Mutations in the retromer complex, which is involved in sorting integral membrane proteins from endosomes to cellular compartments, are associated with atypical parkinsonism, but how these mutations affect retromer function remains unclear. Through a quantitative proteomic analysis of the retromer interactome, McMillan et al. reveal a new mechanism for perturbed endosomal sorting in parkinsonism.
Abstract
The retromer complex acts as a scaffold for endosomal protein complexes that sort integral membrane proteins to various cellular destinations. The retromer complex is a heterotrimer of VPS29, VPS35, and VPS26. Two of these paralogues, VPS26A and VPS26B, are expressed in humans. Retromer dysfunction is associated with neurodegenerative disease, and recently, three VPS26A mutations (p.K93E, p.M112V, and p.K297X) were discovered to be associated with atypical parkinsonism. Here, we apply quantitative proteomics to provide a detailed description of the retromer interactome. By establishing a comparative proteomic methodology, we identify how this interactome is perturbed in atypical parkinsonism-associated VPS26A mutants. In particular, we describe a selective defect in the association of VPS26A (p.K297X) with the SNX27 cargo adaptor. By showing how a retromer mutant leads to altered endosomal sorting of specific PDZ ligand–containing cargo proteins, we reveal a new mechanism for perturbed endosomal cargo sorting in atypical parkinsonism.
Introduction
Retromer is a highly conserved heterotrimer of VPS29, VPS35, and VPS26. Two of these paralogues, VPS26A and VPS26B, are expressed in humans (Seaman, 2012; Burd and Cullen, 2014). Retromer is associated with the cytosolic face of endosomes where it scaffolds a multiprotein complex that orchestrates the sorting of integral membrane proteins (i.e., cargos) into transport carriers destined for the plasma membrane, the trans-Golgi network, and specialized organelles (Seaman et al., 1998; Cullen and Korswagen, 2011; Burd and Cullen, 2014). Defects in retromer are associated with neurological disease. Retromer expression is lowered in brains of patients with Alzheimer’s disease and Parkinson’s disease, and retromer mutations are observed in familial and sporadic forms of these diseases (Small et al., 2005; Muhammad et al., 2008; Vilariño-Güell et al., 2011, 2014; Zimprich et al., 2011; Vardarajan et al., 2012; MacLeod et al., 2013; Shannon et al., 2014; Rovelet-Lecrux et al., 2015). For example, the autosomal dominant Parkinson disease––linked VPS35 (p.D620N) mutation leads to perturbed retromer function by disrupting the association with the actin-nucleating Wiskott-Aldrich syndrome and SCAR homologue (WASH) complex (McGough et al., 2014a; Zavodszky et al., 2014). Further mutations in the VPS26A subunit (VPS26A [p.K93E], VPS26A [p.M112V], and VPS26A [p.K297X]) have also been linked to atypical parkinsonism (Gustavsson et al., 2015). How these mutations perturb retromer function remains unclear. Alterations in retromer accessory proteins are observed in neurological disease, including the retromer cargo adaptor sorting nexin-27 (SNX27) in Down’s syndrome and infantile myoclonic epilepsy (Wang et al., 2013; Damseh et al., 2015). Identifying retromer accessory proteins and how they assemble to form a functional complex is essential in defining the molecular details of retromer activity and in providing insight into the pathoetiology of retromer-associated disease.
Here, we have applied proteomics to provide the first detailed, quantitative description of the retromer interactome. By establishing a comparative proteomic methodology, we identify how this interactome is perturbed in VPS26A mutants in patients with atypical parkinsonism (Gustavsson et al., 2015). In particular, we describe the identification of a selective defect in the association of VPS26A (p.K297X) with SNX27. By establishing that this leads to perturbed endosomal sorting of specific cargo proteins, we reveal a new mechanism for perturbed endosomal trafficking in parkinsonism.
Results and discussion
Quantitative identification of interacting proteins for individual retromer subunits
Retromer’s role in cargo sorting is mediated, in part, through an ability to recruit accessory proteins (Harbour et al., 2010). Limited information is available regarding the range of retromer-interacting proteins. We previously identified VPS35-interacting proteins using stable isotope labeling with amino acids in cell culture (SILAC) proteomics (McGough et al., 2014a,b). Here, we extended this procedure using VPS29, VPS26A, and VPS26B to validate known interactions, to determine whether VPS26A and VPS26B display distinct interactions that may distinguish their functions, and to increase the detection power of our overall analysis. Previously, proteins not strongly enriched in the VPS35 interactome were excluded (McGough et al., 2014b). Such proteins may be relevant to retromer function, with their low enrichment due to weak interaction or through association with a subunit other than VPS35. Comparison of the VPS35 interactome with those for VPS29, VPS26A, and VPS26B would highlight these proteins, despite them being weakly enriched in an individual interactome.
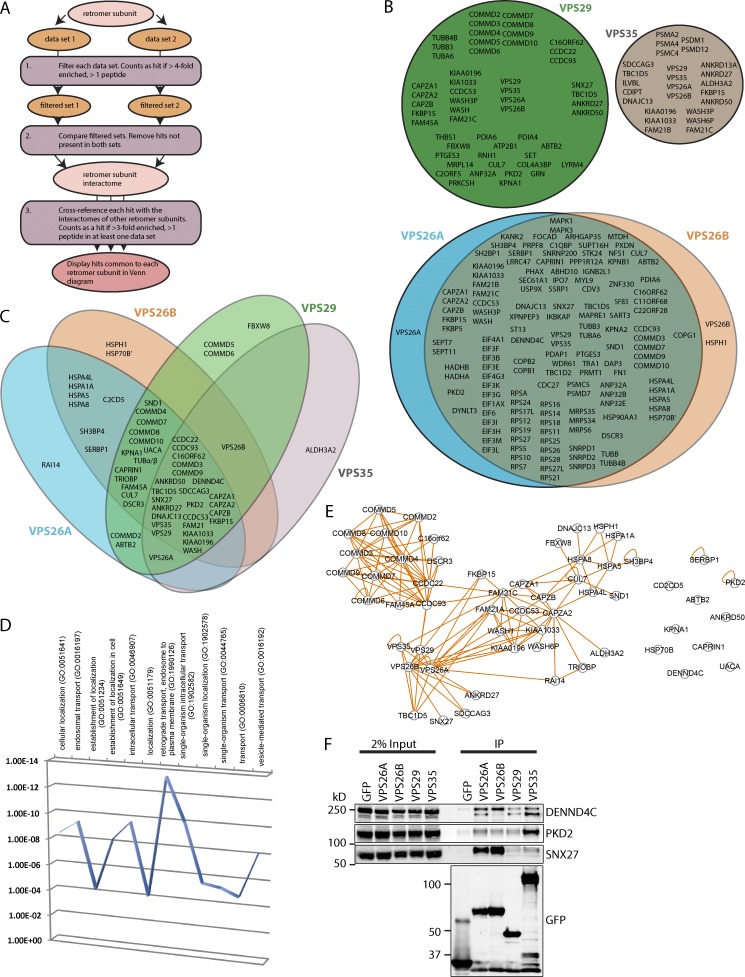
For these SILAC experiments,we lentivirally transduced human retinal pigment epithelial-1 (RPE-1) cells to generate cell populations expressing GFP-tagged VPS26A, VPS26B, or VPS29 (Fig. S1, A–F). For the VPS29 interactome, we grew GFP-VPS29 expressing cells in amino acids of “medium” mass (R6K4), alongside cells expressing GFP grown in unlabeled, “light” amino acids (R0K0). We subjected these cells to GFP-Trap (ChromoTek) immunoprecipitation, resolved the combined coimmunoprecipitates by SDS-PAGE and identified proteins using liquid chromatography–tandem mass spectrometry. From duplicate experiments, a single list of VPS29-interacting proteins was generated by excluding proteins quantified from a single unique peptide and any that were less than fourfold enriched in the GFP-VPS29 immunoprecipitate (Fig. 1 A and Table S1). The two filtered lists were combined by excluding proteins not present in both lists, leading to 53 proteins being identified as the VPS29 interactome (Fig. 1, A and B; and Table S1).
Figure 1.
Identification of the detailed retromer interactome. (A) Schematic describing the filtering of retromer subunit interactomes. (B) Venn diagram showing the proteins that interact with VPS29 (green), VPS35 (gray), VPS26A (blue), and VPS26B (orange). (C) Venn diagram showing retromer accessory proteins. Color code as in B. (D) Proteins identified in C were subjected to gene ontology analysis using the PANTHER classification system. (E) Proteins identified in C were subjected to esyN analysis generating a network of protein interactions. Each node represents a protein, and each connecting line represents an interaction indicated by experimental evidence. (F) RPE-1 cells expressing GFP-tagged VPS26A, VPS26B, VPS29, and VPS35 were subjected to GFP-nanotrap immunoisolation followed by quantitative Western analysis for validation of DENND4C, PKD2, and SNX27 association. Data are from a single experiment representative of three independent experiments.
For VPS26A and VPS26B interactomes, we used triple SILAC, growing GFP, GFP-VPS26A, and GFP-VPS26B expressing cells in R0K0, R6K4, and “heavy” (R10K8) labeled medium, respectively. From duplicate experiments, we again generated a single list of proteins that interacted with each of the VPS26 paralogues (Fig. 1 A and Table S1), with the difference that we considered both the R6K4/R0K0 ratio and count (for VPS26A) and the R10K8/R0K0 ratio and count (for VPS26B) when filtering. If a protein met the required criteria for inclusion in the filtered list for one VPS26 paralogue but not the other, we included that protein to avoid removing proteins that interact with only one paralogue. The identified 141 proteins defined the VPS26 interactome (Fig. 1, A and B; and Table S1).
To gain insight into potential functional differences between VPS26 paralogues, we sorted to identify paralogue-specific interactomes. Here we examined the mean R10K8/R6K4 ratio. For the VPS26B-specific interactome, we excluded proteins with a mean R10K8/R6K4 ratio less than 10, whereas for the VPS26A-specific interactome, we excluded proteins with a mean R10K8/R6K4 ratio greater than 0.1. Using these criteria, all VPS26 interactors were common to VPS26A and VPS26B, with the only exception being VPS26B’s association with a heat shock protein, HSPH1 (Fig. 1 B and Table S1).
Identification of the detailed retromer interactome
In the same parental RPE-1 cells, we previously generated a VPS35 interactome in duplicate (McGough et al., 2014a,b), so we combined these data into a single VPS35 interactome (Fig. 1 A and Table S1). To identify proteins consistently immunoprecipitated by retromer, we identified proteins common to the VPS35, VPS29, and VPS26A and VPS26B interactomes. To avoid excluding proteins that may have been filtered out of the individual interactomes, we cross-referenced each protein in the retromer interactome with the unfiltered lists for each retromer subunit. If a protein was enriched by at least twofold in an unfiltered list, we considered that protein to be present in the interactome of that retromer subunit for our comparison (Fig. 1 C). Gene ontology analysis (PANTHER Classification System; P < 0.05) revealed the majority of interactors had roles in endosomal transport (Fig. 1 D). Network analysis using esyN (Fig. 1 E; Bean et al., 2014) revealed known retromer interactors and the WASH complex and its accessory proteins (Gomez and Billadeau, 2009; Harbour et al., 2010, 2012), including the CCC complex (Phillips-Krawczak et al., 2015) and various other proteins including SNX27 (Temkin et al., 2011; Steinberg et al., 2013), ANKRD27 (VARP; Hesketh et al., 2014; McGough et al., 2014a; Bean et al., 2015), ANKRD50 and SDCCAG3 (McGough et al., 2014a), TBC1D5 (Seaman et al., 2009), and DNAJC13 (Popoff et al., 2007; Shi et al., 2009; Freeman et al., 2014). Most of these core retromer accessory proteins were located within the central region of the Venn diagram (Fig. 1 C). This region contained two proteins not previously shown to interact with retromer, the Rab10 guanine-nucleotide exchange factor DENND4C (Yoshimura et al., 2010) and the polycystin family member PKD2 (Chapin and Caplan, 2010). GFP-nanotrap immunoisolation and quantitative Western analysis confirmed association of retromer with several interactors including SNX27, DENND4C, and PKD2 (Fig. 1 F). DENND4C and PKD2 proteins colocalized with retromer-decorated endosomes (Fig. S2, A and B), and the functional relevance of these interactions is described in Fig. 3 and the Discussion.
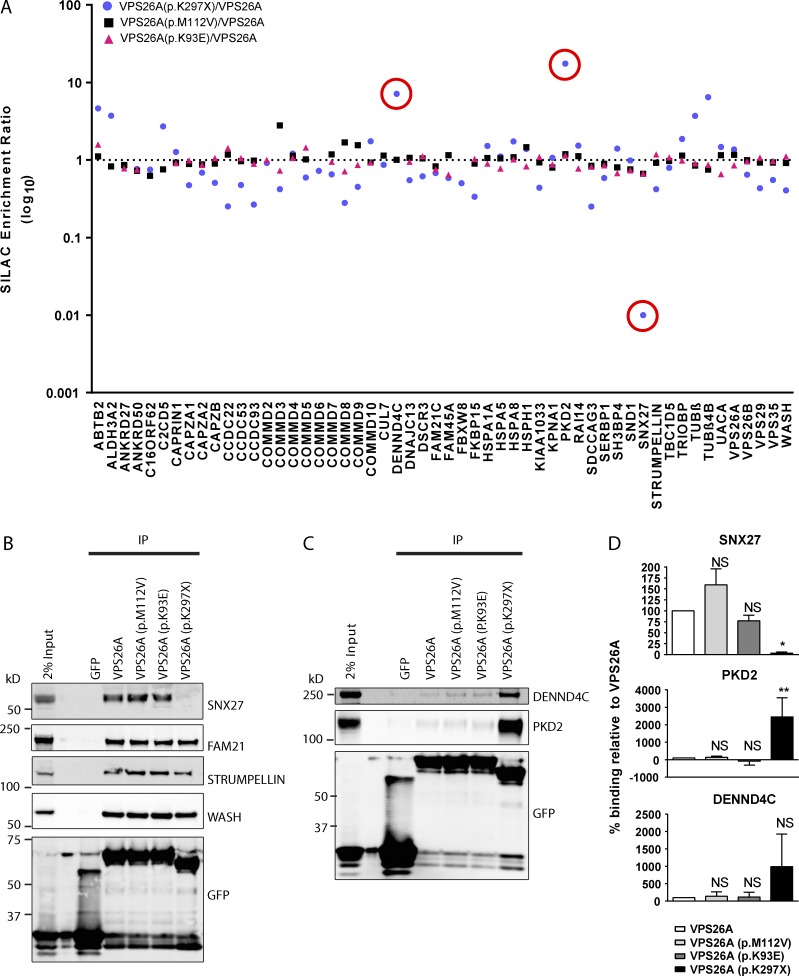
Figure 3.
VPS26A (p.K297X) leads to a loss of and enhancement of VPS26A’s association with specific components of its interactome. (A) Logarithmic graph showing the interactors identified from comparative SILAC proteomics of GFP-VPS26A versus GFP-VPS26A (p.K93E), GFP-VPS26A (p.M112V), or GFP-VPS26A (p.K297X). The SILAC ratio is the fold-enrichment of proteins in GFP-VPS26A mutant over GFP-VPS26A. Red circles indicate either a pronounced enhancement or loss of association with GFP-VPS26A (K297X). Data is mean of n = 2–3 independent experiments. (B and C) Fluorescence-based Western analysis after GFP-Trap immunoprecipitation of GFP-VPS26A, GFP-VPS26A (p.K93E), GFP-VPS26A (p.M112V), and GFP-VPS26A (p.K297X). (D) Quantification of data from three independent experiments. Data are expressed as a percentage of the GFP-VPS26A control and analyzed by a one-way ANOVA followed by a Dunnett posthoc test. Error bars represent mean ± SEM. NS, not significant; *, P < 0.05; **, P < 0.01.
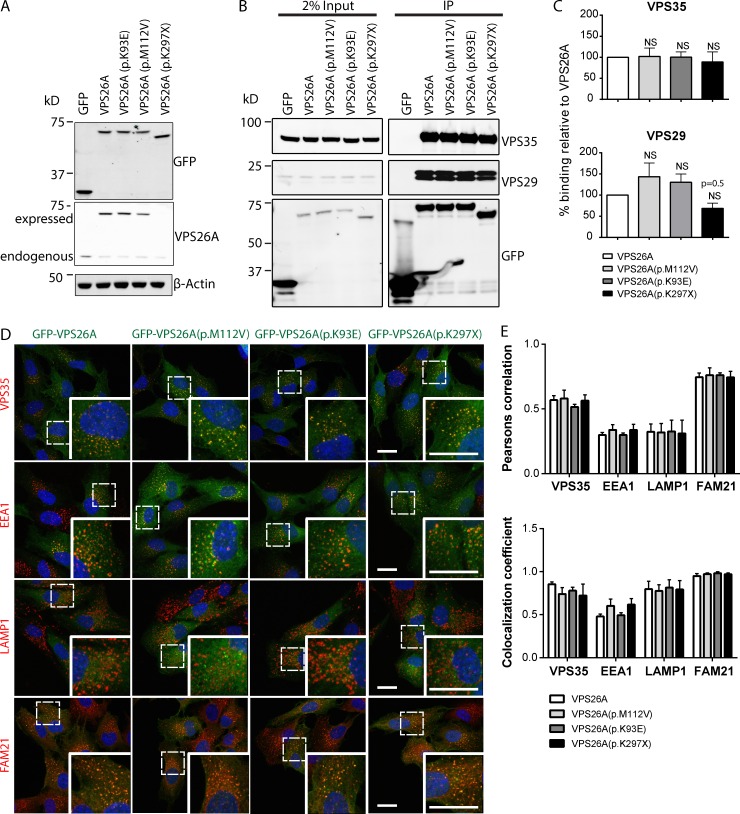
Missense VPS26A mutants do not perturb heterotrimeric retromer assembly or its endosome association
We then turned our attention to how the atypical parkinsonism-associated VPS26A mutants affect assembly of the retromer interactome. Using lentiviruses to transduce RPE-1 cells with constructs encoding for GFP-VPS26A, GFP-VPS26A (p.K93E), GFP-VPS26A (p.M112V), or GFP-VPS26A (p.K297X), we titrated the viral load to produce cell lines where expression of the GFP-tagged transgene approached endogenous levels and was observed to lower the expression of endogenous VPS26A, presumably as a result of competition for inclusion of the GFP-tagged transgene into the stable heterotrimeric complex and resultant destabilization of the noncomplexed endogenous VPS26A (Fig. 2 A). Note that truncation of the last 31 amino acids of VPS26A leads to the molecular weight of the GFP-tagged protein being lower (Fig. 2 A). GFP-nanotrap immunoisolation and quantitative Western analysis revealed that each VPS26A mutant retained the ability to form a heterotrimeric complex (Fig. 2, B and C). Quantitative confocal imaging established that each mutant retained association with endosomes labeled for retromer (VPS35 positive) and the WASH complex (FAM21 positive; Fig. 2, D and E).
Figure 2.
Missense VPS26A mutants do not perturb assembly of the retromer heterotrimer or its endosome association. (A) Fluorescence-based Western blot showing the expression levels of GFP-VPS26A and GFP-VPS26A mutants compared with endogenous VPS26A. Expression of GFP-VPS26A (p.K297X) cannot be seen because of the antibody recognizing the C terminus of VPS26A. (B) Fluorescence-based Western analysis after GFP-Trap immunoprecipitation of GFP-VPS26A and GFP-VPS26A mutants with endogenous VPS35 and VPS29. (C) Quantification of B from three (VPS35) and four (VPS29) independent experiments. Data expressed as a percentage of the GFP-VPS26A control. Error bars represent mean ± SEM. No statistically significant difference was observed. NS, not significant. (D) Immunofluorescence staining of VPS35, EEA1, LAMP1, and FAM21 in RPE-1 cells expressing GFP-VPS26A or the GFP-VPS26A mutants. Bars, 20 µm. (E) Colocalization of GFP-VPS26A or GFP-VPS26A mutants with VPS35, EEA1, LAMP1, or FAM21 from three independent experiments. Error bars represent mean ± SEM. No statistically significant difference was found.
VPS26A (p.K297X) displays a loss of association with the cargo adaptor SNX27 and an enhanced association with PKD2 and DENND4C
To identify loss-of-function and gain-of-function phenotypes associated with each VPS26A mutant, we developed an unbiased, quantitative, and directly comparative proteomic methodology. RPE-1 cells lentivirally transduced with encoding constructs for wild-type GFP-VPS26A or GFP-VPS26A mutants were grown in light and medium-heavy SILAC media. Cells were subjected to GFP-Trap, the immunoprecipitates combined and the proteins resolved by SDS-PAGE and identified by liquid chromatography–tandem mass spectrometry. Data were expressed as a quantified enrichment ratio of the VPS26A mutant over wild-type VPS26A. A ratio for the interacting protein approaching 1.0 was indicative of unperturbed association, whereas a ratio approaching 0.1 or 10.0 was indicative of a reduced or enhanced association to the VPS26A mutant, respectively (Fig. 3 A).
Consistent with the data in Fig. 2 (B and C), all VPS26A mutants retained an unperturbed enrichment with VPS29 and VPS35 as well as with the WASH complex (Fig. 3 A). For the VPS26A (p.K93E) and VPS26A (p.M112V) mutants, no major alterations were observed with any of the core retromer accessory proteins (enrichment ratios between 0.6 and 2.8). In contrast, the VPS26A (p.K297X) mutant showed a dramatic reduction in SNX27 association (ratio of 0.01). An increase in the enrichment ratio of VPS26A (p.K297X) for PKD2 (ratio of 17.172) and DENND4C (ratio of 6.555) was also observed, which we confirmed through quantitative Western analysis (Fig. 3, B–D). Thus, truncation of the last 31 amino acids of VPS26A, as observed in VPS26A (p.K297X), leads to a pronounced loss of and enhancement of VPS26A’s ability to associate with specific retromer accessory proteins.
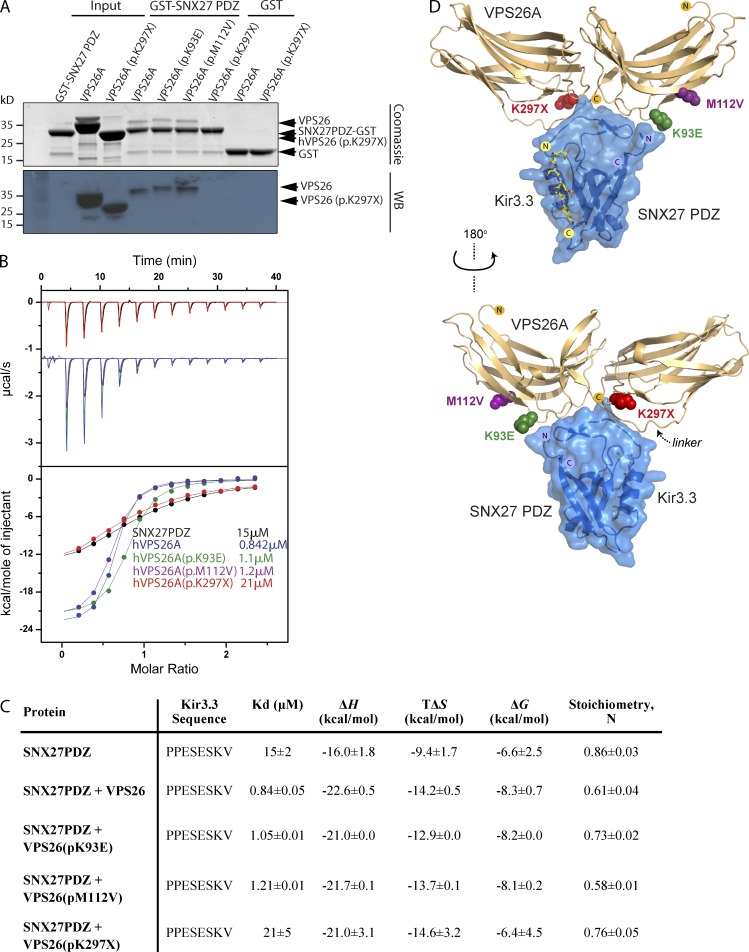
VPS26A (p.K297X) fails to associate with SNX27 in direct binding assays
The PDZ domain of SNX27 binds to VPS26 (Gallon et al., 2014). Using purified proteins, binding of VPS26A to GST-SNX27 was readily observed by Coomassie staining and Western blotting (Fig. 4 A). The same was true for VPS26A (p.K93E) and VPS26A (p.M112V), but binding to VPS26A (p.K297X) was undetectable. Binding of VPS26A to SNX27 enhances SNX27’s affinity for PDZ ligands (Gallon et al., 2014; Chan et al., 2016). In agreement with the pull-down experiments, VPS26A (p.K93E) and VPS26A (p.M112V) enhanced binding of SNX27 to a peptide corresponding to the PDZ ligand of Kir3.3 by more than 10-fold, whereas VPS26A (p.K297X) displayed no ability to enhance this binding affinity (Fig. 4, B and C).
Figure 4.
VPS26A (p.K297X) mutation directly impairs SNX27 interaction. (A) Binding of GST-SNX27 PDZ domain to purified His-tagged VPS26A mutants as detected by Coomassie staining (top) or Western blotting with anti-His antibody (bottom). (B) Binding of the Kir3.3 peptide to the SNX27 PDZ domain was measured by ITC either to SNX27 alone (black circles) or in the presence of wild-type VPS26A (blue circles) or the VPS26A mutants (green through to red circles). Top shows raw data and bottom shows integrated and normalized data. (C) Binding affinities of the Kir3.3 peptide (PPESESKV) to the SNX27 PDZ domain. (D) Structure of VPS26A (gold ribbon) in complex with SNX27 (blue ribbon and surface; Gallon et al., 2014), indicating the sites of VPS26A mutants (colored spheres). The site of Kir3.3 interaction is modeled based on the SNX27-Kir3.3 structure (yellow sticks; Balana et al., 2011).
Structurally, the β-hairpin in the SNX27 PDZ domain binds to a groove in the arrestin-like structure of VPS26A (Gallon et al., 2014). Within this structure, Lys93 and Met112 of VPS26A reside in close proximity to the SNX27 interacting surface, but they do not form direct contacts (Fig. 4 D). Lys297 also does not directly contact SNX27. However, this residue and adjacent C-terminal sequences are important for stabilizing the linker (residues 152–164) between the N- and C-terminal domains. Therefore, the deletion of these C-terminal sequences likely prevents SNX27 interaction by destabilizing the linker region and preventing its binding to the SNX27 β-hairpin.
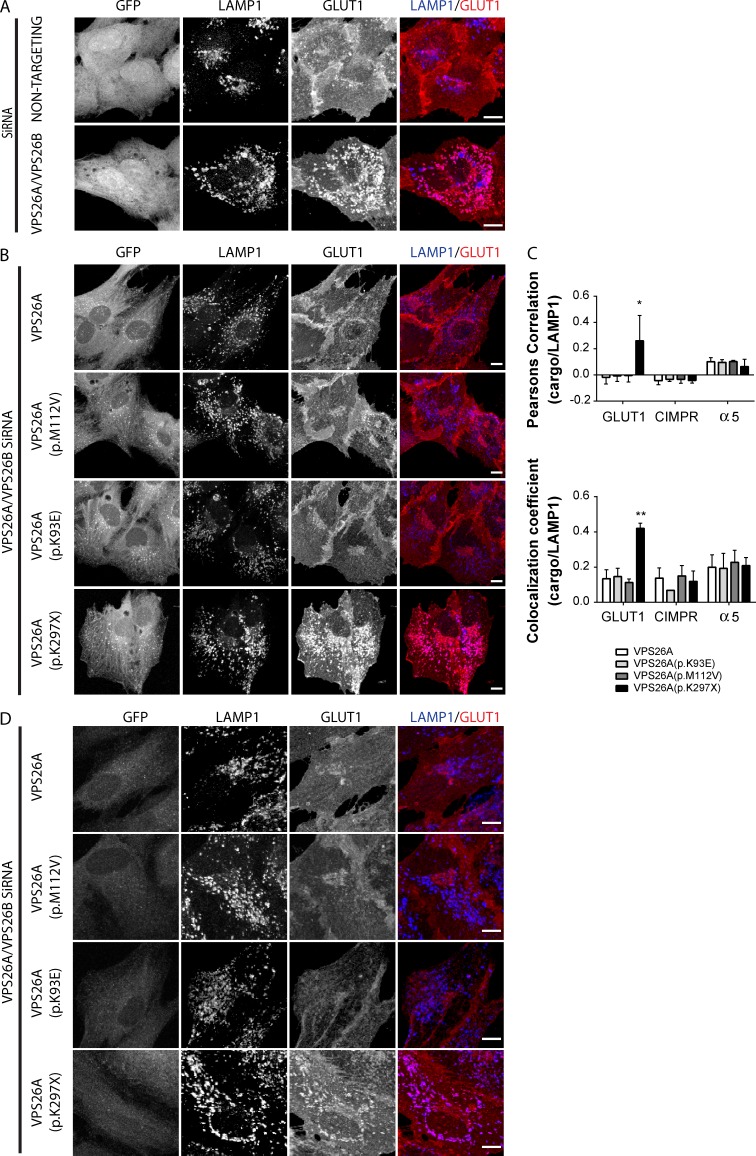
Loss of VPS26A (p.K297X) association to SNX27 leads to defective sorting of SNX27 cargo
More than 100 cell surface integral proteins require SNX27 to prevent their lysosomal degradation and maintain their cell-surface levels (Steinberg et al., 2013). Essential to this sorting is the interaction of VPS26 with SNX27 (Gallon et al., 2014). One cargo is the glucose transporter, GLUT1. Suppression of retromer or SNX27 leads to rerouting of endocytosed GLUT1 to LAMP1-labeled late endosomes/lysosomes (Steinberg et al., 2013). To establish whether the VPS26A mutations rescue the GLUT1 sorting defect observed upon RNAi-mediated suppression of VPS26 expression (Fig. 5 A; Gallon et al., 2014), siRNA-resistant, GFP-tagged VPS26A or mutant VPS26A was expressed in VPS26A- and VPS26B-suppressed RPE-1 cells. Expression of siRNA-resistant GFP-VPS26A, GFP-VPS26A (p.K93E) or GFP-VPS26A (p.M112V) fully rescued the knockdown phenotype (Fig. 5, B and C). In contrast, GFP-VPS26A (p.K297X) failed to rescue the sorting defect (Fig. 5, B and C). This mirrors the GLUT1 sorting defect upon uncoupling of SNX27’s association with retromer (Steinberg et al., 2013) and is consistent with the loss of SNX27 binding to VPS26A (p.K297X).
Figure 5.
Loss of VPS26A (p.K297X) association with SNX27 leads to the mis-trafficking of SNX27-dependent cargo. (A) Immunofluorescence costaining of GLUT1 with LAMP1 after RNAi-mediated suppression of VPS26 expression. A nontargeting RNAi was used as a control. (B) Immunofluorescence costaining of GLUT1 with LAMP1 in RPE-1 cells stably expressing GFP-VPS26 or GFP-VPS26A mutants after RNAi-mediated suppression of VPS26 expression. (C) Quantification of colocalization of GLUT1, CI-MPR, and α5-integrin with LAMP1 from three independent experiments. Data were analyzed by a one-way ANOVA followed by a Dunnett posthoc test compared with the GFP-VPS26A control. Error bars represent mean ± SEM. *, P < 0.05; **, P < 0.01. (D) Immunofluorescence costaining of GLUT1 and LAMP1 in RPE-1 cells overexpressing GFP-VPS26A or the GFP-VPS26A mutants. Bars, 10 µm.
Under these conditions, immunofluorescence staining established that steady-state distribution of the cation-independent mannose 6-phosphate receptor (CI-MPR) was unaltered (Fig. S2 C). Perturbed retromer function is characterized by an alteration in CI-MPR distribution from a normal localization at the trans-Golgi network to peripherally dispersed endosomes (Arighi et al., 2004; Seaman, 2004). Also unaltered by expression of the VPS26A mutants was the steady-state distribution of α5β1-integrin, a cargo that undergoes endosome sorting to the plasma membrane through a retromer-independent pathway (Steinberg et al., 2012; Fig. S2 D). Finally, as each VPS26A mutant was able to assemble into the retromer heterotrimer at the expense of endogenous VPS26A (Fig. 2 A), we considered their potential as function-blocking dominant negatives. In RPE-1 cells, GLUT1 steady-state distribution was significantly perturbed upon overexpression of GFP-VPS26A (p.K297X), whereas a similar phenotype was not observed with wild-type GFP-VPS26A, or GFP-VPS26A (p.K93E) or GFP-VPS26A (p.M112V; Fig. 5 D). Overall, these functional data are consistent with the unperturbed interactomes of VPS26A (p.K93E) and VPS26A (p.M112V) and establish that the inability of VPS26A (p.K297X) to associate with SNX27 manifests as a clear and specific defect in endosomal sorting of SNX27–retromer cargo.
Cargoes for SNX27-mediated sorting include neuronal receptors such as β2-adrenergic receptor, G protein–activated inward rectifying potassium type 2 receptor, serotonin receptor, and parathyroid hormone receptor, as well as N-methyl-d-aspartate and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (Joubert et al., 2004; Lauffer et al., 2010; Balana et al., 2011; Cai et al., 2011; Temkin et al., 2011; Wang et al., 2013; Chan et al., 2016). Loss of VPS26A (p.K297X) association to SNX27 therefore provides further evidence of the importance of the SNX27-retromer-WASH sorting pathway for neuronal health and provides mechanistic insight into how retromer function is perturbed in this form of atypical parkinsonism. In developing an unbiased and comparative proteomic methodology, we provide a powerful tool for defining disease-associated loss-of-function and gain-of-function phenotypes as typified by the enhancement of VPS26A (p.K297X) binding to two newly identified retromer interacting proteins—PKD2 and DENND4C. Although the role of these proteins in retromer-mediated sorting are unclear, our study highlights a novel regulatory feature of the C-terminal tail of VPS26A in the scaffolding function of retromer.
Materials and methods
Antibodies
Antibodies used in this study were rabbit monoclonal α5 integrin (ab150361; Abcam), mouse monoclonal β–actin (A1978; Sigma-Aldrich), rabbit monoclonal CI-MPR (ab124767; Abcam), rabbit polyclonal DENND4C (HPA014917; Sigma-Aldrich), rabbit monoclonal EEA1 (C45B10; Cell Signaling Technology), mouse monoclonal GFP (11814460001; Roche), rabbit monoclonal GLUT1 (115730; Abcam), mouse monoclonal LAMP1 (H4A3; Developmental Studies Hybridoma Bank), rabbit polyclonal PKD2 (sc-25749; Santa Cruz Biotechnology, Inc.), mouse monoclonal SNX27 (ab77799; Abcam), rabbit polyclonal Strumpellin (87442; Santa Cruz Biotechnology, Inc.), rabbit polyclonal VPS35 (97545; Abcam), rabbit monoclonal VPS35 (157220; Abcam), rabbit polyclonal VPS26A (ab137447; Abcam), rabbit polyclonal VPS29 (ab98929; Abcam), and rabbit polyclonal WASH1 and FAM21 (gifts from D.D. Billadeau, Mayo Clinic, Rochester, MN).
Generation of GFP-VPS26A and GFP-VPS26A mutant lentiviral vectors
VPS26A was subcloned from an original plasmid, which was a gift from R. Teasdale (University of Queensland, St. Lucia, Australia; Kerr et al., 2005), into the lentiviral vector pXLG3. To produce siRNA-resistant VPS26A six silent base mismatches (C187T, A189G, A192G, C195T, A198G, and A201G) were introduced into the ORF, generating resistance to VPS26A siRNA oligonucleotide 1 from the ON-TARGET plus human SMART pool (GE Healthcare). To produce siRNA-resistant VPS26B seven silent base mismatches (G795A, C798T, T799A, C800G, G804A, C805A, and C807G) were introduced into the ORF, generating resistance to VPS26B siRNA oligonucleotide 2 from the ON-TARGET plus human SMART pool (GE Healthcare; Gallon et al., 2014). The VPS26A (p.K93E), VPS26A (p.M112V), and VPS26A (p.K297X) mutations were generated using site-directed mutagenesis using the following primers: VPS26A (p.K93E): 5′-GTTCTCCAGGTAAGGCTAGTTCCTCCACTAGGTTTACAAATTCATGA-3′; VPS26A (p.M112V): 5′-GACTCAGAGCAGAAGTTATGATTTTGAATTTGTGCAAGTTGAAAAGCC-3′; VPS26A (p.K297X): 5′-GAAGTCCGGAGTTCACTAAGCATATTAGAAGCATGTTTTAG-3′.
Cell culture and generation of lentiviral vector-expressing stable RPE-1 cell lines
Lentiviral particles were produced in HEK293T cells before being added to RPE-1 cells at near endogenous levels to produce stably expressing cell lines. RPE-1 and HEK293T cells were maintained in DMEM (D5796; Sigma-Aldrich) supplemented with 10% (vol/vol) fetal bovine serum (F7524; Sigma-Aldrich) and penicillin/streptomycin (BRL 15140122; Gibco) under standard conditions.
Immunoprecipitation and Western blot analysis
RPE-1 cell lines stably transduced with the desired GFP-tagged constructs were lysed in Tris-based immunoprecipitation buffer (50 mM Tris-HCl, 0.5% NP-40, and Roche protease inhibitor cocktail) and the GFP immunoprecipitated with GFP-Trap beads (ChromoTek). Immunoblotting was performed using standard procedures. Detection was performed on an Odyssey infrared scanning system (LI-COR Biosciences) using fluorescently labeled secondary antibodies.
SILAC
RPE-1 cell lines stably transduced with the desired GFP-tagged constructs were grown in SILAC DMEM (89985; Thermo Fisher Scientific) supplemented with 10% (vol/vol) dialyzed FBS (F0392; Sigma-Aldrich). Cells were grown in light (R0K0), medium (R6K4), or heavy (R10K8) labeling for at least six passages to achieve full labeling (amino acids were obtained from Sigma-Aldrich, apart from K4, which was from Thermo Fisher Scientific). Cells were lysed in immunoprecipitation buffer (50 mM Tris-HCl, 0.5% NP-40, and Roche protease inhibitor cocktail) and GFP was immunoprecipitated with GFP-trap beads (ChromoTek) for 1 h. Samples were pooled and separated on Nupage 4–12% precast gels (Invitrogen) and subjected to liquid chromatography–tandem mass spectrometry analysis on an Orbitrap Velos mass spectrometer (Thermo Fisher Scientific).
ITC
The rat SNX27 PDZ domain and human VPS26A proteins were purified as described previously (Gallon et al., 2014; Chan et al., 2016). In brief, SNX27 PDZ domain was purified as a GST fusion protein followed by thrombin cleavage to remove the GST tag, and VPS26A was purified via an N-terminal His tag. Proteins were then gel filtered into ITC buffer (50 mM Tris, pH 8, and 100 mM NaCl) using a Superose 200 column. The synthetic Kir3.3 peptide was purchased from GenScript. ITC experiments were performed on a MicroCal iTC200 instrument in ITC buffer. Peptides were titrated into 40 µM SNX27 PDZ domain solutions at 25°C (supplemented with 40 µM hVPS26A proteins when required). Data were processed using ORIGIN to extract the thermodynamic parameters ΔH, Ka (1/Kd) and the stoichiometry n. ΔG and ΔS were derived from the relationships: ΔG = −RTlnKa and ΔG = ΔH − TΔS.
GST pull-down experiments
GST-tagged SNX27 PDZ domain (1 nmol) was mixed with VPS26A proteins (1 nmol) in 500 µl 25 mM Tris, pH 8, 300 mM NaCl, and 1 mM DTT and bound to 25 µl glutathione Sepharose resin. After 2-h incubation at 4°C, the resin was washed four times with pull-down buffer and bound proteins eluted in SDS-PAGE sample buffer. Western blot analysis was done using a nitrocellulose membrane and the iBlot semi-dry transfer system (Thermo Fisher Scientific). His-tagged proteins were detected by ECL on photographic film, using a primary mouse anti-penta-His antibody (34660; QIAGEN) and goat anti-mouse HRP-coupled secondary antibody (A16072; Thermo Fisher Scientific).
RNAi-mediated suppression of endogenous VPS26A and VPS26B
RPE-1 cells stably expressing GFP, wild-type or mutant VPS26A were transfected either with a ON-TARGET plus nontargeting control pool (GE Healthcare; sequences: 5′-UGGUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAUGUUGUGUGA-3′, 5′-UGGUUUACAUGUUUUCUGA-3′, and 5′-UGGUUUACAUGUUUUCCUA-3′) or with the VPS26A suppression oligonucleotide 1 (sequence 5′-GCUAGAACACCAAGGAAUU[DTDT]-3′) and VPS26B suppression oligonucleotide 2 (sequence 5′-GAAGUUCUCUGUGCGCUAU[DTDT]-3′) of the ON-TARGET plus human SMART pool (GE Healthcare) to suppress endogenous VPS26A and VPS26B. Cells were reverse-transfected using DharmaFECT (GE Healthcare), then transfected again 12 h later according to manufacturer’s instructions. 48 h after the second transfection, cells were fixed and stained.
Immunofluorescence staining
Cells were fixed in 4% (vol/vol) paraformaldehyde in PBS for 20 min before being permeabilized with either 0.1% (vol/vol) Triton X-100 or 0.1% (wt/vol) saponin for 5 min. Cells were incubated with 1% (wt/vol) BSA for 10 min followed by incubation for 1 h in the indicated primary antibody. The cells were then incubated with the appropriate Alexa Fluor secondary antibody (405, 488, and 568; Invitrogen) for 1 h with DAPI being used as a nuclear stain before being mounted on coverslips with Fluoromount-G (00–4958-02; eBioscience).
Image acquisition
Microscopy images were recorded at room temperature on a confocal laser-scanning microscope (SP5 AOBS; Leica Biosystems) attached to an inverted epifluorescence microscope (DMI6000; Thermo Fisher Scientific). A 63×, NA 1.4, oil immersion objective (Plan Apochromat BL; Leica Biosystems), and the standard SP5 system acquisition software and detector were used. Images were analyzed with the Volocity software package (PerkinElmer). To filter noise, thresholds were applied uniformly across conditions. The colocalization tool on the Volocity software was used to calculate the Pearson’s correlation and colocalization coefficient. Analysis is based on the quantification of over 100 cells from three independent experiments.
Statistical analysis
All quantified Western blot and confocal colocalization data are the mean of at least three independent experiments. The raw data from the immunoprecipitation Western blotting were first normalized to the GFP to account for differences in amount of protein and then to the VPS26A control. Results are expressed as a percentage of the VPS26A control. Mean and standard error were calculated, followed by a one-way analysis of variance (ANOVA) and posthoc Dunnett test to determine statistical significance. Colocalization data were averaged across individual experiments and the mean and standard error calculated across the three experiments. A one-way ANOVA followed by a posthoc Dunnett test was then used to analyze statistical significance. For all statistical tests, P < 0.05 was considered significant and is indicated by an asterisk.
Plasmids
VPS29 was subcloned from an original plasmid, which was a gift from R. Teasdale. VPS26B and DENND4C were cloned from HeLa cDNA into a pEGFPC1 vector (Takara Bio Inc.) and then subcloned into the XLG lentiviral vector. Full-length mouse PKD2 was a gift from J. Zhou (Harvard Medical School, Boston, MA). The 1–703 PKD2 truncation was cloned from HeLa cDNA into a pEGFPN1 vector (Takara Bio Inc.).
Online supplemental material
Fig. S1 shows the protein expression levels of GFP-tagged VPS26A, VPS26B, and VPS29 in RPE-1 cells as well as GFP-Trap immunoprecipitation and immunofluorescence showing that the GFP-tagged retromer components can still associate into the retromer heterotrimer. Fig. S2 shows that PKD2 and DENND4C can associate with retromer on endosomes by immunofluorescence and that the missense VPS26A mutants do not perturb trafficking of the CI-MPR or α5 integrin. Table S1 contains the SILAC quantified interactomes of the different retromer subunits in RPE-1 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201604057/DC1.
Supplementary Material
Acknowledgments
We thank the Wolfson Bioimaging Facility at the University of Bristol for their support.
This work was supported by the Medical Research Council (MR/K018299/1) and the Wellcome Trust (089928 and 104568) to P.J. Cullen and Australian National Health and Medical Research Council (APP1058734) to B.M. Collins. M. Gallon and I.J. McGough were supported by Wellcome Trust PhD Studentships from the Dynamic Cell Biology program (083474). A.P. Jellett is supported by a Medical Research Council PhD studentship (MR/K501359/1). B.M. Collins is supported by a National Health and Medical Research Council Career Development Fellowship (APP1061574).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ANOVA
- analysis of variance
- CI-MPR
- cation-independent mannose 6 phosphate receptor
- ITC
- isothermal titration calorimetry
- RPE-1
- retinal pigment epithelium 1
- SILAC
- stable isotope labeling with amino acids in cell culture
- SNX-27
- sorting nexin-27
- WASH
- Wiskott-Aldrich syndrome and SCAR homologue
References
- Arighi C.N., Hartnell L.M., Aguilar R.C., Haft C.R., and Bonifacino J.S.. 2004. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165:123–133. 10.1083/jcb.200312055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balana B., Maslennikov I., Kwiatkowski W., Stern K.M., Bahima L., Choe S., and Slesinger P.A.. 2011. Mechanism underlying selective regulation of G protein-gated inwardly rectifying potassium channels by the psychostimulant-sensitive sorting nexin 27. Proc. Natl. Acad. Sci. USA. 108:5831–5836. 10.1073/pnas.1018645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean B.D., Davey M., Snider J., Jessulat M., Deineko V., Tinney M., Stagljar I., Babu M., and Conibear E.. 2015. Rab5-family guanine nucleotide exchange factors bind retromer and promote its recruitment to endosomes. Mol. Biol. Cell. 26:1119–1128. 10.1091/mbc.E14-08-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean D.M., Heimbach J., Ficorella L., Micklem G., Oliver S.G., and Favrin G.. 2014. esyN: network building, sharing and publishing. PLoS One. 9:e106035 10.1371/journal.pone.0106035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C., and Cullen P.J.. 2014. Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6:a016774 10.1101/cshperspect.a016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Loo L.S., Atlashkin V., Hanson B.J., and Hong W.. 2011. Deficiency of sorting nexin 27 (SNX27) leads to growth retardation and elevated levels of N-methyl-D-aspartate receptor 2C (NR2C). Mol. Cell. Biol. 31:1734–1747. 10.1128/MCB.01044-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.S., Clairfeuille T., Landao-Bassonga E., Kinna G., Ng P.Y., Loo L.S., Cheng T.S., Zheng M., Hong W., Teasdale R.D., et al. 2016. Sorting nexin 27 couples PTHR trafficking to retromer for signal regulation in osteoblasts during bone growth. Mol. Biol. Cell. 27:1367–1382. 10.1091/mbc.E15-12-0851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin H.C., and Caplan M.J.. 2010. The cell biology of polycystic kidney disease. J. Cell Biol. 191:701–710. 10.1083/jcb.201006173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., and Korswagen H.C.. 2011. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat. Cell Biol. 14:29–37. 10.1038/ncb2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damseh N., Danson C.M., Al-Ashhab M., Abu-Libdeh B., Gallon M., Sharma K., Yaacov B., Coulthard E., Caldwell M.A., Edvardson S., et al. 2015. A defect in the retromer accessory protein, SNX27, manifests by infantile myoclonic epilepsy and neurodegeneration. Neurogenetics. 16:215–221. 10.1007/s10048-015-0446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman C.L., Hesketh G., and Seaman M.N.. 2014. RME-8 coordinates the activity of the WASH complex with the function of the retromer SNX dimer to control endosomal tubulation. J. Cell Sci. 127:2053–2070. 10.1242/jcs.144659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallon M., Clairfeuille T., Steinberg F., Mas C., Ghai R., Sessions R.B., Teasdale R.D., Collins B.M., and Cullen P.J.. 2014. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. USA. 111:E3604–E3613. 10.1073/pnas.1410552111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T.S., and Billadeau D.D.. 2009. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 17:699–711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson E.K., Guella I., Trinh J., Szu-Tu C., Rajput A., Rajput A.H., Steele J.C., McKeown M., Jeon B.S., Aasly J.O., and Farrer M.J.. 2015. Genetic variability of the retromer cargo recognition complex in parkinsonism. Mov. Disord. 30:580–584. 10.1002/mds.26104 [DOI] [PubMed] [Google Scholar]
- Harbour M.E., Breusegem S.Y., Antrobus R., Freeman C., Reid E., and Seaman M.N.. 2010. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J. Cell Sci. 123:3703–3717. 10.1242/jcs.071472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M.E., Breusegem S.Y., and Seaman M.N.. 2012. Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem. J. 442:209–220. 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- Hesketh G.G., Pérez-Dorado I., Jackson L.P., Wartosch L., Schäfer I.B., Gray S.R., McCoy A.J., Zeldin O.B., Garman E.F., Harbour M.E., et al. 2014. VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev. Cell. 29:591–606. 10.1016/j.devcel.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert L., Hanson B., Barthet G., Sebben M., Claeysen S., Hong W., Marin P., Dumuis A., and Bockaert J.. 2004. New sorting nexin (SNX27) and NHERF specifically interact with the 5-HT4a receptor splice variant: roles in receptor targeting. J. Cell Sci. 117:5367–5379. 10.1242/jcs.01379 [DOI] [PubMed] [Google Scholar]
- Kerr M.C., Bennetts J.S., Simpson F., Thomas E.C., Flegg C., Gleeson P.A., Wicking C., and Teasdale R.D.. 2005. A novel mammalian retromer component, Vps26B. Traffic. 6:991–1001. 10.1111/j.1600-0854.2005.00328.x [DOI] [PubMed] [Google Scholar]
- Lauffer B.E., Melero C., Temkin P., Lei C., Hong W., Kortemme T., and von Zastrow M.. 2010. SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J. Cell Biol. 190:565–574. 10.1083/jcb.201004060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D.A., Rhinn H., Kuwahara T., Zolin A., Di Paolo G., McCabe B.D., Marder K.S., Honig L.S., Clark L.N., Small S.A., and Abeliovich A.. 2013. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 77:425–439. (published erratum appears in Neuron 2013. 77:994) 10.1016/j.neuron.2012.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I.J., Steinberg F., Jia D., Barbuti P.A., McMillan K.J., Heesom K.J., Whone A.L., Caldwell M.A., Billadeau D.D., Rosen M.K., and Cullen P.J.. 2014a Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol. 24:1670–1676. (published erratum appears in Curr. Biol 2014. 14:1678) 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I.J., Steinberg F., Gallon M., Yatsu A., Ohbayashi N., Heesom K.J., Fukuda M., and Cullen P.J.. 2014b Identification of molecular heterogeneity in SNX27-retromer-mediated endosome-to-plasma-membrane recycling. J. Cell Sci. 127:4940–4953. 10.1242/jcs.156299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad A., Flores I., Zhang H., Yu R., Staniszewski A., Planel E., Herman M., Ho L., Kreber R., Honig L.S., et al. 2008. Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Aβ accumulation. Proc. Natl. Acad. Sci. USA. 105:7327–7332. 10.1073/pnas.0802545105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Krawczak C.A., Singla A., Starokadomskyy P., Deng Z., Osborne D.G., Li H., Dick C.J., Gomez T.S., Koenecke M., Zhang J.S., et al. 2015. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol. Biol. Cell. 26:91–103. 10.1091/mbc.E14-06-1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff V., Mardones G.A., Tenza D., Rojas R., Lamaze C., Bonifacino J.S., Raposo G., and Johannes L.. 2007. The retromer complex and clathrin define an early endosomal retrograde exit site. J. Cell Sci. 120:2022–2031. 10.1242/jcs.003020 [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A., Charbonnier C., Wallon D., Nicolas G., Seaman M.N., Pottier C., Breusegem S.Y., Mathur P.P., Jenardhanan P., Le Guennec K., et al. CNR-MAJ collaborators . 2015. De novo deleterious genetic variations target a biological network centered on Aβ peptide in early-onset Alzheimer disease. Mol. Psychiatry. 20:1046–1056. 10.1038/mp.2015.100 [DOI] [PubMed] [Google Scholar]
- Seaman M.N. 2004. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165:111–122. 10.1083/jcb.200312034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N. 2012. The retromer complex - endosomal protein recycling and beyond. J. Cell Sci. 125:4693–4702. 10.1242/jcs.103440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N., McCaffery J.M., and Emr S.D.. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681. 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N., Harbour M.E., Tattersall D., Read E., and Bright N.. 2009. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122:2371–2382. 10.1242/jcs.048686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon B., Soto-Ortolaza A., Rayaprolu S., Cannon H.D., Labbé C., Benitez B.A., Choi J., Lynch T., Boczarska-Jedynak M., Opala G., et al. 2014. Genetic variation of the retromer subunits VPS26A/B-VPS29 in Parkinson’s disease. Neurobiol. Aging. 35:1958.e1–1958.e2. 10.1016/j.neurobiolaging.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Sun L., Banerjee R., Tobin M., Zhang Y., and Grant B.D.. 2009. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 28:3290–3302. 10.1038/emboj.2009.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S.A., Kent K., Pierce A., Leung C., Kang M.S., Okada H., Honig L., Vonsattel J.P., and Kim T.W.. 2005. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann. Neurol. 58:909–919. 10.1002/ana.20667 [DOI] [PubMed] [Google Scholar]
- Steinberg F., Heesom K.J., Bass M.D., and Cullen P.J.. 2012. SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J. Cell Biol. 197:219–230. 10.1083/jcb.201111121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F., Gallon M., Winfield M., Thomas E.C., Bell A.J., Heesom K.J., Tavaré J.M., and Cullen P.J.. 2013. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15:461–471. 10.1038/ncb2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P., Lauffer B., Jäger S., Cimermancic P., Krogan N.J., and von Zastrow M.. 2011. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13:715–721. 10.1038/ncb2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardarajan B.N., Bruesegem S.Y., Harbour M.E., Inzelberg R., Friedland R., St George-Hyslop P., Seaman M.N., and Farrer L.A.. 2012. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol. Aging. 33:2231.e15–2231.e30. 10.1016/j.neurobiolaging.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C., Wider C., Ross O.A., Dachsel J.C., Kachergus J.M., Lincoln S.J., Soto-Ortolaza A.I., Cobb S.A., Wilhoite G.J., Bacon J.A., et al. 2011. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 89:162–167. 10.1016/j.ajhg.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C., Rajput A., Milnerwood A.J., Shah B., Szu-Tu C., Trinh J., Yu I., Encarnacion M., Munsie L.N., Tapia L., et al. 2014. DNAJC13 mutations in Parkinson disease. Hum. Mol. Genet. 23:1794–1801. 10.1093/hmg/ddt570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao Y., Zhang X., Badie H., Zhou Y., Mu Y., Loo L.S., Cai L., Thompson R.C., Yang B., et al. 2013. Loss of sorting nexin 27 contributes to excitatory synaptic dysfunction by modulating glutamate receptor recycling in Down’s syndrome. Nat. Med. 19:473–480. 10.1038/nm.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., and Barr F.A.. 2010. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191:367–381. 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M.N., Moreau K., Jimenez-Sanchez M., Breusegem S.Y., Harbour M.E., and Rubinsztein D.C.. 2014. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5:3828 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A., Benet-Pagès A., Struhal W., Graf E., Eck S.H., Offman M.N., Haubenberger D., Spielberger S., Schulte E.C., Lichtner P., et al. 2011. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 89:168–175. 10.1016/j.ajhg.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.