate transmembrane cargo away from soluble factors, sorting different types of receptors into distinct tubules (Puthenveedu et al., 2010; van Weering and Cullen, 2014). These endosomal tubules are shaped by an array of SNX-BAR proteins, including SNX1, which coincidentally detects the presence of PI(3)P and a curved membrane topology (Fig. 3 A; Carlton et al., 2004). SNX-BAR proteins can heterodimerize and directly collaborate with cytoskeletal motor proteins (van Weering et al., 2012; Hunt et al., 2013). Actin is involved in limiting the diffusion of cargo, and myosin VI and kinesin are implicated in tubule formation by pulling the tubule along cytoskeletal networks (Fig. 3 H; Roux et al., 2002; Delevoye et al., 2014).
For cargo destined for degradation or to be released as exosomes, endosomes invaginate into their own lumen, forming multivesicular bodies (MVBs; Fig. 1 H). If the invagination mechanism was orchestrated from inside the endosome, in an equivalent way to endocytosis, the machinery would be trapped in the MVBs and degraded. Consequently, the mechanisms used are distinct from those considered so far. Late endosomes are highly enriched in an unconventional phospholipid isomer of phosphatidylglycerol, lysobisphosphatidic acid. In vitro, this lipid induces the formation of internal vesicles into giant unilamellar vesicles (GUVs), implying the use of a lipid-based mechanism for membrane bending in MVB formation (Fig. 2 A; Matsuo et al., 2004). The adapter protein Alix may play a role at the interface of lipid-mediated lysobisphosphatidic acid mechanisms and the protein machinery that forms MVBs, the ESCRTs (endosomal sorting complexes required for transport; McCullough et al., 2008). In vitro, three subunits of the ESCRT are sufficient to form intraluminal vesicles into GUVs (Wollert et al., 2009). ESCRT-III subunits polymerize into spiral-shaped filaments (Hanson et al., 2008; Henne et al., 2012; Cashikar et al., 2014; McCullough et al., 2015) that promote membrane constriction and fission without entering the forming vesicle (Fig. 3 D). The mechanical constraints that accommodate a rigid spiral on an inclined membrane surface are proposed to form a conical invagination to push membranes away from the cytoplasm (Barelli and Antonny, 2009). Recently, it has been demonstrated that polymerization of the ESCRT-III filament stores energy that might be used to deform the membrane or catalyze scission (Chiaruttini et al., 2015).
The endosomal pathway features particular constraints that dictate the choice of curvature mechanism: the need to sort different types of cargo and to sort cargo away from the machinery of membrane budding. Membrane-extrinsic mechanisms are therefore of particular importance.
Protrusions from and connections between cells
The generation of negative curvature at protrusions is the same topology as MVBs, but no budding is needed. Scaffolding mechanisms, curved proteins, and cytoskeletal mechanisms all contribute, as with PM invaginations. In plants and fungi, cellular protrusions are important despite the presence of the cell wall, although these are typically on the order of 5 µm in diameter. For a discussion of pollen tubes and root hairs (Fig. 1), which use cell biological mechanisms rather than molecular mechanisms for their formation, we refer readers to another recent review (Rounds and Bezanilla, 2013).
In metazoans, thin, actin-rich structures of 100–500-nm diameter called filopodia protrude outward from the PM (Fig. 1 D). These are ubiquitously found during development, and their roles include cell navigation, adhesion, synapse formation, and giving directionality to morphogen signals (Mattila and Lappalainen, 2008). Recent studies suggest that the inverse BAR (I-BAR) proteins IRSp53 (insulin receptor substrate protein of 53 kD) and MIM (missing in metastasis) and the F-BAR protein srGAP2 (SLIT-ROBO Rho GTPase-activating protein 2) are recruited during early filopodia formation to deform the membrane before actin polymerization occurs (Fig. 3 B; Mattila et al., 2007; Saarikangas et al., 2008, 2009; Guerrier et al., 2009). Many I-BAR proteins, which are recruited preferentially to areas of negative curvature, also contain other domains that are expected to play roles in filopodia formation. These include GTPase-activating protein (GAP) activity and the SH3 domain for recruiting other proteins (Chen et al., 2015; Prévost et al., 2015). The interactions between formin-like proteins and Cdc42 also creates a curved scaffold that can bend a membrane outward, and this activity is expected to work in conjunction with the formin activity of elongating actin filaments to protrude the membrane (Fig. 3, A and F; Kühn et al., 2015). In GUVs, filopodia-like protrusions can be reconstituted by localizing the Arp2/3 complex to the surface, with protrusions formed by elastic forces from the membrane and by intense actin polymerization, similar to the processes observed in vivo (Fig. 3 F; Liu et al., 2008). Furthermore, some transmembrane proteins that localize preferentially to filopodia are shaped to induce negative curvature, such as the dopamine transporter (Fig. 2 B; Caltagarone et al., 2015).
To invade rice epidermal cells, the pathogenic fungus Magnaporthe oryzae forms a structure called the appressorium (Fig. 1 E). Before penetration, the cortical network inside the appressorium is strengthened, and a septin-mediated F-actin ring forms around the future penetration site. High turgor pressure inside the fungal cell bends the membrane inside the septin ring outward, and, together with localized enzymatic degradation of the cell wall, a protrusion is formed. To enforce this effect, the BAR domain containing protein Rvs167 (reduced viability upon starvation protein 167) and Las17 (an Arp2/3 complex nucleation promoting factor) localizes to the F-actin ring (Fig. 3 B), forming a dense protein environment facilitated by the barrier function of septins (Dagdas et al., 2012). Septins themselves localize preferentially to and polymerize at curved membranes and promote curvature alone and together with actin (Bezanilla et al., 2015; Bridges et al., 2016).
In plants, ∼50-nm-diameter tunnels called plasmodesmata interconnect cells and mediate cell-to-cell communication and material exchange (Fig. 1 F; Maule et al., 2012). They are a highly curved membrane structure characterized by the close association of two cellular membrane systems: a cylindrical sleeve formed by a continuum of the PM (the plasmodesma itself) and an entrapped nanometer-thin tube of ER, reaching from one cell to the next, the so-called desmotubule. The lipid composition of plasmodesmata and desmotubules is believed to be important for their structure and function. The PM of plasmodesmata has a protein and lipid composition that is distinct from the cell’s PM (Tilsner et al., 2011; Grison et al., 2015) and also constitutes a barrier for lipid diffusion (Grabski et al., 1993). It is enriched in sterols and in sphingolipids with very long-chain saturated fatty acids (Grison et al., 2015) and accumulates glycosylphosphatidylinositol-anchored proteins (Fig. 2 A; Fernandez-Calvino et al., 2011). Thus, plasmodesmata may be the first example of a specialized membrane domain that resembles a liquid-ordered phase similar to that of model membranes, in which the enrichment of particular lipids drives curvature stress (Bagatolli and Mouritsen, 2013). The molecular mechanisms of lateral lipid segregation and stabilization remain unclear, although it likely results from lipid corralling via protein scaffolding and external factors, such as the cytoskeleton and cell wall reinforcements through the plasmodesmata-localized polysaccharide callose (Simpson et al., 2009).
The desmotubule’s small (10–15-nm diameter) size has fueled interest in the mechanisms behind the maintenance of such a uniquely high curvature. Several proteins from the reticulon (RTN) family, first characterized at the ER (which we consider next), are present in the plasmodesmata proteome (Fernandez-Calvino et al., 2011), and these introduce curvature via their transmembrane domain (Fig. 2 C, a). Two of the proteins, RTNLB3 and RTNLB6, are recruited to the cell plate at late telophase, when primary plasmodesmata form, and they remain associated with primary plasmodesmata in the mature cell wall (Knox et al., 2015; Kriechbaumer et al., 2015). However, as the desmotubule is far narrower than the surrounding ER, comparable to the size of a vesicle fission stalk, other mechanisms must contribute. In electron microscopy images (Ding et al., 1992), the so-called “central rod” of energy-dense material could represent the lipid head groups in a virtually lumenless tubule (Bayer et al., 2014). It has been hypothesized that the desmotubule membrane has an unusual composition, giving rise to a highly curved hexagonal lipid phase (Fig. 2 A; Jouhet, 2013).
While all three sets of PM protrusion that we consider here involve generating negative curvature, many mechanisms appear to be used to different extents. Plasmodesmata illustrate how lipid composition can give rise to high spontaneous curvatures, whereas filopodia formation uses analogous mechanisms to those in endocytosis and the endosomal pathway with BAR domain proteins and forces from cytoskeletal polymerization. The micrometer-scale curvatures of fungal protrusions appear to use BAR domain proteins to activate the actin cytoskeleton and also the curvatures of septin scaffolds.
The secretory pathway
The ER in both plants and animals consists of a continuum of structurally diverse sheets and tubules (Fig. 1 J; Baumann and Walz, 2001; Voeltz et al., 2006). It is extremely dynamic, with tubules undergoing constant fusion and fission (Prinz et al., 2000). In mammals, the ER is often surrounded by microtubules, with tubules generated and reorganized with help from microtubule-associated motor proteins pulling the membrane (Fig. 3 H; Waterman-Storer and Salmon, 1998). In Saccharomyces cerevisiae, the cytoskeletal role is taken over by actin filaments, with myosin Myo4p being implicated in ER shaping (Estrada et al., 2003). Actomyosin is also involved in ER remodeling in plants (Quader et al., 1989; Liebe and Menzel, 1995; Sparkes et al., 2009). However, as indicated by the persistence of ER architecture even after treatment with cytoskeleton-depolymerizing factors in yeast, other ER-shaping mechanisms are at work (Prinz et al., 2000). These roles are fulfilled by RTNs and DP1/YOP1 (defective in polyposis/yeast orthologue) proteins in animals, fungi, and plants (Voeltz et al., 2006). Although the exact structure of the hairpin they form remains undetermined, it is thought to insert shallowly into the outer monolayer (Fig. 2 C, a), acting similarly to an amphipathic helix (Fig. 2 D). Scaffolding mechanisms via oligomerization have been proposed to enhance tubulation (Fig. 3 C; Hu et al., 2008; Shibata et al., 2008). Loss of RTN1, RTN2, and YOP1 completely depletes yeast of ER tubules (Voeltz et al., 2006). Similar effects are seen on deleting the three RTN proteins in mammals and Caenorhabditis elegans (Audhya et al., 2007; Anderson and Hetzer, 2008). In vitro, purified yeast YOP1 and RTN1P tubulate liposomes, indicating that they are necessary and sufficient to shape the ER membrane into tubules (Hu et al., 2008). Dynamin-like proteins called atlastins in mammals and Sey1 in yeast (Hu et al., 2009) interact with both RTNs and DP1/YOP1. They are proposed to make three-way junctions in the ER by inserting hydrophobic domains into the bilayer and using the energy of GTP hydrolysis to mediate ER fusion (Figs. 2 C and 3 C).
From the ER, vesicles bud for traffic to the Golgi apparatus for secretion. Vesicle budding at the ER and Golgi use similar mechanisms to each other, and these are also well-studied pathways for elucidating the principles of membrane curvature, so we discuss these mechanisms together. The related small GTPases Arf1 and Sar1 orchestrate coatomer protein complex I and II (COPI and COPII) budding from the Golgi and ER, respectively (Fig. 1 I). Both proteins have N-terminal amphipathic helices, which fold upon GTP binding (Fig. 2 D). Arf1 and Sar1 binding are implicated in deforming the membrane, leading to tubulation at high protein concentrations in vitro (Lee et al., 2005; Beck et al., 2008; Krauss et al., 2008). Similar to the clathrin lattice, the COP coats oligomerize around the vesicles, stabilizing curvature as they bind cargo (Fig. 3 C). Insertion of an amphipathic helix on ArfGAP1 stimulates GAP activity, and the interaction between the curvature-sensitive GAP activity and membrane penetration by Arf1 is predicted to form specific domains of curvature-dependent Arf1 activation (Fig. 2 E; Bigay et al., 2003; Antonny et al., 2005). Curvature generation by the Sar1 amphipathic helix is proposed to mediate the scission of vesicles from the parent membrane (Lee et al., 2005), and higher membrane curvature seems to elevate Sar1 GTPase activity (Hanna et al., 2016), raising the possibility of positive feedback between curvature, GTPase activity, and membrane deformation. Dimerization might contribute to membrane bending by Arf1 and Sar1 (Beck et al., 2008; Hariri et al., 2014) by forming a protein scaffold at the membrane surface (Fig. 3 C). In reconstitutions of COPI-mediated vesicle formation on GUVs, the presence of COPI proteins in addition to Arf1 increases membrane deformation (Manneville et al., 2008), and this is similarly true for Sar1 and the COPII coat (Fig. 3 C; Lee et al., 2005). The ability of the amphipathic helix of Arf1 to deform the membrane (Fig. 2 D) also acts together with lipid composition and phase (Fig. 2 A). Budding increases as tension decreases and lipid disorder increases (Manneville et al., 2008). In COPII-mediated budding, the Sec23/24 complex has a curved shape, corresponding to the size of the vesicle (Fig. 3 A; Bi et al., 2002). Cargo interactions, protein crowding, and the rigidity of the Sec13/31 coat are proposed to contribute to membrane curvature during COPII vesicle budding (Copic et al., 2012). ER and Golgi budding display similar principles of membrane curvature to other membrane trafficking processes: collaboration of multiple molecular mechanisms and the integration of curvature sensing and generation. Again, this is likely to contribute to a highly controlled, reversible and error-free process.
Mitochondria, chloroplasts, and the nucleus
The major function of mitochondria is to provide energy to cells through oxidative phosphorylation (McBride et al., 2006). Invaginations, called cristae membranes, form at the inner membrane toward the mitochondrial matrix and greatly increase the total surface area of the inner membrane for chemical reactions (Fig. 1 K). Lamellar, tubular, and discoid cristae shapes are observed at junctions with the flat inner membrane, termed cristae junctions, which have regular 15–40-nm tubular openings. The mitochondrial contact site and cristae organizing system (MICOS) complex localizes there, and its absence causes unattached stacks of membranes to form in the matrix (Pfanner et al., 2014). Mic10, a core subunit of the MICOS complex, forms tubular membranes in vitro, with diameters between 10 and 30 nm (Barbot et al., 2015), and its depletion in vivo leads to cristae junction disappearance (Harner et al., 2011). Molecularly, Mic10 forms a hairpin-like structure, with two transmembrane segments that span the inner mitochondrial membrane. Oligomerization of Mic10 via two GxxxG motifs in its transmembrane domains is also required for membrane tubulation in vitro and for cristae junction formation in vivo (Fig. 2 C). Similarly, a GxxxG motif is involved in the dimerization of the F1Fo–ATPase complexes that localize to the highly curved ridges of cristae and are the final step of oxidative phosphorylation (Harner et al., 2011; Alkhaja et al., 2012). In vitro reconstitution has recently demonstrated that the F1Fo–ATPase monomer induces a kink in the membrane, and on dimerization, the resulting angle between monomers induces high local curvature (Fig. 2 C, b; Jiko et al., 2015), linking a core mitochondrial enzyme with membrane curvature generation.
Plant chloroplasts, as well as cyanobacteria, create energy via photosynthesis, which relies on a H+ gradient across thylakoid membranes to drive ATP synthase. In higher plants, these specialized membranes surround tightly stacked coin-shaped compartments inside the chloroplasts called grana (Fig. 1 L). Similar to mitochondria, this increases the surface area available for chemical reactions. The height of a single grana disk averages ∼24 nm, with a diameter of 300–600 nm (Mustárdy and Garab, 2003; Mullineaux, 2005). The extremely tight bends of the membrane at thylakoid edges depend on the levels of curvature thylakoid 1 (CURT1) family proteins. In Arabidopsis thaliana, all four CURT1 proteins have been predicted to share an N-terminal chloroplast-targeting peptide, two transmembrane domains, and an N-terminal amphipathic helix (Fig. 2 E). Mutants lacking any of these four proteins exhibit wider thylakoid grana, with fewer stacked disks, whereas double, triple, and quadruple curt1 mutants are completely devoid of grana (Armbruster et al., 2013). The overexpression of CURT1A increases stack heights and produces disks with smaller lumens. Purified CURT1A protein shows intrinsic membrane-bending capability in liposome tubulation experiments (Armbruster et al., 2013). The sack-shaped thylakoids of cyanobacteria lack the distinct shape of the grana of higher plants, and most of their CURT1 homologues lack the key amphipathic helices (Armbruster et al., 2013). Hence, organelle architecture and thus photosynthetic control needed for higher plant function are reliant on specific curvature proteins.
The nucleus is the largest organelle and despite its double membrane is not typically thought of as a membranous structure. However, at nuclear pores, membranes need to be tightly curved for the close apposition between the inner and outer membranes and for assembly of the nuclear pore complexes, which regulate transport of molecules into and out of the nucleus (Fig. 1 M). Nuclear pore complexes are comprised of ∼30 different nucleoporins (Nups; Hoelz et al., 2011). Peripheral Nups select cargo and are anchored by structural Nups to the nuclear membrane. Amphipathic helices are present in several structural Nups (e.g., Nup133 and Nup53, among others), where they sense and generate membrane curvature during the assembly of the nuclear pore complex (Fig. 2 F; Doucet and Hetzer, 2010; Vollmer et al., 2012). Furthermore, the vertebrate Nup Nup153, a peripheral filament Nup that was not expected to interact with the nuclear envelope, also contains an amphipathic helix at its N terminus and is essential for nuclear pore complex assembly during interphase (Mészáros et al., 2015; Vollmer et al., 2015).
In intracellular organelles that are not hubs of membrane traffic, we see that membrane-intrinsic mechanisms dominate, in particular the penetration of amphipathic helices and hairpins into the bilayer, which are part of wider protein scaffolds.
Conclusions and future directions
Tremendous progress has been made in the last 10–15 yr in elucidating how membranes are shaped, both in dynamic and stable contexts. Especially at sites of highly dynamic yet tightly regulated membrane curvature, membrane deformation is sensed, generated, and stabilized in a concerted and cooperative manner. Both membrane-intrinsic and membrane-extrinsic factors are used, the latter being especially important for changing curvature when quick association and dissociation are needed. The cytoskeleton plays a fundamental role during the formation of membrane tubules: its high turnover ensures timely reactions. ER tubules, for example, extend at rates of several micrometers per second (Hamada et al., 2014). In contrast, some organelle shapes, such as thylakoids and nuclear pores, need to be maintained for hours, days, or indefinitely (Iwai et al., 2014; Kabachinski and Schwartz, 2015). Here, the use of specialized lipid domains and protein complexes containing membrane-penetrating segments and formation of higher order oligomers guarantee the stability of the membrane architectures.
In some of the examples of membrane curvature generation we have discussed, the collaborative interplay of diverse molecular mechanisms has been extensively studied. In others, there are many gaps to fill, especially in less studied organisms like plants, fungi, or protists, which bring their own challenges. Plants appear not to have BAR domain proteins, though structurally or functionally homologue proteins may yet be discovered. However, other mechanisms may dominate, and organisms with a cell wall and no body heat render local lipid asymmetries a lot more likely than they are for mammals.
Parasitic protists pose other intriguing questions. As many parasites divide within a single host cell, their structures are miniaturized, and specific curvature proteins will be needed for this adaptation. Moreover, additional specially adapted organelles are necessary for their infection mechanisms and life cycles. Thus, the principles of membrane curvature, so far largely unraveled in mammals and yeast, are set to have a wide impact in understanding diverse cell physiologies.
Acknowledgments
We thank Dr. R. Waller, Dr. J. Carlton, Dr. D. Owen, and members of the Gallop laboratory for critical reading of the manuscript. J.L. Gallop and I.K. Jarsch are supported by a Wellcome Trust Research Career Development Fellowship (grant WT095829AIA). F. Daste is supported by a European Research Council Starting Grant (281971) awarded to J.L. Gallop. Gurdon Institute funding is provided by the Wellcome Trust (grant 092096) and Cancer Research UK (grant C6946/A14492).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BAR
- Bin-amphiphysin-Rvs
- CALM
- clathrin assembly lymphoid myeloid leukemia
- CME
- clathrin-mediated endocytosis
- ESCRT
- endosomal sorting complex required for transport
- GAP
- GTPase-activating protein
- GUV
- giant unilamellar vesicle
- MVB
- multivesicular body
- Nup
- nucleoporin
- PM
- plasma membrane
- RTN
- reticulon
References
- Alkhaja A.K., Jans D.C., Nikolov M., Vukotic M., Lytovchenko O., Ludewig F., Schliebs W., Riedel D., Urlaub H., Jakobs S., and Deckers M.. 2012. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell. 23:247–257. 10.1091/mbc.E11-09-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.J., and Hetzer M.W.. 2008. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 182:911–924. 10.1083/jcb.200805140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B., Bigay J., Casella J.F., Drin G., Mesmin B., and Gounon P.. 2005. Membrane curvature and the control of GTP hydrolysis in Arf1 during COPI vesicle formation. Biochem. Soc. Trans. 33:619–622. 10.1042/BST0330619 [DOI] [PubMed] [Google Scholar]
- Armbruster U., Labs M., Pribil M., Viola S., Xu W., Scharfenberg M., Hertle A.P., Rojahn U., Jensen P.E., Rappaport F., et al. 2013. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell. 25:2661–2678. 10.1105/tpc.113.113118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Desai A., and Oegema K.. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178:43–56. 10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avinoam O., Schorb M., Beese C.J., Briggs J.A., and Kaksonen M.. 2015. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science. 348:1369–1372. 10.1126/science.aaa9555 [DOI] [PubMed] [Google Scholar]
- Bagatolli L.A., and Mouritsen O.G.. 2013. Is the fluid mosaic (and the accompanying raft hypothesis) a suitable model to describe fundamental features of biological membranes? What may be missing? Front. Plant Sci. 4:457 10.3389/fpls.2013.00457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbot M., Jans D.C., Schulz C., Denkert N., Kroppen B., Hoppert M., Jakobs S., and Meinecke M.. 2015. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 21:756–763. 10.1016/j.cmet.2015.04.006 [DOI] [PubMed] [Google Scholar]
- Barelli H., and Antonny B.. 2009. Cell biology: Detached membrane bending. Nature. 458:159–160. 10.1038/458159a [DOI] [PubMed] [Google Scholar]
- Baumann O., and Walz B.. 2001. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205:149–214. 10.1016/S0074-7696(01)05004-5 [DOI] [PubMed] [Google Scholar]
- Bayer E.M., Mongrand S., and Tilsner J.. 2014. Specialized membrane domains of plasmodesmata, plant intercellular nanopores. Front. Plant Sci. 5:507 10.3389/fpls.2014.00507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K., Sinning I., Hurt E., Brügger B., Béthune J., and Wieland F.. 2008. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc. Natl. Acad. Sci. USA. 105:11731–11736. 10.1073/pnas.0805182105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Gladfelter A.S., Kovar D.R., and Lee W.L.. 2015. Cytoskeletal dynamics: a view from the membrane. J. Cell Biol. 209:329–337. 10.1083/jcb.201502062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Corpina R.A., and Goldberg J.. 2002. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 419:271–277. 10.1038/nature01040 [DOI] [PubMed] [Google Scholar]
- Bigay J., Gounon P., Robineau S., and Antonny B.. 2003. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 426:563–566. 10.1038/nature02108 [DOI] [PubMed] [Google Scholar]
- Boucrot E., Ferreira A.P., Almeida-Souza L., Debard S., Vallis Y., Howard G., Bertot L., Sauvonnet N., and McMahon H.T.. 2015. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 517:460–465. 10.1038/nature14067 [DOI] [PubMed] [Google Scholar]
- Boulant S., Kural C., Zeeh J.C., Ubelmann F., and Kirchhausen T.. 2011. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 13:1124–1131. 10.1038/ncb2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges A.A., Jentzsch M.S., Oakes P.W., Occhipinti P., and Gladfelter A.S.. 2016. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J. Cell Biol. 213:23–32. 10.1083/jcb.201512029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch D.J., Houser J.R., Hayden C.C., Sherman M.B., Lafer E.M., and Stachowiak J.C.. 2015. Intrinsically disordered proteins drive membrane curvature. Nat. Commun. 6:7875 10.1038/ncomms8875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caltagarone J., Ma S., and Sorkin A.. 2015. Dopamine transporter is enriched in filopodia and induces filopodia formation. Mol. Cell. Neurosci. 68:120–130. 10.1016/j.mcn.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Peter B.J., Oorschot V.M., Rutherford A., Mellor H., Klumperman J., McMahon H.T., and Cullen P.J.. 2004. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 14:1791–1800. 10.1016/j.cub.2004.09.077 [DOI] [PubMed] [Google Scholar]
- Cashikar A.G., Shim S., Roth R., Maldazys M.R., Heuser J.E., and Hanson P.I.. 2014. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife. 10.7554/eLife.02184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Ileto B., Frere S.G., Chan R.B., Voronov S.V., Roux A., and Di Paolo G.. 2011. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev. Cell. 20:206–218. 10.1016/j.devcel.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie J.S., Mears J.A., Fang S., Leonard M., Schmid S.L., Milligan R.A., Hinshaw J.E., and Dyda F.. 2011. A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell. 147:209–222. 10.1016/j.cell.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Shi Z., and Baumgart T.. 2015. Regulation of membrane-shape transitions induced by I-BAR domains. Biophys. J. 109:298–307. 10.1016/j.bpj.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.P., Mendoza-Topaz C., Howard G., Chadwick J., Shvets E., Cowburn A.S., Dunmore B.J., Crosby A., Morrell N.W., and Nichols B.J.. 2015. Caveolae protect endothelial cells from membrane rupture during increased cardiac output. J. Cell Biol. 211:53–61. 10.1083/jcb.201504042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N., Redondo-Morata L., Colom A., Humbert F., Lenz M., Scheuring S., and Roux A.. 2015. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell. 163:866–879. 10.1016/j.cell.2015.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A., Warrington A., Taylor K.A., and Svitkina T.. 2011. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr. Biol. 21:1167–1175. 10.1016/j.cub.2011.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copic A., Latham C.F., Horlbeck M.A., D’Arcangelo J.G., and Miller E.A.. 2012. ER cargo properties specify a requirement for COPII coat rigidity mediated by Sec13p. Science. 335:1359–1362. 10.1126/science.1215909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas Y.F., Yoshino K., Dagdas G., Ryder L.S., Bielska E., Steinberg G., and Talbot N.J.. 2012. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science. 336:1590–1595. 10.1126/science.1222934 [DOI] [PubMed] [Google Scholar]
- Daumke O., Lundmark R., Vallis Y., Martens S., Butler P.J., and McMahon H.T.. 2007. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 449:923–927. 10.1038/nature06173 [DOI] [PubMed] [Google Scholar]
- Day C.A., Baetz N.W., Copeland C.A., Kraft L.J., Han B., Tiwari A., Drake K.R., De Luca H., Chinnapen D.J., Davidson M.W., et al. 2015. Microtubule motors power plasma membrane tubulation in clathrin-independent endocytosis. Traffic. 16:572–590. 10.1111/tra.12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C., Miserey-Lenkei S., Montagnac G., Gilles-Marsens F., Paul-Gilloteaux P., Giordano F., Waharte F., Marks M.S., Goud B., and Raposo G.. 2014. Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Reports. 6:445–454. 10.1016/j.celrep.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Haudenshield J.S., Hull R.J., Wolf S., Beachy R.N., and Lucas W.J.. 1992. Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell. 4:915–928. 10.1105/tpc.4.8.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C.M., and Hetzer M.W.. 2010. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 119:469–477. 10.1007/s00412-010-0289-2 [DOI] [PubMed] [Google Scholar]
- Estrada P., Kim J., Coleman J., Walker L., Dunn B., Takizawa P., Novick P., and Ferro-Novick S.. 2003. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 163:1255–1266. 10.1083/jcb.200304030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L., Faulkner C., Walshaw J., Saalbach G., Bayer E., Benitez-Alfonso Y., and Maule A.. 2011. Arabidopsis plasmodesmal proteome. PLoS One. 6:e18880 10.1371/journal.pone.0018880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.G., Mills I.G., Peter B.J., Vallis Y., Praefcke G.J., Evans P.R., and McMahon H.T.. 2002. Curvature of clathrin-coated pits driven by epsin. Nature. 419:361–366. 10.1038/nature01020 [DOI] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S.C., Kirchhausen T., and Walz T.. 2004. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 432:573–579. 10.1038/nature03079 [DOI] [PubMed] [Google Scholar]
- Frick M., Bright N.A., Riento K., Bray A., Merrified C., and Nichols B.J.. 2007. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 17:1151–1156. 10.1016/j.cub.2007.05.078 [DOI] [PubMed] [Google Scholar]
- Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E.H., De Camilli P., and Unger V.M.. 2008. Structural basis of membrane invagination by F-BAR domains. Cell. 132:807–817. 10.1016/j.cell.2007.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop J.L., Jao C.C., Kent H.M., Butler P.J., Evans P.R., Langen R., and McMahon H.T.. 2006. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 25:2898–2910. 10.1038/sj.emboj.7601174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabski S., De Feijter A.W., and Schindler M.. 1993. Endoplasmic reticulum forms a dynamic continuum for lipid diffusion between contiguous soybean root cells. Plant Cell. 5:25–38. 10.1105/tpc.5.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grison M.S., Brocard L., Fouillen L., Nicolas W., Wewer V., Dörmann P., Nacir H., Benitez-Alfonso Y., Claverol S., Germain V., et al. 2015. Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell. 27:1228–1250. 10.1105/tpc.114.135731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N.V., Chen K., Jin W.L., Frost A., and Polleux F.. 2009. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 138:990–1004. 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T., Ueda H., Kawase T., and Hara-Nishimura I.. 2014. Microtubules contribute to tubule elongation and anchoring of endoplasmic reticulum, resulting in high network complexity in Arabidopsis. Plant Physiol. 166:1869–1876. 10.1104/pp.114.252320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M.G. IV, Mela I., Wang L., Henderson R.M., Chapman E.R., Edwardson J.M., and Audhya A.. 2016. Sar1 GTPase activity is regulated by membrane curvature. J. Biol. Chem. 291:1014–1027. 10.1074/jbc.M115.672287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.G., Bright N.A., Howard G., and Nichols B.J.. 2009. SDPR induces membrane curvature and functions in the formation of caveolae. Nat. Cell Biol. 11:807–814. 10.1038/ncb1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.G., Howard G., and Nichols B.J.. 2011. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J. Cell Sci. 124:2777–2785. 10.1242/jcs.084319 [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth R., Lin Y., and Heuser J.E.. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180:389–402. 10.1083/jcb.200707031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H., Bhattacharya N., Johnson K., Noble A.J., and Stagg S.M.. 2014. Insights into the mechanisms of membrane curvature and vesicle scission by the small GTPase Sar1 in the early secretory pathway. J. Mol. Biol. 426:3811–3826. 10.1016/j.jmb.2014.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., and Neupert W.. 2011. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30:4356–4370. 10.1038/emboj.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich W. 1973. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 28:693–703. [DOI] [PubMed] [Google Scholar]
- Helfrich P., and Jakobsson E.. 1990. Calculation of deformation energies and conformations in lipid membranes containing gramicidin channels. Biophys. J. 57:1075–1084. 10.1016/S0006-3495(90)82625-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Kent H.M., Ford M.G., Hegde B.G., Daumke O., Butler P.J., Mittal R., Langen R., Evans P.R., and McMahon H.T.. 2007. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 15:839–852. 10.1016/j.str.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Boucrot E., Meinecke M., Evergren E., Vallis Y., Mittal R., and McMahon H.T.. 2010. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 328:1281–1284. 10.1126/science.1188462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., Zhao Y., and Emr S.D.. 2012. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 151:356–371. 10.1016/j.cell.2012.08.039 [DOI] [PubMed] [Google Scholar]
- Hoelz A., Debler E.W., and Blobel G.. 2011. The structure of the nuclear pore complex. Annu. Rev. Biochem. 80:613–643. 10.1146/annurev-biochem-060109-151030 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., and Prinz W.A.. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250. 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., and Blackstone C.. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561. 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.D., Townley A.K., Danson C.M., Cullen P.J., and Stephens D.J.. 2013. Microtubule motors mediate endosomal sorting by maintaining functional domain organization. J. Cell Sci. 126:2493–2501. 10.1242/jcs.122317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isas J.M., Ambroso M.R., Hegde P.B., Langen J., and Langen R.. 2015. Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding. Structure. 23:873–881. 10.1016/j.str.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M., Yokono M., and Nakano A.. 2014. Visualizing structural dynamics of thylakoid membranes. Sci. Rep. 4:3768 10.1038/srep03768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C.C., Hegde B.G., Gallop J.L., Hegde P.B., McMahon H.T., Haworth I.S., and Langen R.. 2010. Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. J. Biol. Chem. 285:20164–20170. 10.1074/jbc.M110.127811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiko C., Davies K.M., Shinzawa-Itoh K., Tani K., Maeda S., Mills D.J., Tsukihara T., Fujiyoshi Y., Kühlbrandt W., and Gerle C.. 2015. Bovine F1Fo ATP synthase monomers bend the lipid bilayer in 2D membrane crystals. eLife. 4:e06119 10.7554/eLife.06119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J. 2013. Importance of the hexagonal lipid phase in biological membrane organization. Front. Plant Sci. 4:494 10.3389/fpls.2013.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabachinski G., and Schwartz T.U.. 2015. The nuclear pore complex--structure and function at a glance. J. Cell Sci. 128:423–429. 10.1242/jcs.083246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K., Wang P., Kriechbaumer V., Tilsner J., Frigerio L., Sparkes I., Hawes C., and Oparka K.. 2015. Putting the squeeze on plasmodesmata: A role for reticulons in primary plasmodesmata formation. Plant Physiol. 168:1563–1572. 10.1104/pp.15.00668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M., Jia J.Y., Roux A., Beck R., Wieland F.T., De Camilli P., and Haucke V.. 2008. Arf1-GTP-induced tubule formation suggests a function of Arf family proteins in curvature acquisition at sites of vesicle budding. J. Biol. Chem. 283:27717–27723. 10.1074/jbc.M804528200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V., Botchway S.W., Slade S.E., Knox K., Frigerio L., Oparka K., and Hawes C.. 2015. Reticulomics: protein-protein interaction studies with two plasmodesmata-localized reticulon family proteins identify binding partners enriched at plasmodesmata, endoplasmic reticulum, and the plasma membrane. Plant Physiol. 169:1933–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S., Erdmann C., Kage F., Block J., Schwenkmezger L., Steffen A., Rottner K., and Geyer M.. 2015. The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation. Nat. Commun. 6:7088 10.1038/ncomms8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Suzuki K.G., Kasai R.S., Ritchie K., and Fujiwara T.K.. 2011. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 36:604–615. 10.1016/j.tibs.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Lee M.C., Orci L., Hamamoto S., Futai E., Ravazzola M., and Schekman R.. 2005. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 122:605–617. 10.1016/j.cell.2005.07.025 [DOI] [PubMed] [Google Scholar]
- Lewellyn E.B., Pedersen R.T., Hong J., Lu R., Morrison H.M., and Drubin D.G.. 2015. An engineered minimal WASP-myosin fusion protein reveals essential functions for endocytosis. Dev. Cell. 35:281–294. 10.1016/j.devcel.2015.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe S., and Menzel D.. 1995. Actomyosin-based motility of endoplasmic reticulum and chloroplasts in Vallisneria mesophyll cells. Biol. Cell. 85:207–222. 10.1016/0248-4900(96)85282-8 [DOI] [PubMed] [Google Scholar]
- Liu A.P., Richmond D.L., Maibaum L., Pronk S., Geissler P.L., and Fletcher D.A.. 2008. Membrane-induced bundling of actin filaments. Nat. Phys. 4:789–793. 10.1038/nphys1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet A., Gallop J.L., Burden J.J., Camdere G., Chandra P., Vallis Y., Hopkins C.R., Lagnado L., and McMahon H.T.. 2011. Endophilin drives the fast mode of vesicle retrieval in a ribbon synapse. J. Neurosci. 31:8512–8519. 10.1523/JNEUROSCI.6223-09.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R., Doherty G.J., Howes M.T., Cortese K., Vallis Y., Parton R.G., and McMahon H.T.. 2008. The GTPase-activating protein GRAF1 regulates the CLIC/GEEC endocytic pathway. Curr. Biol. 18:1802–1808. 10.1016/j.cub.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manneville J.B., Casella J.F., Ambroggio E., Gounon P., Bertherat J., Bassereau P., Cartaud J., Antonny B., and Goud B.. 2008. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc. Natl. Acad. Sci. USA. 105:16946–16951. 10.1073/pnas.0807102105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M., Takeda S., Sone M., Ohki T., Mori H., Kamioka Y., and Mochizuki N.. 2006. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 25:2889–2897. 10.1038/sj.emboj.7601176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N.S., Matile S., Dubochet J., Sadoul R., et al. 2004. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 303:531–534. 10.1126/science.1092425 [DOI] [PubMed] [Google Scholar]
- Mattila P.K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454. 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- Mattila P.K., Pykäläinen A., Saarikangas J., Paavilainen V.O., Vihinen H., Jokitalo E., and Lappalainen P.. 2007. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 176:953–964. 10.1083/jcb.200609176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A., Faulkner C., and Benitez-Alfonso Y.. 2012. Plasmodesmata “in Communicado”. Front. Plant Sci. 3:30 10.3389/fpls.2012.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M., Neuspiel M., and Wasiak S.. 2006. Mitochondria: more than just a powerhouse. Curr. Biol. 16:R551–R560. 10.1016/j.cub.2006.06.054 [DOI] [PubMed] [Google Scholar]
- McCullough J., Fisher R.D., Whitby F.G., Sundquist W.I., and Hill C.P.. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA. 105:7687–7691. 10.1073/pnas.0801567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Clippinger A.K., Talledge N., Skowyra M.L., Saunders M.G., Naismith T.V., Colf L.A., Afonine P., Arthur C., Sundquist W.I., et al. 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 350:1548–1551. 10.1126/science.aad8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., and Boucrot E.. 2015. Membrane curvature at a glance. J. Cell Sci. 128:1065–1070. 10.1242/jcs.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., and Gallop J.L.. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438:590–596. 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- Mészáros N., Cibulka J., Mendiburo M.J., Romanauska A., Schneider M., and Köhler A.. 2015. Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev. Cell. 33:285–298. 10.1016/j.devcel.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.E., Mathiasen S., Bright N.A., Pierre F., Kelly B.T., Kladt N., Schauss A., Merrifield C.J., Stamou D., Höning S., and Owen D.J.. 2015. CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev. Cell. 33:163–175. 10.1016/j.devcel.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S., Parton R.G., Vogel F., Behlke J., Henske A., and Kurzchalia T.V.. 1995. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell. 6:911–927. 10.1091/mbc.6.7.911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morén B., Shah C., Howes M.T., Schieber N.L., McMahon H.T., Parton R.G., Daumke O., and Lundmark R.. 2012. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol. Biol. Cell. 23:1316–1329. 10.1091/mbc.E11-09-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot S., Galli V., Klein M., Chiaruttini N., Manzi J., Humbert F., Dinis L., Lenz M., Cappello G., and Roux A.. 2012. Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell. 151:619–629. 10.1016/j.cell.2012.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow I.C., and Parton R.G.. 2005. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 6:725–740. 10.1111/j.1600-0854.2005.00318.x [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W. 2005. Function and evolution of grana. Trends Plant Sci. 10:521–525. 10.1016/j.tplants.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Mustárdy L., and Garab G.. 2003. Granum revisited. A three-dimensional model--where things fall into place. Trends Plant Sci. 8:117–122. 10.1016/S1360-1385(03)00015-3 [DOI] [PubMed] [Google Scholar]
- Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J., Evans P.R., and McMahon H.T.. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 303:495–499. 10.1126/science.1092586 [DOI] [PubMed] [Google Scholar]
- Pfanner N., van der Laan M., Amati P., Capaldi R.A., Caudy A.A., Chacinska A., Darshi M., Deckers M., Hoppins S., Icho T., et al. 2014. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J. Cell Biol. 204:1083–1086. 10.1083/jcb.201401006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost C., Zhao H., Manzi J., Lemichez E., Lappalainen P., Callan-Jones A., and Bassereau P.. 2015. IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat. Commun. 6:8529 10.1038/ncomms9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W.A., Grzyb L., Veenhuis M., Kahana J.A., Silver P.A., and Rapoport T.A.. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461–474. 10.1083/jcb.150.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Lauffer B., Temkin P., Vistein R., Carlton P., Thorn K., Taunton J., Weiner O.D., Parton R.G., and von Zastrow M.. 2010. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 143:761–773. 10.1016/j.cell.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quader H., Hofmann A., and Schnepf E.. 1989. Reorganization of the endoplasmic reticulum in epidermal cells of onion bulb scales after cold stress: Involvement of cytoskeletal elements. Planta. 177:273–280. 10.1007/BF00392816 [DOI] [PubMed] [Google Scholar]
- Renard H.F., Simunovic M., Lemière J., Boucrot E., Garcia-Castillo M.D., Arumugam S., Chambon V., Lamaze C., Wunder C., Kenworthy A.K., et al. 2015. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature. 517:493–496. 10.1038/nature14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer W., Berland L., Chambon V., Gaus K., Windschiegl B., Tenza D., Aly M.R., Fraisier V., Florent J.C., Perrais D., et al. 2007. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 450:670–675. 10.1038/nature05996 [DOI] [PubMed] [Google Scholar]
- Rounds C.M., and Bezanilla M.. 2013. Growth mechanisms in tip-growing plant cells. Annu. Rev. Plant Biol. 64:243–265. 10.1146/annurev-arplant-050312-120150 [DOI] [PubMed] [Google Scholar]
- Roux A., Cappello G., Cartaud J., Prost J., Goud B., and Bassereau P.. 2002. A minimal system allowing tubulation with molecular motors pulling on giant liposomes. Proc. Natl. Acad. Sci. USA. 99:5394–5399. 10.1073/pnas.082107299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J., Hakanen J., Mattila P.K., Grumet M., Salminen M., and Lappalainen P.. 2008. ABBA regulates plasma-membrane and actin dynamics to promote radial glia extension. J. Cell Sci. 121:1444–1454. 10.1242/jcs.027466 [DOI] [PubMed] [Google Scholar]
- Saarikangas J., Zhao H., Pykäläinen A., Laurinmäki P., Mattila P.K., Kinnunen P.K., Butcher S.J., and Lappalainen P.. 2009. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 19:95–107. 10.1016/j.cub.2008.12.029 [DOI] [PubMed] [Google Scholar]
- Saarikangas J., Kourdougli N., Senju Y., Chazal G., Segerstråle M., Minkeviciene R., Kuurne J., Mattila P.K., Garrett L., Hölter S.M., et al. 2015. MIM-induced membrane bending promotes dendritic spine initiation. Dev. Cell. 33:644–659. 10.1016/j.devcel.2015.04.014 [DOI] [PubMed] [Google Scholar]
- Schmid S.L., and Mettlen M.. 2013. Cell biology: Lipid switches and traffic control. Nature. 499:161–162. 10.1038/nature12408 [DOI] [PubMed] [Google Scholar]
- Senju Y., Itoh Y., Takano K., Hamada S., and Suetsugu S.. 2011. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J. Cell Sci. 124:2032–2040. 10.1242/jcs.086264 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., and Voeltz G.K.. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904. 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., and Maule A.J.. 2009. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell. 21:581–594. 10.1105/tpc.108.060145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S.J. 1972. A fluid lipid-globular protein mosaic model of membrane structure. Ann. N.Y. Acad. Sci. 195:16–23. 10.1111/j.1749-6632.1972.tb54780.x [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Frigerio L., Tolley N., and Hawes C.. 2009. The plant endoplasmic reticulum: a cell-wide web. Biochem. J. 423:145–155. 10.1042/BJ20091113 [DOI] [PubMed] [Google Scholar]
- Stachowiak J.C., Schmid E.M., Ryan C.J., Ann H.S., Sasaki D.Y., Sherman M.B., Geissler P.L., Fletcher D.A., and Hayden C.C.. 2012. Membrane bending by protein-protein crowding. Nat. Cell Biol. 14:944–949. 10.1038/ncb2561 [DOI] [PubMed] [Google Scholar]
- Stachowiak J.C., Brodsky F.M., and Miller E.A.. 2013. A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat. Cell Biol. 15:1019–1027. 10.1038/ncb2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Amari K., and Torrance L.. 2011. Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma. 248:39–60. 10.1007/s00709-010-0217-6 [DOI] [PubMed] [Google Scholar]
- van Weering J.R., and Cullen P.J.. 2014. Membrane-associated cargo recycling by tubule-based endosomal sorting. Semin. Cell Dev. Biol. 31:40–47. 10.1016/j.semcdb.2014.03.015 [DOI] [PubMed] [Google Scholar]
- van Weering J.R., Sessions R.B., Traer C.J., Kloer D.P., Bhatia V.K., Stamou D., Carlsson S.R., Hurley J.H., and Cullen P.J.. 2012. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 31:4466–4480. 10.1038/emboj.2012.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Vollmer B., Schooley A., Sachdev R., Eisenhardt N., Schneider A.M., Sieverding C., Madlung J., Gerken U., Macek B., and Antonin W.. 2012. Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 31:4072–4084. 10.1038/emboj.2012.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer B., Lorenz M., Moreno-Andrés D., Bodenhöfer M., De Magistris P., Astrinidis S.A., Schooley A., Flötenmeyer M., Leptihn S., and Antonin W.. 2015. Nup153 recruits the Nup107-160 complex to the inner nuclear membrane for interphasic nuclear pore complex assembly. Dev. Cell. 33:717–728. 10.1016/j.devcel.2015.04.027 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., and Salmon E.D.. 1998. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 8:798–807. 10.1016/S0960-9822(98)70321-5 [DOI] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., and Hurley J.H.. 2009. Membrane scission by the ESCRT-III complex. Nature. 458:172–177. 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]