Mutated N-Ras causes leukemia and melanoma when properly delivered to cellular membranes. Here, VPS35, a component of the retromer, is shown to carry N-Ras from one membrane compartment to another. Thus, VPS35 may be a target for anticancer drug discovery.
Abstract
Ras guanosine triphosphatases (GTPases) regulate signaling pathways only when associated with cellular membranes through their C-terminal prenylated regions. Ras proteins move between membrane compartments in part via diffusion-limited, fluid phase transfer through the cytosol, suggesting that chaperones sequester the polyisoprene lipid from the aqueous environment. In this study, we analyze the nature of the pool of endogenous Ras proteins found in the cytosol. The majority of the pool consists of farnesylated, but not palmitoylated, N-Ras that is associated with a high molecular weight (HMW) complex. Affinity purification and mass spectrographic identification revealed that among the proteins found in the HMW fraction is VPS35, a latent cytosolic component of the retromer coat. VPS35 bound to N-Ras in a farnesyl-dependent, but neither palmitoyl- nor guanosine triphosphate (GTP)–dependent, fashion. Silencing VPS35 increased N-Ras’s association with cytoplasmic vesicles, diminished GTP loading of Ras, and inhibited mitogen-activated protein kinase signaling and growth of N-Ras–dependent melanoma cells.
Introduction
Ras proteins are peripheral membrane proteins that secondarily associate with the cytoplasmic leaflet of cellular membranes by virtue of a series of posttranslational modifications. The three mammalian RAS genes encode four proteins, N-Ras, H-Ras, K-Ras4A, and K-Ras4B; the latter two are splice variants of a single locus. Ras proteins are 95% identical in their first 165 amino acids, which fold into the guanine nucleotide-binding domain, but differ dramatically in their final 23–24 amino acids, which constitute the hypervariable region (HVR). It is the HVR that undergoes posttranslational modification and thereby directs membrane association and trafficking of the proteins. All Ras HVRs end in a CAAX sequence that is modified by farnesylation, AAX proteolysis and prenylcysteine carboxylmethylation (Wright and Philips, 2006). CAAX processing is necessary, but not sufficient, to deliver Ras proteins to the plasma membrane (PM). A second signal in the HVR is also required (Hancock et al., 1990; Choy et al., 1999; Apolloni et al., 2000). In the case of N-Ras and H-Ras, that signal is one or two cysteines that become modified with a palmitoyl lipid. In the case of K-Ras4B, the second signal is a polylysine motif that forms an electrostatic interaction with the negatively charged inner leaflet of the PM. The second signal of K-Ras4A is a hybrid of the two (Tsai et al., 2015).
The enzymes that further process farnesylated CAAX sequences, Rce1 (Ras converting enzyme 1) and Icmt (isoprenylcysteine carboxyl methyltransferase), are ER-restricted polytopic membrane proteins, and DHHC9/GCP16, a palmitoylacyltransferase that modifies N-Ras and H-Ras, is a Golgi-resident enzyme (Swarthout et al., 2005). Mature, palmitoylated Ras proteins are subject to esterase-catalyzed depalmitoylation (Goodwin et al., 2005; Rocks et al., 2005; Lin and Conibear, 2015) and can undergo a cycle of palmitoylation/depalmitoylation that allows them to cycle between endomembrane and PM (Rocks et al., 2010). Thus, nascent Ras proteins traffic to the cytosolic face of the endomembrane system en route to and from the PM.
Because farnesylation is irreversible, the intracellular trafficking of Ras proteins between membrane compartments requires that the lipidated form of the proteins traverse the aqueous environment of the cytosol. Live-cell imaging of GFP-tagged N-Ras and H-Ras has revealed that some of the trafficking, presumably that of palmitoylated species, is via vesicular transport with the Ras protein riding on the cytoplasmic face of transport vesicles and endosomes (Choy et al., 1999; Apolloni et al., 2000). However, FRAP has revealed that, at least in the case of retrograde trafficking to the Golgi apparatus that follows depalmitoylation, the kinetics are consistent with diffusion-limited, fluid phase transfer (Goodwin et al., 2005; Rocks et al., 2005). Thus, current models of Ras trafficking predict that there should be a pool of Ras in the fluid phase of the cytosol and raise the question of how a farnesylated protein remains soluble in the aqueous environment of that compartment. One way in which farnesylated Ras proteins could remain soluble in the cytosol is by binding to chaperones that sequester the lipid moiety. Indeed, RhoGDI (Michaelson et al., 2001) and RabGDI (Seabra and Wasmeier, 2004) are proteins that perform just such a function for their cognate small GTPases that are modified with one or two 20-carbon geranylgeranyl polyisoprene lipids. Several farnesyl-protein binding proteins have been described (Figueroa et al., 2001; Rotblat et al., 2004; Berg et al., 2010), and recently, PDE6δ has been characterized as a polyisoprene-binding transport chaperone for prenylated small GTPases, including Ras (Hanzal-Bayer et al., 2002; Nancy et al., 2002; Chandra et al., 2011; Ismail et al., 2011).
To test the hypothesis that a significant pool of Ras exists in the cytosol of cells, we studied the subcellular localization of Ras proteins by live-cell imaging and subcellular fractionation with and without metabolic labeling to reveal modification status. We found a large pool of N-Ras in the cytosol that was resolved by chromatography into a prenyl-dependent high molecular weight (HMW) fraction as well as a low molecular weight (LMW) fraction. We identified VPS35 as a cytosolic protein that binds N-Ras in a prenyl-dependent fashion and regulates its subcellular localization. We found that a melanoma cell line that depends on mutant N-Ras also depended on VPS35 for growth.
Results
Cellular N-Ras is largely cytosolic
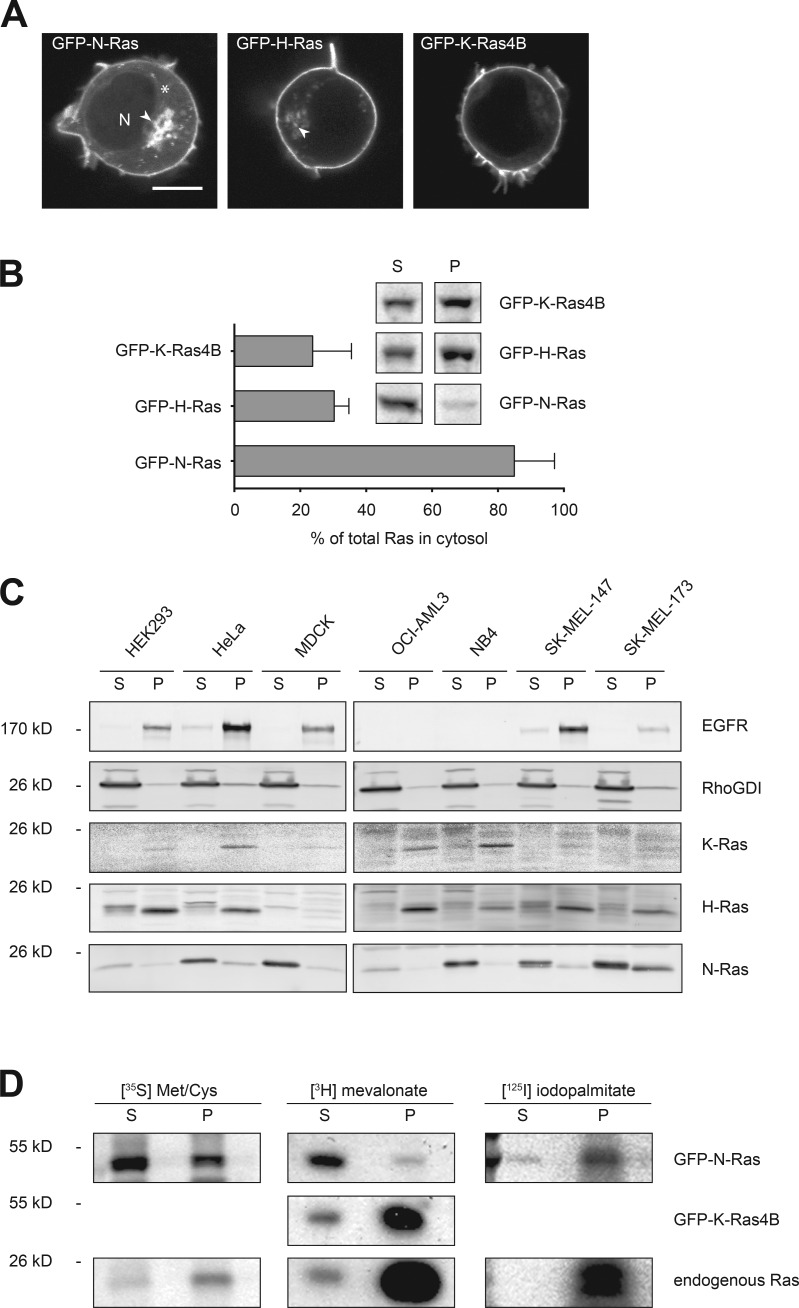
We have previously used live imaging of cells expressing GFP-tagged Ras proteins to study their steady-state subcellular localization and trafficking (Choy et al., 1999). The palmitoylated isoforms, N-Ras and H-Ras, differed from K-Ras4B in that they are expressed on endomembranes, including the Golgi apparatus, in addition to the PM (Choy et al., 1999). Another difference was that expression of GFP-N-Ras resulted in a higher level of cytosolic fluorescence than that observed with other isoforms. The unique, cytosolic localization of GFP-N-Ras was particularly evident in Jurkat lymphocytes, which afford a favorable, spherical geometry for confocal analysis (Fig. 1 A). To confirm that a pool of cellular N-Ras is cytosolic, we stably expressed GFP-tagged N-Ras, H-Ras, and K-Ras4B in MDCK cells at levels equivalent those of endogenous Ras (Fig. S1 A), metabolically labeled the cells with [35S]methionine/cysteine, disrupted the cells with nitrogen cavitation, generated P350 and S350 fractions by ultracentrifugation at 350,000 g, immunoprecipitated GFP-tagged Ras proteins, and quantified the relative amounts of each protein in the two fractions (Fig. 1 B). Whereas 69.7 ± 2.6% and 76.2 ± 6.8% of GFP–H-Ras and GFP–K-Ras4B were recovered in the P350, respectively, only 15.0 ± 7.0% of GFP–N-Ras was recovered in that fraction, thus revealing a marked difference in the behavior of N-Ras relative to the other Ras isoforms.
Figure 1.
Prenylated N-Ras is largely cytosolic. (A) Jurkat T cells were transfected with GFP-tagged N-, H-, or K-Ras4B and imaged alive with a laser-scanning confocal microscope. N, nucleus. Arrowheads indicate Golgi, and the asterisk indicates cytosol. Bar, 10 µm. (B) S350 (S) and P350 (P) fractions were prepared from MDCK cells expressing the indicated GFP-tagged Ras protein that were metabolically labeled with [35S]methionine/cysteine for 17 h. GFP-tagged Ras proteins were immunoprecipitated from detergent extracts of the two fractions. Protein levels were analyzed by phosphorimager and the cytosolic fractions plotted as percentages of total Ras; mean ± SEM, n = 3. A representative radiograph is shown as an inset. (C) The indicated cell types were fractionated as in B. Cell equivalents from each fraction were loaded and analyzed for the indicated proteins by immunoblot. Epidermal growth factor receptor (EGFR) and RhoGDI serve as markers for membrane and cytosol, respectively. Representative data are shown (n = 3). (D) S350 and P350 fractions were generated from MDCK cells stably expressing GFP–N-Ras or GFP–K-Ras4B at endogenous levels that were metabolically labeled with [35S]methionine/cysteine, [3H]melavonate, or [125I]iodopalmitate, and immunoprecipitated Ras was analyzed by fluorimetry (3H) or phosphorimager (35S and 125I).
To confirm that N-Ras makes up the bulk of the pool of soluble, endogenous Ras, we studied membrane association of endogenous Ras isoforms in various cell lines by subcellular fractionation and immunoblot analysis (Fig. 1 C). Whereas endogenous H-Ras and K-Ras were enriched in the P350 fraction, the bulk of N-Ras was recovered in the S350 fraction. A small component of endogenous H-Ras observed in the S350 fraction migrated slower than that in the P350, consistent with a minor pool of soluble protein not yet CAAX processed. In contrast, the major pool of N-Ras observed in the S350 comigrated with the minor pool in the P350, consistent with full CAAX processing.
Cytosolic N-Ras is prenylated
To investigate the state of HVR modification of cytosolic N-Ras, we performed metabolic labeling with radioisotope labeled methionine/cysteine, mevalonate, and palmitate and examined both endogenous Ras as well as GFP–N-Ras and GFP–K-Ras4B stably expressed at endogenous levels (Fig. S1 A) in MDCK cells (Fig. 1 D). [35S]methionine/cysteine-labeled GFP–N-Ras was distributed like endogenous N-Ras in both fractions, with the majority of labeled protein in the S350 fraction. [3H]mevalonate-labeled GFP–N-Ras was also observed in both fractions, with most of the label recovered in the S350 fraction, demonstrating that soluble GFP–N-Ras is prenylated. The P350/S350 ratio of [3H]mevalonate-labeled GFP–K-Ras4B and endogenous Ras was inverted relative to that of GFP–N-Ras, confirming that CAAX processed N-Ras differs from that of other Ras isoforms with regard to localization of the lipidated form of the protein in the cytosol. Whereas the majority of [3H]mevalonate-labeled GFP–N-Ras was recovered in the S350 fraction, the vast majority of [125I]iodopalmitate-labeled GFP–N-Ras was in the P350 fraction along with endogenous [125I]iodopalmitate-labeled Ras. We conclude that a substantial portion of the cellular pool of N-Ras is modified with a farnesyl, but not palmitoyl, lipid and, despite prenylation, remains cytosolic.
Farnesylated, cytosolic N-Ras is a component of an HMW complex
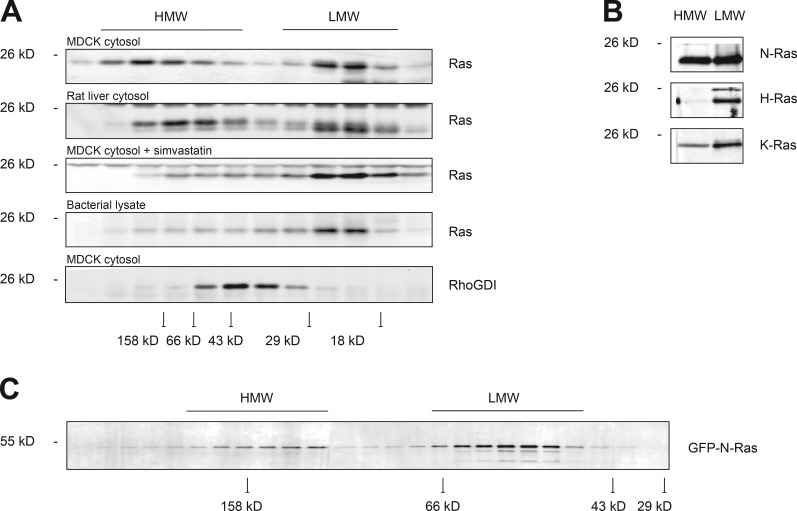
The existence of a substantial pool of farnesylated but soluble N-Ras suggested that either N-Ras can remain soluble in a prenylated state or that there is one or more cytosolic chaperones that can shield the prenyl moiety from the aqueous environment of the cytosol in a manner similar to the way in which RhoGDI sequesters the geranylgeranyl modification of Rho proteins (Hoffman et al., 2000; Michaelson et al., 2001). To determine whether cytosolic Ras is associated with protein complexes, we performed gel filtration chromatography on detergent-free cytosol (S350) harvested from MDCK cells disrupted by nitrogen cavitation (Fig. 2 A). We detected two peaks of endogenous Ras, one migrating where a p21 is expected (LMW) and one migrating in a broad range centered on 150 kD (HMW). We observed two similar peaks when we examined detergent free cytosol harvested from rat liver by Dounce homogenization, arguing that the phenomenon is not an artifact of tissue culture. H-Ras expressed in bacteria and therefore unprocessed was soluble and migrated only with the LMW peak. Importantly, when we examined S350 harvested from MDCK cells that had been treated with simvastatin to diminish protein prenylation, we observed only an LMW peak. As a control, we studied RhoGDI and found that it chromatographed with an apparent molecular mass of ∼45 kD, consistent with it serving as a chaperone for Rho, Rac, and Cdc42 proteins by forming a 1:1 complex, as previously described (Michaelson et al., 2001). Immunoblot analysis demonstrated that whereas the LMW pool of endogenous Ras contained all three Ras isoforms, the HMW pool was composed predominantly of N-Ras, with a small component of K-Ras (Fig. 2 B). Like endogenous N-Ras, cytosolic GFP–N-Ras eluted from gel filtration in two peaks (Fig. 2 C), suggesting that the GFP tag did not affect protein–protein interactions with cytosolic chaperones. We conclude that the cytosolic pool of N-Ras consists of both free p21 and also N-Ras associated with other cytosolic proteins in a prenylation-dependent manner.
Figure 2.
Prenylated N-Ras forms HMW complexes. (A) Cytosol (S350) from MDCK cells (±simvastatin) or rat liver homogenate or bacterially expressed H-Ras were subjected to Superdex 75 size-exclusion chromatography, and endogenous protein levels in each fraction were analyzed by immunoblot. (B) Protein levels of endogenous Ras isoforms in HMW and LMW fractions from A were analyzed by immunoblot. (C) Superdex 200 chromatography of S350 from MDCK cells stably expressing GFP–N-Ras. In A and C, peak elution of various molecular weight standards are indicated at the bottom.
Farnesylated, cytosolic N-Ras associates with VPS35
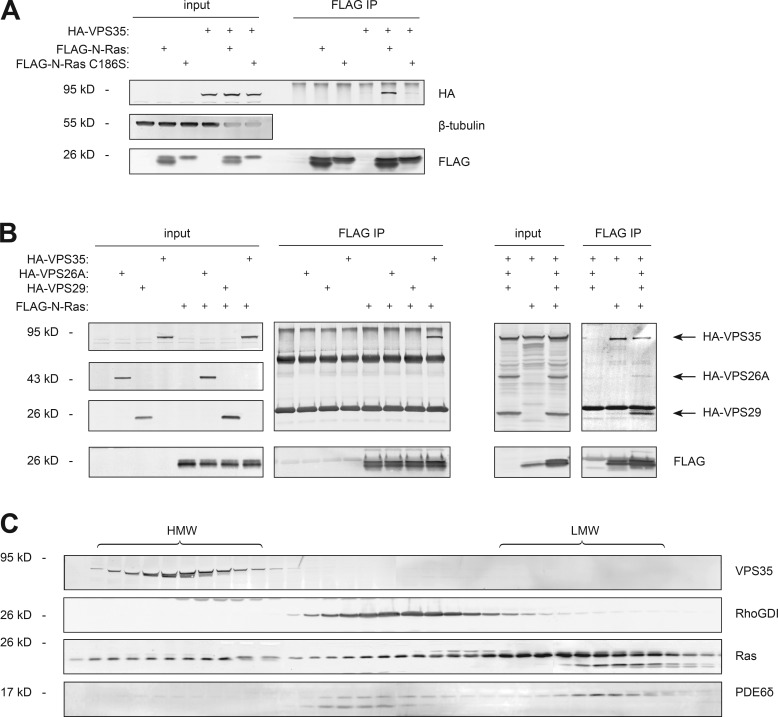
To identify cytosolic proteins associated with N-Ras in a prenyl-dependent manner, we stably expressed tandem affinity peptide (TAP) tagged N-Ras or N-RasC186S (prenylation deficient) in a tetracycline-inducible system in HEK293 cells and induced expression to match endogenous levels of Ras (Fig. S1 B). We then prepared detergent-free cytosol (S350) by nitrogen cavitation, affinity purified TAP-tagged N-Ras and subjected affinity purified proteins to analysis by mass spectroscopy (Table S1). Neither RhoGDI nor PDE6δ was detected. Among the proteins identified that were enriched in the TAP–N-Ras relative to the TAP–N-RasC186S preparation were all three components of the cargo recognition unit of the retromer (Trousdale and Kim, 2015): VPS35, VPS29, and VPS26A. To validate the interaction we expressed FLAG–N-Ras and HA-VPS35 in HEK293 cells and found that HA-VPS35 coimmunoprecipitated with FLAG–N-Ras, but not FLAG–N-RasC186S (Fig. 3 A). When coexpressed individually with FLAG–N-Ras, we were able to coimmunoprecipitate neither HA-VPS26A nor HA-VPS29 (Fig. 3 B), suggesting that it is VPS35 that binds N-Ras. However, when HA-tagged versions of all three components of the retromer cargo recognition unit (Hierro et al., 2007) were expressed together, all three were coimmunoprecipitated by FLAG–N-Ras (Fig. 3 B). Endogenous VPS35 chromatographed with the HMW pool of Ras in MDCK cell cytosol (Fig. 3 C), consistent with participation in a Ras-containing complex. Interestingly, the validated prenyl-sequestering Ras chaperone PDE6δ (Chandra et al., 2011) chromatographed predominantly with the p21 pool of Ras, consistent with an Apo form. A relatively small fraction of endogenous PDE6δ comigrated with RhoGDI, suggesting that, whereas PDE6δ may form a 1:1 complex with a minor pool of cytosolic Ras, it is not a component of the complexes that contribute to the HMW pool.
Figure 3.
VPS35 interacts with N-Ras in a prenyl-dependent fashion. (A and B) HEK293 cells were transiently transfected with VPS35 and/or N-Ras (wild type or C186S; A); N-Ras and either VPS35, VPS26A, or VPS29 (B); or all three. Whole-cell lysates (input) were immunoprecipitated with an anti-FLAG antibody (FLAG immunoprecipitation [IP]), and samples were analyzed by immunoblot. Data shown are representative of four independent experiments. (C) The cytosol (S350) of MDCK cells was subjected to Superdex 75 chromatography, and the indicated endogenous protein levels were analyzed by immunoblot. Data shown are representative of at least three independent experiments.
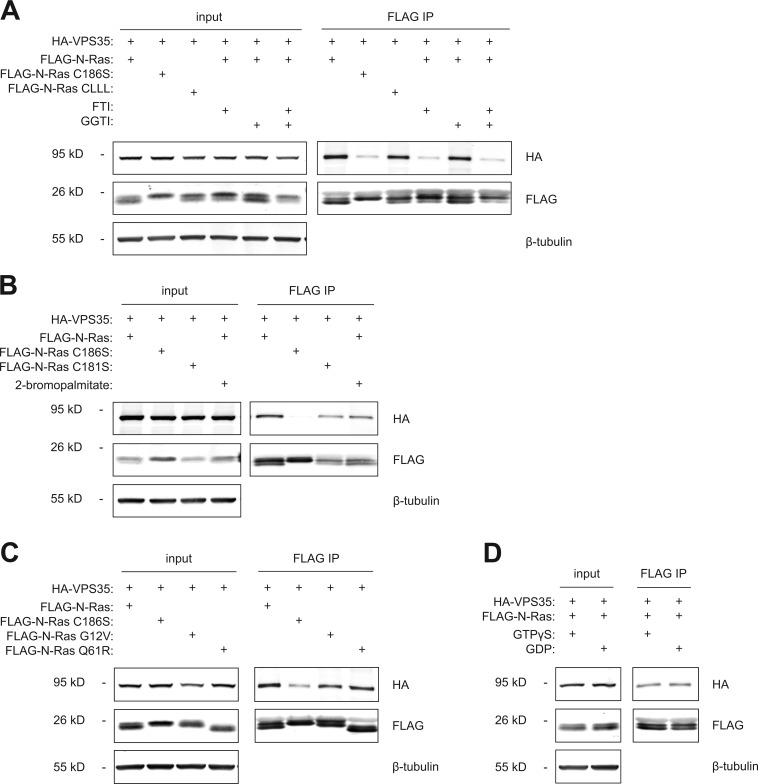
To further explore the nature of the interaction between N-Ras and VPS35, we extended analysis of prenylation dependence by testing the effects of inhibition of prenyltransferases as well as the identity of the prenyl lipid (Fig. 4 A). A farnesyltransferase inhibitor, but not a geranylgeranyltransferase inhibitor, blocked the interaction. We found that a geranylgeranylated form of N-Ras (N-Ras-CLLL) bound VPS35 almost as efficiently as did N-Ras with a native, farneslylated C terminus, demonstrating that if geranylgeranylation is forced, the two proteins retain the ability to interact. To test dependence on palmitoylation, we expressed wild-type N-Ras ± 2-bromopalmitate (an inhibitor of protein palmitoylation) or N-RasC181S (a palmitoylation-deficient mutant) and found that in both cases, the interaction was preserved (Fig. 4 B). To determine whether VPS35 plays a role in promoting acylation by delivering the substrate (farnesyl–N-Ras) to the protein acyltransferase on the Golgi (DHHC9/GCP16), we measured N-Ras palmitoylation by metabolic labeling with [3H]palmitate with and without silencing of VPS35 and found no difference (Fig. S2). To explore the guanine nucleotide dependence of the interaction, we compared FLAG-tagged wild type with the oncogenic (constitutively GTP bound) mutants N-RasG12V or N-RasQ61R (Fig. 4 C) and found similar binding. In addition, no difference was observed in the capacity of FLAG–N-Ras to interact with HA-VPS35 when the GTPase was loaded with GTP-γS versus GDP (Fig. 4 D). Thus, the interaction of N-Ras with VPS35 is independent of the GTP-binding state of the protein.
Figure 4.
The VPS35–N-Ras interaction is dependent on prenylation, but not palmitoylation or GTP binding. (A–D) HEK293 cells were transiently transfected with plasmids directing expression of the indicated proteins and analyzed by immunoprecipitation and immunoblot as in Fig. 3. Before lysis, cells were treated with farnesyltransferase inhibitor (FTI) and/or geranylgeranyltransferase inhibitor (GGTI; A) or 2-bromopalmitate (B). (D) Lysates were treated with GTP-γS or GDP before immunoprecipitation (IP). Data shown are representative of at least three independent experiments.
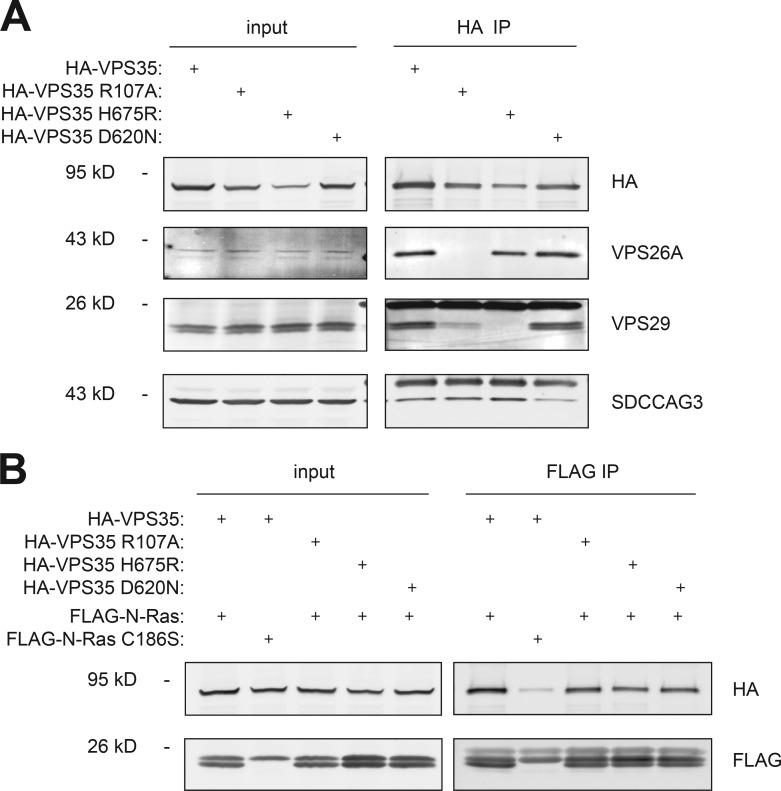
To determine whether VPS26A and VPS29 are required for the association of VPS35 with N-Ras, we used mutations of VPS35 reported to interfere with the formation of the VPS26–VPS35–VPS29 complex (Gokool et al., 2007; Helfer et al., 2013). We confirmed the effects of these mutations on subunit binding (Fig. 5 A). Neither an R107A nor an H675R substitution in VPS35, mutations that affect the binding of VPS26A and VPS29, respectively, reduced the ability of VPS35 to interact with N-Ras (Fig. 5 B), nor did a Parkinson’s disease–associated D620N substitution previously shown to block the interaction of VPS35 with the WASH complex (Zavodszky et al., 2014) and with seriologically defined colon cancer antigen 3 (SDCCAG3; McGough et al., 2014). These results suggest that the interaction of VPS35 with N-Ras is independent of the function of VPS35 on the retromer. Indeed, the vast majority of endogenous VPS35 protein was found in the cytosol (Fig. S3), as expected for a coat protein.
Figure 5.
Retromer-deficient VPS35 mutants associate with N-Ras. HEK293 cells were transiently transfected with plasmids directing expression of the indicated proteins. Whole-cell lysates (input) were immunoprecipitated by anti-HA antibody (HA immunoprecipitation [IP]; A) or anti-FLAG antibody (FLAG immunoprecipitation; B), and samples were analyzed by immunoblot. Data shown are representative of at least three independent experiments.
VPS35 interacts with N-Ras in intact cells
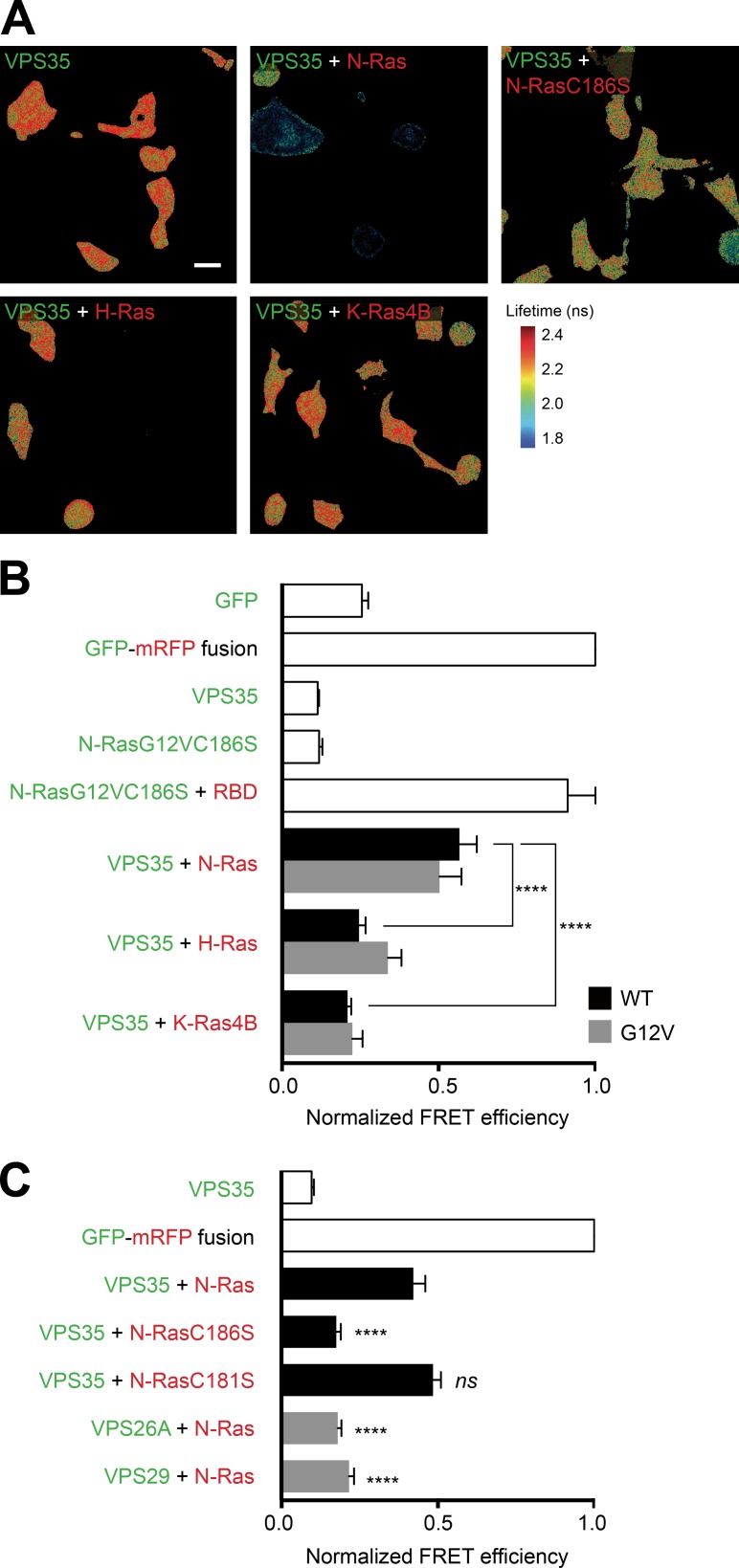
Coimmunoprecipitation experiments with FLAG-tagged Ras proteins showed some affinity of VPS35 for all three Ras isoforms (Fig. S4 A). However, VPS35 was not a universal binding partner for prenylated proteins because among four Ras-related small GTPases tested (Rac1, Rac2, Rab5, and Rab7), only Rac2 behaved like N-Ras by interacting with VPS35 (Fig. S4 B). Because coimmunoprecipitation can report ex vivo interactions, we confirmed our biochemical data with a fluorescence resonance energy transfer (FRET)–based assay that reports protein–protein interactions in intact cells (Fig. 6). We tagged VPS35 with GFP as a FRET donor and Ras proteins with mRFP as a FRET acceptor, coexpressed the tagged proteins, and measured FRET by fluorescence lifetime imaging (FLIM) in fixed BHK cells. As a positive control, we coexpressed GFP–N-RasG12V,C186S, a GTP-bound soluble form of N-Ras, with mRFP-RBD, a probe that avidly binds GTP-bound Ras proteins, and observed a signal of the same magnitude as that measured by expressing a GFP-mRFP fusion protein predicted to give maximal FRET (Fig. 6 B). Whereas GFP-VPS35 coexpressed with mRFP–N-Ras gave a signal approximately half that of the positive control, the FLIM–FRET signal did not differ from background when GFP-VPS35 was coexpressed with either mRFP–H-Ras or mRFP–K-Ras4B (Fig. 6 B), suggesting that the interaction is N-Ras specific in intact cells. FLIM-FRET analysis of N-RasC186S and N-RasC181S confirmed that the interaction between VPS35 and N-Ras is dependent on prenylation, but not palmitoylation (Fig. 6 C). Substituting neither GFP-VPS26A nor GFP-VPS29 for GFP-VPS35 gave a FLIM–FRET signal with mRFP–N-Ras (Fig. 6 C), suggesting that either VPS35 binds N-Ras without its retromer complex proteins or that the geometry of the multimolecular complex is such that GFP and mRFP are not brought close enough to allow for FRET.
Figure 6.
VPS35 associates with prenylated N-Ras, but not H- or K-Ras4B in intact cells. BHK cells were transiently transfected with the indicated constructs. GFP-tagged proteins are indicated in green, and mRFP-tagged proteins in red. The next day, cells were serum starved for 2 h before fixation, and FRET between the two tagged proteins was measured by FLIM. (A) Images of GFP lifetime represented in pseudocolor. Bar, 30 µm. (B and C) FRET efficiency values relative to the value of a GFP-mRFP fusion protein. Values plotted are mean ± SEM, n = 3 (>30 cells examined for each condition). ****, P < 0.0001; ns, not significant (compared with GFP-VPS35 plus mRFP–N-Ras). WT, wild type.
VPS35 is required for proper subcellular trafficking of N-Ras
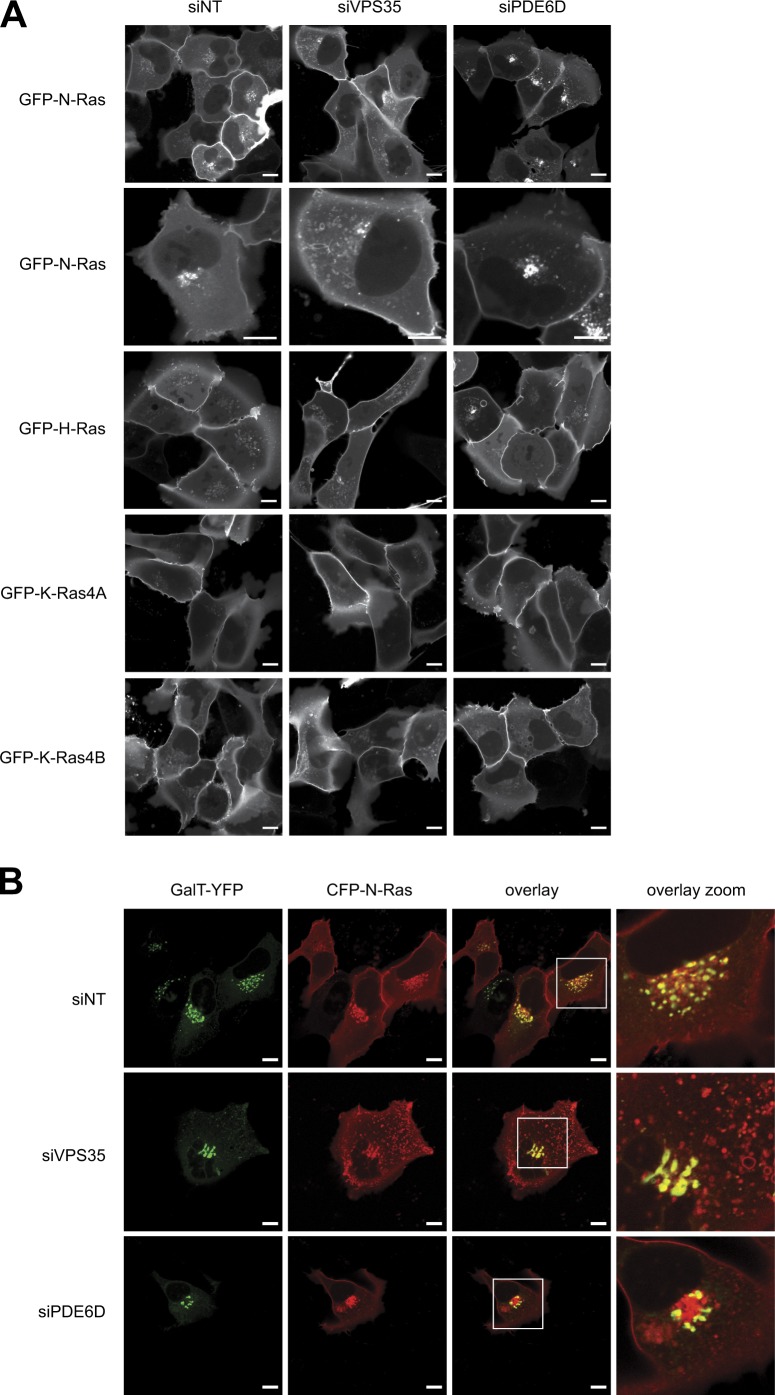
To determine the role of VPS35 on the subcellular distribution of N-Ras, we used live-cell imaging to study GFP-Ras proteins in cells with and without knockdown of VPS35 (Fig. 7). As a control, we also silenced PDE6D (Fig. S5 A). As previously reported, at steady-state GFP–N-Ras was distributed among the PM, Golgi apparatus, and cytosol, with a paucity of vesicles. Upon silencing, VPS35 vesicles and tubulovesicular structures appeared that were decorated with GFP–N-Ras (Fig. 7 A). Although the morphology of the trans-Golgi was altered by VPS35 knockdown as revealed by GalT-YFP (Fig. 7 B), the Golgi structures retained decoration with GFP–N-Ras. In contrast, silencing VPS35 had no effect on the steady-state distribution of GFP–H-Ras, GFP–K-Ras4B, or GFP–K-Ras-4A (Fig. 7 A). Thus, VPS35 regulates specifically the subcellular trafficking of N-Ras. In contrast to one previous study (Chandra et al., 2011), we observed no inhibition of delivery of any Ras protein to the PM upon silencing PDE6D, although, interestingly, knockdown of this chaperone did result in subtle changes in the relative distribution of GalT-YFP–positive and negative structures in the paranuclear region containing the Golgi apparatus (Fig. 7 B).
Figure 7.
Silencing VPS35 promotes a vesicular distribution of GFP–N-Ras. (A and B) U2OS cells were transfected with untargeted siRNA (NT) or a validated pool targeting VPS35 or PDE6D. After 4 d, the cells were transfected with plasmids directing expression of the indicated fluorescent proteins and imaged live the next day. Representative images of U2OS cells expressing one of the four Ras isoforms (A) or coexpressing both N-Ras and β-1,4-galactosyltransferase (GalT; B). Bars, 10 µm.
VPS35 is required for proliferation of N-Ras mutant melanoma cells
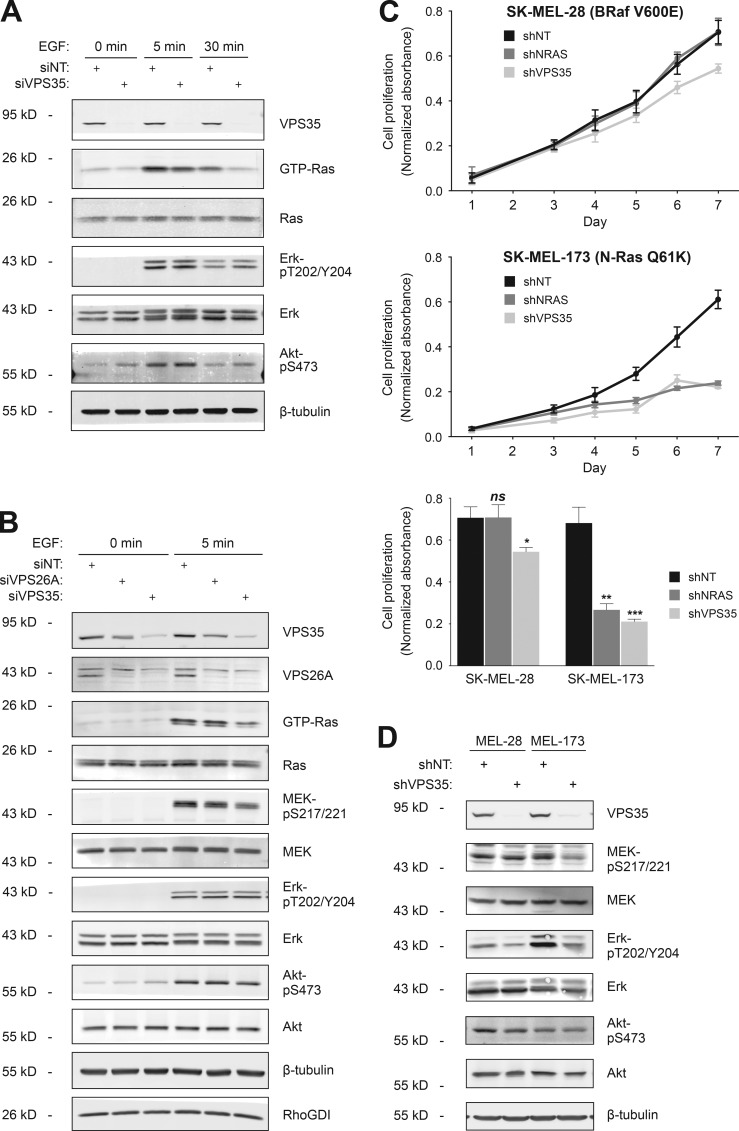
To determine the effect of VPS35 on Ras signaling, we silenced expression of the protein in HEK293 cells and studied the biochemical response of serum-starved cells to stimulation with epidermal growth factor (Fig. 8 A). Silencing VPS35 was accompanied by a marked decrease in GTP-loading of endogenous Ras, both at the 5-min peak and at 30 min after stimulation. Similar effects on GTP loading were not observed when VPS26A was silenced (Fig. 8 B), suggesting that either the effect of VPS35 knockdown was not dependent on retromer function or the endogenous paralogue, VPS26B, is able to fully substitute for VPS26A. Curiously, in HEK293 cells, diminished GTP loading of Ras in response to VPS35 knockdown was accompanied by diminished phospho-MEK, but not phospho-Erk or phospho-Akt (Fig. 8 B). To clarify the role of VPS35 in Ras signaling, we turned to melanoma cell lines that are driven by either oncogenic N-Ras or BRaf. From these, we generated cell lines that express, in a tetracycline-inducible fashion, shRNAs capable of silencing either NRAS or VPS35 (Fig. S5 B). Whereas silencing either NRAS or VPS35 markedly inhibited cell proliferation of SK-MEL-173 cells that harbor oncogenic N-RasQ61K, the proliferation of SK-MEL-28 cells that harbor wild-type N-Ras and oncogenic BRafV600E was unaffected by silencing NRAS and only minimally slowed by silencing VPS35 (Fig. 8 C). Concordant with the effects on proliferation, silencing VPS35 diminished phospho-MEK and phospho-Erk to a greater degree in SK-MEL-173 than in SK-MEL-28 cells (Fig. 8 D). Thus, an NRAS-dependent tumor cell line also depends on VPS35 for optimal MAPK signaling and proliferation.
Figure 8.
VPS35 is required for GTP loading of Ras and proliferation of N-Ras mutant melanoma cells. (A and B) HEK293 cells were transfected with untargeted siRNA (NT) or a validated pool targeting VPS35 or VPS26A. After 3 d, the cells were serum starved overnight and stimulated with EGF for the indicated time. The indicated proteins in cell lysates were quantified by immunoblot, and GTP loading was analyzed by GST-RBD pull down. Data shown are representative of four independent experiments. (C) Proliferation as measured by an MTS assay of SK-MEL-28 and -173 melanoma cell lines (BRAF or NRAS mutant, respectively) stably transfected with a tetracycline-inducible shRNA targeting NRAS or VPS35. shRNA was induced on the day of plating. Data shown are relative MTS values from day 1 to day 7 (top two panels) or at day 7 (bottom) after plating (mean ± SEM, n = 4; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant compared with untargeted siRNA). (D) Immunoblots show MEK, Erk, and Akt phosphorylation in the melanoma cell lines used in C.
Discussion
Ras proteins were assigned to the inner leaflet of the PM 35 yr ago on the basis of electron microscopic immunocytochemical analysis of the brush border of H-Ras transformed epithelial cells (Willingham et al., 1980). Careful quantitative localization of Ras by subcellular fractionation is lacking from the extensive literature that followed, although prenylated endogenous Ras proteins were observed in the S100 fraction in at least one study (Casey et al., 1989). We found that Ras was not limited to the PM and decorated the cytoplasmic face of endomembranes, including the Golgi (Choy et al., 1999). In the same study, we observed that pulse-labeled GFP–N-Ras and endogenous Ras persisted in the cytosol of MDCK cells for at least 4 h (Choy et al., 1999). The results reported here confirm a significant pool of endogenous Ras in the cytosol of a variety of cell types, including human cancer cells. Importantly, metabolic labeling revealed that the pool of cytosolic Ras proteins includes prenylated proteins, but not palmitoylated forms. This result validates the predictions from current models of Ras trafficking that hold that depalmitoylated N-Ras and H-Ras traffic between membrane compartments, in part, by diffusion through the cytosol (Goodwin et al., 2005; Rocks et al., 2005). Similarly, there is strong evidence that K-Ras4B can move rapidly from one membrane compartment to another (Silvius et al., 2006; Schmick et al., 2014). Whereas N-Ras was observed predominantly in the S350, most cellular H-Ras was found in the P350 fraction. This striking difference may be explained by mono- versus dipalmitoylation of N-Ras versus H-Ras, respectively. If, as predicted, palmitoylation fixes Ras proteins in membranes and the cytosolic pool of farnesylated Ras proteins are devoid of palmitate, one would expect a greater proportion of N-Ras because only a single depalmitoylation event is required to render that isoform free of acyl chains.
Rho family Ras-related GTPases are held in the cytosol by binding to RhoGDI, which shields the 20-carbon geranylgeranyl polyisoprene lipid from the aqueous environment (Hoffman et al., 2000; Michaelson et al., 2001). Whereas it is possible that Ras proteins modified with the less hydrophobic 15-carbon farnesyl polyisoprene might be soluble in an aqueous environment, it is more likely that they too require a chaperone to shield the farnesyl lipid. Indeed, several farnesyl-protein binding proteins have been described, including PRA1 (Figueroa et al., 2001), PDE6δ (Hanzal-Bayer et al., 2002; Nancy et al., 2002; Chandra et al., 2011), galectin-1 (Rotblat et al., 2004), and smgGDS (Berg et al., 2010). Among these, PDE6δ is particularly interesting, because it is structurally similar to RhoGDI (Hanzal-Bayer et al., 2002; Ismail et al., 2011). Any or all of these might serve the function of solubilizing farnesylated N-Ras. Indeed, we have found that overexpression of PDE6δ can extract GFP-tagged N-Ras and K-Ras4B, but not H-Ras or K-Ras4A, from membranes (Tsai et al., 2015). Our analysis of endogenous cytosolic Ras by gel filtration chromatography revealed two pools, HMW and LMW, the former composed predominantly of N-Ras. The dependence of the HMW pool on prenylation suggests that it is composed of Ras in association with one or more proteins, at least one of which is a prenyl-binding protein. The presence of all three Ras isoforms in the LMW pool suggests that it consists of nascent, unprocessed protein. Interestingly, no PDE6δ was detected in the HMW fractions, suggesting an alternate chaperone. Affinity purification of proteins that associate with N-Ras in a prenyl-dependent fashion allowed us to identify VPS35 as one such protein. VPS35 chromatographed with the HMW pool of cytosolic N-Ras. Thus, VPS35 appears to be among the prenyl-binding proteins that interact with farnesylated cytosolic N-Ras.
Because VPS35 is one of three proteins that forms the cargo recognition unit of the retromer (Hierro et al., 2007), and because N-Ras traffics retrograde from the PM and endosomes to the trans-Golgi network (Goodwin et al., 2005; Rocks et al., 2005), it is appealing to consider the retromer as a platform for retrograde N-Ras trafficking by vesicular transport. However, the kinetics of N-Ras retrograde transport as measured by FRAP were inconsistent with vesicular trafficking (Goodwin et al., 2005). Moreover, several aspects of our findings argue against this model. First, the VPS35–N-Ras interaction was detected in the S350 fraction, which is devoid of membrane-bound organelles. Second, mutations in VPS35 known to block interaction with VPS26A or VPS29 and prevent association with endosomes (Gokool et al., 2007) did not affect the ability of VPS35 to interact with N-Ras. Third, GTP loading of Ras was affected by silencing VPS35, but not VPS26A. Finally, at baseline, we did not observe GFP–N-Ras on the tubulovesicular structures characteristic of the retromer. Although VPS35 complexes on membranes with VPS26A, VPS29, and two of four relevant sorting nexins to form the organelle that is the retromer, VPS35 is, by definition, a vesicular coat protein, and the membrane-bound form is in equilibrium with a much larger pool in the cytosol (Trousdale and Kim, 2015). Our data are most consistent with the cytosolic form of VPS35 serving as a farnesyl–N-Ras binding partner. Nevertheless, the accumulation of GFP–N-Ras on vesicles upon silencing VPS35 suggests that this component of the retromer is involved in removing N-Ras from endosomes and perhaps delivering the protein “retrograde” to the trans-Golgi, a function that would parallel the function of the fully formed retromer for transmembrane receptors.
Whereas the N-Ras–VPS35 interaction was not affected by the GTP-binding state of the GTPase, there was an isoform selectivity for N-Ras observed in intact cells by FRET. These results suggest that it is the unique HVR of N-Ras that is particularly sensitive to depalmitoylation that favors the interaction with VPS35. The crystal structure of the C-terminal portion of VPS35 reveals a horseshoe-shaped, right-handed, α-helical solenoid with five hydrophobic grooves displayed on one surface (Hierro et al., 2007). One possible model for the interaction is the association of the modified C terminus of N-Ras with one or more of these grooves. Because the interaction is unaffected by the absence of palmitoylation, if a lipid modification alone, or in part, mediates this interaction, it must be the farnesyl polyisoprene, although our results suggest that a geranylgeranyl lipid will also support binding. Whereas the prenyl binding pocket of RhoGDI makes contact with amino acids in the C terminus of its clients (Cdc42, Rho, and Rac), explaining specificity (Hoffman et al., 2000), the PDE6δ pocket contacts only the polyisoprene lipid and is therefore promiscuous with regard to prenyl-protein binding (Ismail et al., 2011). The fact that VPS35 binds N-Ras, but not other Ras isoforms, in intact cells as measured by FRET and that some, but not all, prenylated Ras superfamily family GTPases interact suggests that VPS35 is more like RhoGDI in this regard and can discriminate among prenylated binding partners.
If VPS35 functions as a cytosolic chaperone for N-Ras that is involved in the subcellular trafficking of the GTPase, it is likely to regulate to some extent the signaling function of N-Ras. Curiously, in HEK293 cells, we found that whereas silencing VPS35 markedly diminished GTP loading of Ras, neither signaling to Erk nor signaling to Akt was affected. This suggests that either the residual degree of GTP loading of Ras is sufficient for downstream signaling or that Ras-independent signaling to Erk and Akt comes into play in the setting of VPS35 deficiency. In contrast to HEK293 cells, silencing VPS35 did diminish pErk levels in SK-MEL-173 melanoma cells, demonstrating some degree of cell-type dependence. Because the retromer is responsible for recycling numerous receptors, it is conceivable that inhibition of retromer function by knockdown of VPS35 somehow alters signaling differentially in various cells. Importantly, the EGF receptor has been reported to be insensitive to retromer function (Steinberg et al., 2013). Our observation that, in contrast to knockdown of VPS35, silencing VPS26A does not affect GTP loading of Ras argues against a retromer-based effect rather than a direct effect of VPS35 deficiency. Because the readout of Ras signaling through phosphorylation of signaling intermediates is difficult to interpret in the setting of feedback loops, the final common readout of cell proliferation may be a better way to assess the role of VPS35. We found that silencing VPS35 in a mutant N-Ras–dependent melanoma cell line inhibited proliferation as much as did silencing N-Ras itself, whereas it had relatively little effect on the growth of melanoma cells driven by oncogenic BRaf. Thus, VPS35 is required for maximal proliferation in an N-Ras–driven tumor cell. This suggests that VPS35 might be a therapeutic target for these tumors.
VPS35 has received a great deal of attention over the past decade because of associations with neurodegenerative disorders (Wang and Bellen, 2015). Relatively low expression levels of the VPS35 message have been associated with Alzheimer’s disease (Small et al., 2005; Muhammad et al., 2008; Wen et al., 2011), and mutations in the VPS35 gene have been linked to late-onset Parkinson’s disease (Vilariño-Güell et al., 2011; Zimprich et al., 2011). The mechanism is thought to relate to retromer dysfunction causing inefficient processing of amyloid precursor protein in Alzheimer’s disease (Sullivan et al., 2011) and diminished recycling of Wntless in Parkinson’s disease (Franch-Marro et al., 2008). Whether the function of cytosolic VPS35 as a chaperone for N-Ras plays any role in the pathogenesis of these diseases remains to be determined.
Materials and methods
Plasmids and RNA interference
Full-length or partial coding regions of each cDNA were amplified by PCR and cloned in-frame into the indicated vectors, including pEGFP-C1, pmCherry-C1, pEYFP-N1, pECFP-C1 (Takara Bio Inc.), pcDNA5/FRT/TO, pcDNA3.1+ (Invitrogen), pTRIPz (GE Healthcare), pGEX-2T (GE Healthcare), pET-14b (EMD Millipore), and pCGN (Fiordalisi et al., 2001). Amino acid substitutions were generated by site-directed mutagenesis using the QuikChange XL Site-Directed Mutagenesis kit (Agilent Technologies). All plasmids were verified by bidirectional sequencing.
For siRNA knockdown, cells were transfected with ON-TARGETplus SMARTpools (GE Healthcare) targeting VPS35, PDE6D or nontargeting control using DharmaFECT 1 reagent according to the manufacturer’s instructions. For shRNA knockdown, targeting sequences for VPS35 were inserted into pTRIPz lentiviral vector by PCR, and lentiviral particles were generated with standard protocols (http://tcf.epfl.ch/files/content/sites/tcf/files/shared/LV_production.pdf). pTripz-based NRAS-targeting lentivirus vectors (Grabocka et al., 2014) were provided by D. Bar-Sagi (New York University Langone Medical Center, New York, NY). Stable cells lines were generated by lentiviral infection and selection with puromycin for 5 d. Knockdown was induced by 0.4 µg/ml doxycycline for at least 72 h.
Cell culture and transfection
HEK293, COS-1, HeLa, 293T, MDCK, and U2OS cells were from ATCC. Flp-In T-Rex 293 cells were from Thermo Fisher Scientific. SK-MEL-28, SK-MEL-147, and SK-MEL-173 melanoma cells were obtained from E. Hernando (New York University Langone Medical Center). These cells were cultured in DMEM supplemented with 10% FBS. Jurkat (ATCC) and OCI-AML3 and NB4 (from K. Shannon, University of California, San Francisco, San Francisco, CA) cells were cultured in RPMI-1640 medium supplemented with 10% FBS. All media were supplemented with 50 U/ml penicillin G and 50 µg/ml streptomycin. All cells used in our study were maintained at 37°C in 5% CO2 incubator and tested negative for mycoplasma. Transfection was performed using Lipofectamine 2000 or 3000 (Invitrogen) or SuperFect (QIAGEN) according to the manufacturer’s instructions.
Cell fractionation
Adherent cells were washed with cold PBS and scraped off tissue culture plates in prechilled potassium homogenization buffer (150 mM KCl, 0.2 mM MgSO4, 0.2 mM CaCl2, and 40 mM Tris, pH 7.4). Cell suspensions were spun in a bomb (Parr Instruments) pressurized with nitrogen to 450 psi for 20 min at 4°C and then released dropwise to room atmosphere and collected in tubes containing protease inhibitors. Cavitates were collected and centrifuged at 500 g for 10 min at 4°C to remove nuclei and unbroken cells. Postnuclear supernatant was further centrifuged with a TLA100.1 rotor (Beckman Coulter) at 90,000 rpm (350,000 g) for 1 h. Supernatant (S350 cytosolic fraction) was collected without disturbing pellet (P350 membrane fraction). Pellet was washed twice with cold PBS and resuspended in RIPA buffer (150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and 50 mM Tris, pH 8.0). Extracted proteins from the P350 were clarified by centrifugation at 23,000 g. For immunoprecipitation experiments, the S350 was adjusted with 10× RIPA buffer to achieve detergent equivalence with the extracted P350.
Metabolic labeling
For [35S]methionine/cysteine labeling, MDCK cells stably expressing GFP–N-Ras were incubated in methionine/cysteine-free DMEM with 10% dialyzed FBS for 1 h and labeled with 50 mCi/ml [35S]methionine/cysteine (PerkinElmer) overnight. For [3H]mevalonate labeling, MDCK cells stably expressing GFP–N- or K-Ras4B were pretreated for 20 min with 50 mM mevastatin in DMEM with 10% FBS and incubated for 17 h in the presence of 100 mCi/ml [3H]mevalanonlactone (PerkinElmer) in the continued presence of mevastatin. For [125I]iodopalmitate ([125I]iodohexadecanoic acid; gift from M. Resh, Memorial Sloan-Kettering Cancer Center, New York, NY) labeling, MDCK cells stably expressing GFP–N-Ras were preincubated in serum-free DMEM supplemented with 100 mg/ml fatty acid–free BSA for 1 h and labeled with 50 mCi/ml [125I]iodopalmitate for 12 h.
Cells were washed with cold PBS, suspended in homogenization buffer, and fractioned into cytosol and membrane as described in the Cell fractionation paragraph. GFP–N-/K-Ras4B and endogenous Ras were immunoprecipitated by GFP and pan-Ras antibodies, respectively. [3H]mevalonate-labeled Ras was visualized by fluorography with KODAK BioMax film exposed for 48 h. [35S]methionine/cysteine- and [125I]iodopalmitate-labeled proteins were analyzed with a Typhoon phosphorimager (GE Healthcare). [3H]palmitate labeling of HEK293 cells expressing FLAG–N- and H-Ras constructs followed by FLAG immunoprecipitation, SDS-PAGE, and fluorography were performed as described previously (Tsai et al., 2015).
Immunoprecipitation and immunoblot analysis
Cells were transfected with the relevant constructs. For pharmacological treatments, 2-bromopalmitate was used at 25 µM for 3 h, and farnesyltransferase inhibitor (FTI, L-744,832; Enzo Life Sciences) and geranylgernayltransferase inhibitor (GGTI, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole; Enzo Life Sciences) were used at 20 µM and 15 µM, respectively, for 24 h.
48 h after transfection, cells were washed twice with cold PBS and lysed in buffer (150 mM NaCl, 1% NP-40, 50 mM Tris, pH 7.5, 1 mM EDTA, and 10 mM NaF) supplemented with Roche cOmplete, Mini EDTA-free protease inhibitor cocktail tablet, PhosSTOP EASYpack, phosphatase inhibitor cocktail tablet (Roche), 2 mM Pefabloc SC, and 1 mM DTT. Lysates were clarified by centrifugation at 20,000 g at 4°C for 10 min. Protein were precipitated by the addition of HA antibody and protein A agarose (Invitrogen), or antibody directly conjugated agarose, and incubated for 1–4 h with gentle rotation at 4°C. Antibodies used for immunoprecipitation include anti-HA (F-7; Santa Cruz Biotechnology, Inc.), anti-GFP agarose (RQ2; Medical & Biological Laboratories), anti-Ras Y13-259AC preconjugated (259, Santa Cruz Biotechnology, Inc.), and EZview red anti-FLAG affinity gel (M2; Sigma-Aldrich). Protein–antibody complexes were collected by centrifugation at 2,500 g for 5 min and washed twice with lysis buffer. Supernatant was discarded and beads were resuspended in 2× Laemmli sample buffer containing 5% β-mercaptoethanol.
For immunoblots, samples were resolved on 4–20%, 10%, or 14% Tris-glycine polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Primary antibodies were used at 1:1,000 dilution unless otherwise indicated and include those directed against pan-Ras (Ab-3) from EMD Millipore; Ras (EP1125Y), VPS26A, and VPS35 from Epitomics/Abcam; VPS29 and FLAG (M2) from Sigma-Aldrich; β-tubulin (E7, 1:5,000) and hLAMP1 (H4A3) from Developmental Studies Hybridoma Bank; GFP (7.1 and 13.1) from Roche; Kras (F234, 1:200), Nras (F155, 1:200), Hras (C-20, 1:200), ERK 1 (K-23, 1:3,000), HA (F-7), RhoGDI (A-20), PDE6δ (N-15), and SDCCAG3 (E-2) from Santa Cruz Biotechnology, Inc.; and phospho-Erk1/2 (Thr202/Tyr204;E10), phospho-Akt (Ser473), MEK1/2 (L38C12), phospho-MEK1/2 (Ser217/221), and epidermal growth factor receptor (D38B1) from Cell Signaling Technology. Secondary antibodies include IRDye 680LT or IRDye 800CW conjugated goat anti–mouse, goat anti–rabbit, or donkey anti–goat secondary antibodies (LI-COR) at 1:20,000 dilution. Blots were scanned and bands of interest quantified using the Odyssey Infrared Imaging system (LI-COR Biosciences).
Size-exclusion chromatography
Cytosol (S350) of MDCK cells with or without treatment with simvastatin (10 µM × 24 h) and with or without stable expression of GFP–N-Ras was prepared as described in the Cell fractionation paragraph and was concentrated by centrifuging at 4,700 g at 4°C in Centricon centrifugal filter units with cutoff limit of 10 kD molecular weight (EMD Millipore). Rat liver cytosol was a gift from J.-P. Simon (New York University School of Medicine, New York, NY). Concentrated samples were centrifuged at 22,000 g at 4°C for 10 min to remove protein aggregates, and supernatants were injected onto a Superdex 75 5/150 GL or 200 10/300 GL size-exclusion column (GE Healthcare Life Sciences) pre-equilibrated with running buffer (20 mM Hepes, 1 mM MgCl2, and 0.3 M sucrose). Flow rate was set at 0.5 ml/min, and the first 40 eluted fractions were collected at 0.5 ml per fraction. The following protein molecular weight markers were used: dextran blue, >2,000 kD; alcohol dehydrogenase, 150 kD; albumin, 66 kD; ovalbumin, 43 kD; carbonic anhydrase, 29 kD; and myoglobin, 18 kD.
Tandem affinity purification and mass spectrometry
Flp-In T-Rex 293 stables expressing in an inducible fashion pcDNA5/FRT/TO-TAP-N-Ras wild-type or C186S mutant were harvested at 95% confluency after 4 h of 2 µg/ml doxycycline treatment. Cytosol (S350) was prepared as described in the Cell fractionation paragraph, and TAP-tagged N-Ras was purified with the Interplay TAP Purification kit (Agilent Technologies) according to the manufacturer’s instructions, with the exception that Laemmli buffer elution was substituted for EGTA as the final step of elution to increase yield. Eluted samples were subjected to SDS-PAGE and proteins eluted from the gel were digested with trypsin and fragments were subjected to analysis with a Thermo Fisher Scientific Q Exactive Orbitrap Mass Spectrometer with nano-flow HPLC.
FRET measurement by FLIM microscopy
BHK cells were transfected with 1 µg plasmid DNA encoding GFP-tagged VPS35 (or VPS26A or VPS29) and 1 µg plasmid DNA encoding mRFP-tagged N-, H-, or K-Ras4B (wild type or mutant) at 85% confluency on glass coverslips in 6-well plates with DMEM supplemented with 10% fetal calf serum. 24 h after transfection, cells were incubated in serum-free DMEM for 2 h and then washed and fixed with 4% paraformaldehyde. GFP lifetime was measured using a Lambert FLIM unit (Roden) attached to a wide-field Nikon microscope. Fluorescein at 1 µM was used as a lifetime reference standard. The raw FRET fraction data were normalized against the FRET fraction values of GFP in cells expressing GFP–mRFP fusion. Three independent experiments were performed for each sample, and data from a total of 35–50 cells were pooled to calculate mean and SEM. Tests for significant differences from GFP-VPS35/mRFP–N-Ras were performed by two-tailed paired Student’s t test.
EGF stimulation and Ras activity assay
For EGF stimulation, cells were transfected with the relevant constructs and serum-starved (0.1% FBS) overnight. The next day, cells were stimulated with 10 ng/ml EGF for 5 or 30 min. After stimulation, cells were washed with cold PBS once and lysed in GST-RBD pull-down buffer (25 mM Hepes, pH 7.5, 125 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1% NP-40, and 10% glycerol). Clarified lysates were incubated with GST-RBD glutathione agarose beads for 1 h with gentle rotation at 4°C. Protein–bead complexes were collected by centrifugation at 2,500 g for 5 min and washed twice with GST-RBD pull-down buffer. Supernatant was discarded and beads were resuspended in 2× Laemmli buffer containing 5% β-mercaptoethanol. Samples were subjected to SDS-PAGE and immunoblot analysis.
Cell proliferation assay
Each SK-MEL-28 and SK-MEL-173 stable cell line was seeded with six replicates on 96-well plates at a density of 2 × 103 cells per well and cultured in the presence or absence of 0.4 µg/ml doxycycline with doxycycline replenishment every 48 h. Cell proliferation was measured on indicated days using CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer’s instructions. Four independent experiments were performed for each sample to calculate mean and SEM. Tests for significant differences were performed by two-tailed paired Student’s t test.
Confocal microscopy
Live U2OS cells were seeded in 35-mm dishes containing no. 0 glass coverslips over 15-mm cutouts (MatTek Corporation) and transfected with GFP–N-Ras. Cells were imaged with an LSM 800 with Airyscan (ZEISS) using a 63×, N.A. 1.4 objective and a humidified metabolic chamber maintained at 37°C and 5% CO2 (Pecon GmbH). Images were processed with Photoshop CS6 (Adobe).
Online supplemental material
Fig. S1 shows by immunoblot the expression levels of GFP- or TAP-tagged N-Ras relative to endogenous Ras. Fig. S2 shows that silencing VPS35 does not affect N-Ras palmitoylation as determined by metabolic labeling with [3H]palmitate. Fig. S3 shows that the majority of cellular VPS35 is found in the cytosol (S350). Fig. S4 shows that VPS35 could be coimmunoprecipitated by all four Ras isoforms in contrast to FLIM-FRET analysis in live cells, which revealed a specificity for N-Ras (Fig. 6) and that whereas Rac2 mimics Ras in coimmunoprecipitation with VPS35, three other Ras-related GTPases (Rac1, Rab5, and Rab7) do not. Fig. S5 shows by immunoblot the efficiency of knockdown of VPS35 or PDE6δ by siRNA in the U2OS cells used for imaging studies (Fig. 7) and VPS35 or N-Ras by inducible shRNA in the melanoma cell lines used for signaling and proliferation assays (Fig. 8). Table S1, included as an Excel file, shows the complete results of the mass spectrometry identification of proteins affinity purified from HEK293 cell lysates with TAP-tagged N-Ras or N-RasC186S. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201604061/DC1.
Supplementary Material
Acknowledgments
We thank Adam Mor for his assistance imaging Jurkat cells, Matthew Seaman for advice on the retromer, Jean-Pierre Simon for supplying rat liver cytosol, and Michelle Krogsgaard for assistance with fast protein liquid chromatography.
This work was funded by National Institutes of Health grants GM055279, CA116034, and CA163489 (to M.R. Philips) and by grant CPRIT RP130059 (to J.F. Hancock).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- FLIM
- fluorescence lifetime imaging
- FRET
- fluorescence resonance energy transfer
- HMW
- high molecular weight
- HVR
- hypervariable region
- LMW
- low molecular weight
- PM
- plasma membrane
- TAP
- tandem affinity peptide
References
- Apolloni A., Prior I.A., Lindsay M., Parton R.G., and Hancock J.F.. 2000. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475–2487. 10.1128/MCB.20.7.2475-2487.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T.J., Gastonguay A.J., Lorimer E.L., Kuhnmuench J.R., Li R., Fields A.P., and Williams C.L.. 2010. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285:35255–35266. 10.1074/jbc.M110.129916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey P.J., Solski P.A., Der C.J., and Buss J.E.. 1989. p21ras is modified by a farnesyl isoprenoid. Proc. Natl. Acad. Sci. USA. 86:8323–8327. 10.1073/pnas.86.21.8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A., Grecco H.E., Pisupati V., Perera D., Cassidy L., Skoulidis F., Ismail S.A., Hedberg C., Hanzal-Bayer M., Venkitaraman A.R., et al. 2011. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 14:148–158. 10.1038/ncb2394 [DOI] [PubMed] [Google Scholar]
- Choy E., Chiu V.K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I.E., and Philips M.R.. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 98:69–80. 10.1016/S0092-8674(00)80607-8 [DOI] [PubMed] [Google Scholar]
- Figueroa C., Taylor J., and Vojtek A.B.. 2001. Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J. Biol. Chem. 276:28219–28225. 10.1074/jbc.M101763200 [DOI] [PubMed] [Google Scholar]
- Fiordalisi J.J., Johnson R.L. II, Ulkü A.S., Der C.J., and Cox A.D.. 2001. Mammalian expression vectors for Ras family proteins: generation and use of expression constructs to analyze Ras family function. Methods Enzymol. 332:3–36. 10.1016/S0076-6879(01)32189-4 [DOI] [PubMed] [Google Scholar]
- Franch-Marro X., Wendler F., Guidato S., Griffith J., Baena-Lopez A., Itasaki N., Maurice M.M., and Vincent J.P.. 2008. Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat. Cell Biol. 10:170–177. 10.1038/ncb1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokool S., Tattersall D., Reddy J.V., and Seaman M.N.. 2007. Identification of a conserved motif required for Vps35p/Vps26p interaction and assembly of the retromer complex. Biochem. J. 408:287–295. 10.1042/BJ20070555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J.S., Drake K.R., Rogers C., Wright L., Lippincott-Schwartz J., Philips M.R., and Kenworthy A.K.. 2005. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 170:261–272. 10.1083/jcb.200502063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabocka E., Pylayeva-Gupta Y., Jones M.J., Lubkov V., Yemanaberhan E., Taylor L., Jeng H.H., and Bar-Sagi D.. 2014. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 25:243–256. 10.1016/j.ccr.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J.F., Paterson H., and Marshall C.J.. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 63:133–139. 10.1016/0092-8674(90)90294-O [DOI] [PubMed] [Google Scholar]
- Hanzal-Bayer M., Renault L., Roversi P., Wittinghofer A., and Hillig R.C.. 2002. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 21:2095–2106. 10.1093/emboj/21.9.2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer E., Harbour M.E., Henriot V., Lakisic G., Sousa-Blin C., Volceanov L., Seaman M.N., and Gautreau A.. 2013. Endosomal recruitment of the WASH complex: active sequences and mutations impairing interaction with the retromer. Biol. Cell. 105:191–207. 10.1111/boc.201200038 [DOI] [PubMed] [Google Scholar]
- Hierro A., Rojas A.L., Rojas R., Murthy N., Effantin G., Kajava A.V., Steven A.C., Bonifacino J.S., and Hurley J.H.. 2007. Functional architecture of the retromer cargo-recognition complex. Nature. 449:1063–1067. 10.1038/nature06216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar N., and Cerione R.A.. 2000. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 100:345–356. 10.1016/S0092-8674(00)80670-4 [DOI] [PubMed] [Google Scholar]
- Ismail S.A., Chen Y.-X., Rusinova A., Chandra A., Bierbaum M., Gremer L., Triola G., Waldmann H., Bastiaens P.I.H., and Wittinghofer A.. 2011. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7:942–949. 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- Lin D.T., and Conibear E.. 2015. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife. 4:e11306 10.7554/eLife.11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I.J., Steinberg F., Gallon M., Yatsu A., Ohbayashi N., Heesom K.J., Fukuda M., and Cullen P.J.. 2014. Identification of molecular heterogeneity in SNX27-retromer-mediated endosome-to-plasma-membrane recycling. J. Cell Sci. 127:4940–4953. 10.1242/jcs.156299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Silletti J., Murphy G., D’Eustachio P., Rush M., and Philips M.R.. 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152:111–126. 10.1083/jcb.152.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad A., Flores I., Zhang H., Yu R., Staniszewski A., Planel E., Herman M., Ho L., Kreber R., Honig L.S., et al. 2008. Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc. Natl. Acad. Sci. USA. 105:7327–7332. 10.1073/pnas.0802545105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nancy V., Callebaut I., El Marjou A., and de Gunzburg J.. 2002. The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J. Biol. Chem. 277:15076–15084. http://dx.doi.org/10.1074/jbc.M109983200 [DOI] [PubMed] [Google Scholar]
- Rocks O., Peyker A., Kahms M., Verveer P.J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., and Bastiaens P.I.. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 307:1746–1752. 10.1126/science.1105654 [DOI] [PubMed] [Google Scholar]
- Rocks O., Gerauer M., Vartak N., Koch S., Huang Z.P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., et al. 2010. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 141:458–471. 10.1016/j.cell.2010.04.007 [DOI] [PubMed] [Google Scholar]
- Rotblat B., Niv H., André S., Kaltner H., Gabius H.J., and Kloog Y.. 2004. Galectin-1(L11A) predicted from a computed galectin-1 farnesyl-binding pocket selectively inhibits Ras-GTP. Cancer Res. 64:3112–3118. 10.1158/0008-5472.CAN-04-0026 [DOI] [PubMed] [Google Scholar]
- Schmick M., Vartak N., Papke B., Kovacevic M., Truxius D.C., Rossmannek L., and Bastiaens P.I.. 2014. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 157:459–471. 10.1016/j.cell.2014.02.051 [DOI] [PubMed] [Google Scholar]
- Seabra M.C., and Wasmeier C.. 2004. Controlling the location and activation of Rab GTPases. Curr. Opin. Cell Biol. 16:451–457. 10.1016/j.ceb.2004.06.014 [DOI] [PubMed] [Google Scholar]
- Silvius J.R., Bhagatji P., Leventis R., and Terrone D.. 2006. K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol. Biol. Cell. 17:192–202. 10.1091/mbc.E05-05-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S.A., Kent K., Pierce A., Leung C., Kang M.S., Okada H., Honig L., Vonsattel J.P., and Kim T.W.. 2005. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann. Neurol. 58:909–919. 10.1002/ana.20667 [DOI] [PubMed] [Google Scholar]
- Steinberg F., Gallon M., Winfield M., Thomas E.C., Bell A.J., Heesom K.J., Tavaré J.M., and Cullen P.J.. 2013. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15:461–471. 10.1038/ncb2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C.P., Jay A.G., Stack E.C., Pakaluk M., Wadlinger E., Fine R.E., Wells J.M., and Morin P.J.. 2011. Retromer disruption promotes amyloidogenic APP processing. Neurobiol. Dis. 43:338–345. 10.1016/j.nbd.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout J.T., Lobo S., Farh L., Croke M.R., Greentree W.K., Deschenes R.J., and Linder M.E.. 2005. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 280:31141–31148. 10.1074/jbc.M504113200 [DOI] [PubMed] [Google Scholar]
- Trousdale C., and Kim K.. 2015. Retromer: Structure, function, and roles in mammalian disease. Eur. J. Cell Biol. 94:513–521. 10.1016/j.ejcb.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Tsai F.D., Lopes M.S., Zhou M., Court H., Ponce O., Fiordalisi J.J., Gierut J.J., Cox A.D., Haigis K.M., and Philips M.R.. 2015. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl. Acad. Sci. USA. 112:779–784. 10.1073/pnas.1412811112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C., Wider C., Ross O.A., Dachsel J.C., Kachergus J.M., Lincoln S.J., Soto-Ortolaza A.I., Cobb S.A., Wilhoite G.J., Bacon J.A., et al. 2011. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 89:162–167. 10.1016/j.ajhg.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., and Bellen H.J.. 2015. The retromer complex in development and disease. Development. 142:2392–2396. 10.1242/dev.123737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Tang F.L., Hong Y., Luo S.W., Wang C.L., He W., Shen C., Jung J.U., Xiong F., Lee D.H., et al. 2011. VPS35 haploinsufficiency increases Alzheimer’s disease neuropathology. J. Cell Biol. 195:765–779. 10.1083/jcb.201105109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham M.C., Pastan I., Shih T.Y., and Scolnick E.M.. 1980. Localization of the src gene product of the Harvey strain of MSV to plasma membrane of transformed cells by electron microscopic immunocytochemistry. Cell. 19:1005–1014. 10.1016/0092-8674(80)90091-4 [DOI] [PubMed] [Google Scholar]
- Wright L.P., and Philips M.R.. 2006. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47:883–891. 10.1194/jlr.R600004-JLR200 [DOI] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M.N., Moreau K., Jimenez-Sanchez M., Breusegem S.Y., Harbour M.E., and Rubinsztein D.C.. 2014. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5:3828 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A., Benet-Pagès A., Struhal W., Graf E., Eck S.H., Offman M.N., Haubenberger D., Spielberger S., Schulte E.C., Lichtner P., et al. 2011. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 89:168–175. 10.1016/j.ajhg.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.