Abstract
Most mitochondrial proteins are imported through the TIM23 translocation channel, the structure and molecular nature of which are still unclear. In this issue, Ramesh et al. (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201602074) show that the TIM23 subunit Tim17 contains a disulfide bond that is crucial for protein translocation and channel gating.
Mitochondria are dynamic organelles that play a central role in essential cellular processes ranging from ATP production to apoptosis. Though mitochondria contain their own genome, ∼1,000 mitochondrial proteins are encoded by nuclear DNA and synthesized on cytosolic ribosomes with specific mitochondrial targeting signals. About 70% of mitochondrial proteins are targeted to the organelle by positively charged N-terminal extensions called presequences. They are recognized by receptors of the TOM complex on the cytosolic surface of the mitochondrial outer membrane and are translocated through the translocation channels in the outer and inner mitochondrial membranes in a reaction driven by the TIM23 complex in the inner membrane (Marom et al., 2011). The TIM23 complex consists of at least 11 subunits, but the high-resolution structure and even the molecular nature of the translocation channel in its core still remain unclear. Several studies established Tim23 as a constituent of the translocation channel of the TIM23 complex (Truscott et al., 2001; Martinez-Caballero et al., 2007; Alder et al., 2008; Malhotra et al., 2013). Like Tim23, Tim17 is predicted to span the inner membrane with four α-helical transmembrane segments. The transmembrane segments of Tim17 and Tim23 contain glycine motifs that, in Tim23, play essential roles in the structural integrity of the TIM23 complex (Demishtein-Zohary et al., 2015). Such an analysis has not yet been performed with Tim17. Despite the certain degree of sequence similarity in their transmembrane segments, Tim17 and Tim23 cannot substitute for each other (Ryan et al., 1994). The experiments performed during the 20 years since its identification revealed that Tim17 is essential for translocation of proteins through the TIM23 complex. It is found in close vicinity to translocating proteins (Berthold et al., 1995), and it affects the behavior of the endogenous TIM23 channels in electrophysiological analyses (Martinez-Caballero et al., 2007). Recent data suggest that Tim17 may be a major recruitment point of the import motor of the TIM23 complex (Banerjee et al., 2015). Still, the exact function of Tim17 within the TIM23 complex remains unclear. In this issue, Ramesh et al. identify a disulfide bond in yeast Tim17 and show it is important for efficient protein translocation through the TIM23 complex and for dynamic gating of its translocation channel, thereby providing insights into Tim17 function.
The authors investigated the oxidation status of the four Tim17 cysteine residues. To do so, they used an alkylating reagent that reacts with reduced but not oxidized thiols, causing characteristic mass shifts visible on a gel. This analysis revealed that two cysteine residues in Tim17 are reduced and two form a disulfide bond. Through a mutational approach, they delineated the oxidized cysteines and demonstrated that Tim17 contains a stable disulfide bond between two highly conserved cysteine residues, C10 and C77. According to Tim17 topology models, C10 and C77 are exposed to the intermembrane space (IMS), with C10 localized N-terminally to the first transmembrane segment and C77 localized C-terminally to the second transmembrane segment. The disulfide bond thus locks transmembrane segments 1 and 2 of Tim17 to each other.
Ramesh et al. (2016) go on to demonstrate that the disulfide bond in Tim17 is essential for viability of yeast cells at higher temperatures and plays an important role during translocation of proteins through the TIM23 complex. Using electrophysiological approaches, they found that the gating properties of the TIM23 channel were severely compromised in the absence of oxidized Tim17. Furthermore, the rapid switching between opened and closed states (“flickering”) of the TIM23 channels induced by presequence peptides was absent in mutant channels lacking the disulfide bond in Tim17. Rather, presequence peptides abruptly and irreversibly blocked the mutant channels.
Tim17 and Tim23 belong to the same protein family as Tim22, the central component of the TIM22 complex involved in insertion of hydrophobic carrier proteins into the inner mitochondrial membrane. Intriguingly, the presence of an intramolecular disulfide bond in yeast Tim22 has recently been reported (Wrobel et al., 2013; Okamoto et al., 2014). Lack of this disulfide bond leads to severe destabilization of the entire TIM22 complex, and mutant Tim22 proteins are easily degraded at higher temperatures (Okamoto et al., 2014). In an independent study, Wrobel et al. (2016) recently showed that the disulfide bonds present in Tim17 and in Tim22 are evolutionarily conserved in higher eukaryotes. In contrast, Tim23 does not appear to contain a disulfide bond in any of the organisms tested (Okamoto et al., 2014; Wrobel et al., 2016), in agreement with the lack of evolutionary conservation of its cysteine residues.
Does the disulfide bond in Tim17 play a regulatory or a structural role? Ramesh et al. (2016) could not detect yeast Tim17 in a fully reduced form under any of the conditions tested and therefore concluded that this disulfide bond does not serve a regulatory but rather a structural role (Fig. 1). Wrobel et al. (2016) similarly propose that the disulfide bonds in the Tim17/Tim22/Tim23 protein family properly position weak transmembrane domains in the bilayer of the inner mitochondrial membrane, thereby supporting the assembly of the protein translocation complexes. Various prediction servers do not predict the transmembrane segments of Tim23 as more hydrophobic or stable than the ones in Tim17 or Tim22. However, Tim17 and Tim23 heterodimerize, and the disulfide bond in Tim17 may be sufficient to stabilize both proteins. It is also likely that locking of transmembrane segments 1 and 2 of Tim23 would be detrimental to the function of this protein, in view of recent findings revealing these segments at least as part of the voltage-sensing mechanism in the TIM23 complex (Malhotra et al., 2013). Lack of the disulfide bond in Tim17 did not affect membrane association of Tim17 itself. However, Wrobel et al. (2016) observed that both Tim23 and Tim50, the major receptor of the TIM23 complex, could be partially extracted from mitochondrial membranes from Tim17 mutant cells. Ramesh et al. (2016) performed a thorough analysis of the effects of the disulfide bond in Tim17 on the TIM23 complex. The typical pattern of the Tim17–Tim23 core of the TIM23 complex observed by blue native PAGE was altered in mutant mitochondria (Ramesh et al., 2016). Such changes are indicative of extensive remodeling of the TIM23 complex (Chacinska et al., 2005; Banerjee et al., 2015). It will be interesting to study the effects of the lack of the disulfide bond in Tim17 on the assembly and conformation of the entire TIM23 complex, in particular with regard to the association of the import motor with the translocation channel.
Figure 1.
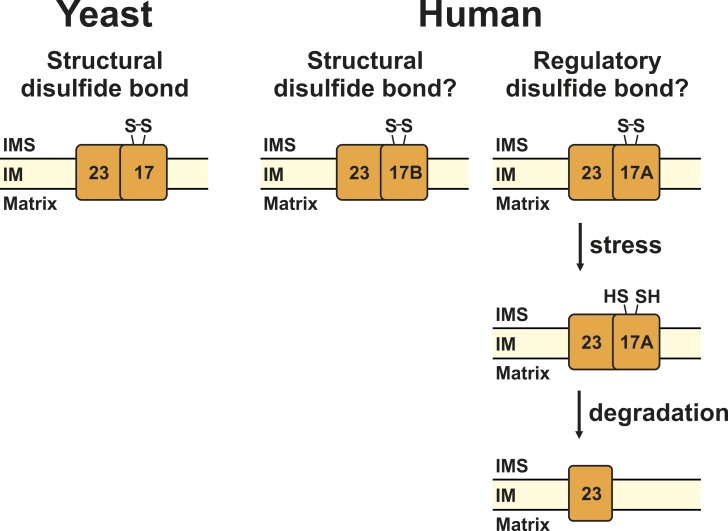
A disulfide bond in Tim17. In this issue, Ramesh et al. (2016) identify a disulfide bond in yeast Tim17 that likely plays a structural role in the TIM23 complex (left). In an independent study, Wrobel et al. (2016) demonstrate that this disulfide bond is not only present in yeast, but also in human Tim17B (right). Humans have an additional Tim17 protein, Tim17A, which is degraded under stress conditions. Could the degradation of Tim17A be regulated via the redox state of its cysteine residues? IMS, intermembrane space; IM, inner membrane.
Yeast has one Tim17 protein, whereas in humans, two homologues are present, Tim17A and Tim17B (Bauer et al., 1999). Though both human proteins are ubiquitously expressed in various tissues, Tim17B appears to have a more fundamental housekeeping role, whereas Tim17A is degraded under stress conditions and thus may be a stress-regulated protein (Rainbolt et al., 2013). Both Tim17A and Tim17B contain the conserved cysteine pair present in yeast Tim17. Wrobel et al. (2016) show that these two cysteines form a disulfide bond in Tim17B. However, the oxidation status of cysteine residues in Tim17A has not been analyzed so far. It is tempting to speculate that the disulfide bond in Tim17A, if present, does not play a structural role, but rather has a regulatory role. Since yeast Tim22 mutants unable to form the disulfide bond are easily degraded (Okamoto et al., 2014), it will be interesting to see whether degradation of Tim17A under stress conditions (Rainbolt et al., 2013) is regulated via the oxidation state of its two conserved cysteine residues (Fig. 1).
What is the mechanism of disulfide bond formation in Tim17? The presence of disulfide bonds in the IMS of mitochondria was long considered impossible, as this compartment is continuous with the cytosol. This is true at least for small molecules, the passage of which occurs in an undisturbed manner through porins in the outer membrane. Numerous reducing enzymes and high concentrations of reduced glutathione in the cytosol prevent formation of disulfide bonds. Although the environment of the IMS is indeed reducing (Kojer et al., 2012), it became clear about a decade ago that a number of proteins in this compartment contain disulfide bonds, which are inserted by the mitochondrial disulfide relay system during their import into mitochondria (Stojanovski et al., 2008). The mitochondrial disulfide relay system consists of the oxidoreductase Mia40 (Tim40) and the sulfhydryl oxidase Erv1. Mia40 oxidizes cysteine residues of the typical substrates of this pathway. The formation of disulfide bonds leads to stable folding of these proteins, thereby preventing their escape back into the cytosol. The oxidized form of Mia40 is regenerated by Erv1. However, the mitochondrial disulfide relay system might be more complex than initially thought, as Mia40 is also involved in the import of proteins even when all of their cysteines are removed (Weckbecker et al., 2012). Recent experiments suggest that binding of the incoming polypeptide chains in the hydrophobic pocket of Mia40 may be more important for import than introduction of disulfide bonds (Peleh et al., 2016), and it seems that this alternate function for Mia40 might be responsible for Tim17’s import as well. In vitro import of radiolabelled Tim17 was reduced when mitochondria depleted of Mia40 were used (Ramesh et al., 2016). When using mitochondria isolated from a temperature-sensitive mutant of Mia40, mia40-4, Ramesh et al. (2016) observed that the import defect was similar for wild-type and Tim17 mutant unable to form a disulfide bond, indicating that the disulfide bond is not necessary for Tim17 import, but Mia40 is. In line with this, both wild-type and mutant Tim17 bound to Mia40 during import. Interestingly, only mutations in the hydrophobic pocket of Mia40, but not the ones abolishing its oxidase function, impaired the import of Tim17 (Ramesh et al., 2016; Wrobel et al., 2016). All of these experiments suggest that Mi40 is essential for the import of Tim17, but not for its oxidation (Ramesh et al., 2016; Wrobel et al., 2016). Ramesh et al. (2016) provide evidence that newly imported Tim17 forms a mixed disulfide with sulfhydryl oxidase Erv1. In addition, the authors show that newly imported Tim17 remained in fully reduced form upon import into erv1 mutant mitochondria. Together, these data suggest that Erv1 may be the enzyme oxidizing Tim17. Such a mechanism has so far not been observed with any other substrate of the mitochondrial disulfide relay system. Whether Tim17 is unique in this respect, or whether other disulfide bonds in the IMS can be directly introduced by Erv1, awaits future experiments.
The presence of the disulfide bond in Tim17 identified by Ramesh et al. (2016) is an unexpected feature of the TIM23 complex. Though the molecular function of Tim17 within the TIM23 complex still remains unclear, this surprising twist will undoubtedly spark future structural and functional studies of the TIM23 complex on the way to understand the mysteries of the mitochondrial protein import machineries.
Acknowledgments
I am grateful to Rupa Banerjee, Umut Günsel, and Walter Neupert for the comments on this manuscript and to Deutsche Forschungsgemeinschaft for financial support (grant MO1944/1-1). I apologize to many colleagues whose work I could not cite because of space limitations.
The author declares no competing financial interests.
References
- Alder N.N., Jensen R.E., and Johnson A.E.. 2008. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 134:439–450. 10.1016/j.cell.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Banerjee R., Gladkova C., Mapa K., Witte G., and Mokranjac D.. 2015. Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein. eLife. 4:e11897 10.7554/eLife.11897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M.F., Gempel K., Reichert A.S., Rappold G.A., Lichtner P., Gerbitz K.D., Neupert W., Brunner M., and Hofmann S.. 1999. Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol. 289:69–82. 10.1006/jmbi.1999.2751 [DOI] [PubMed] [Google Scholar]
- Berthold J., Bauer M.F., Schneider H.C., Klaus C., Dietmeier K., Neupert W., and Brunner M.. 1995. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell. 81:1085–1093. 10.1016/S0092-8674(05)80013-3 [DOI] [PubMed] [Google Scholar]
- Chacinska A., Lind M., Frazier A.E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H.E., Truscott K.N., Guiard B., et al. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 120:817–829. 10.1016/j.cell.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Demishtein-Zohary K., Marom M., Neupert W., Mokranjac D., and Azem A.. 2015. GxxxG motifs hold the TIM23 complex together. FEBS J. 282:2178–2186. 10.1111/febs.13266 [DOI] [PubMed] [Google Scholar]
- Kojer K., Bien M., Gangel H., Morgan B., Dick T.P., and Riemer J.. 2012. Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 31:3169–3182. 10.1038/emboj.2012.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra K., Sathappa M., Landin J.S., Johnson A.E., and Alder N.N.. 2013. Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat. Struct. Mol. Biol. 20:965–972. 10.1038/nsmb.2613 [DOI] [PubMed] [Google Scholar]
- Marom M., Azem A., and Mokranjac D.. 2011. Understanding the molecular mechanism of protein translocation across the mitochondrial inner membrane: still a long way to go. Biochim. Biophys. Acta. 1808:990–1001. 10.1016/j.bbamem.2010.07.011 [DOI] [PubMed] [Google Scholar]
- Martinez-Caballero S., Grigoriev S.M., Herrmann J.M., Campo M.L., and Kinnally K.W.. 2007. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 282:3584–3593. 10.1074/jbc.M607551200 [DOI] [PubMed] [Google Scholar]
- Okamoto H., Miyagawa A., Shiota T., Tamura Y., and Endo T.. 2014. Intramolecular disulfide bond of Tim22 protein maintains integrity of the TIM22 complex in the mitochondrial inner membrane. J. Biol. Chem. 289:4827–4838. 10.1074/jbc.M113.543264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleh V., Cordat E., and Herrmann J.M.. 2016. Mia40 is a trans-site receptor that drives protein import into the mitochondrial intermembrane space by hydrophobic substrate binding. eLife. 5:e16177 10.7554/eLife.16177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt T.K., Atanassova N., Genereux J.C., and Wiseman R.L.. 2013. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab. 18:908–919. 10.1016/j.cmet.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A., Peleh V., Martinez-Caballero S., Wollweber F., Sommer F., van der Laan M., Schroda M., Alexander R.T., Campo M.L., and Herrmann J.M.. 2016. A disulfide bond in the TIM23 complex is crucial for voltage gating and mitochondrial protein import. J. Cell Biol. 10.1083/jcb.201602074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.R., Menold M.M., Garrett S., and Jensen R.E.. 1994. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol. Biol. Cell. 5:529–538. 10.1091/mbc.5.5.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Müller J.M., Milenkovic D., Guiard B., Pfanner N., and Chacinska A.. 2008. The MIA system for protein import into the mitochondrial intermembrane space. Biochim. Biophys. Acta. 1783:610–617. 10.1016/j.bbamcr.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Truscott K.N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A.J., Rassow J., Pfanner N., and Wagner R.. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082. 10.1038/nsb726 [DOI] [PubMed] [Google Scholar]
- Weckbecker D., Longen S., Riemer J., and Herrmann J.M.. 2012. Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J. 31:4348–4358. 10.1038/emboj.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L., Trojanowska A., Sztolsztener M.E., and Chacinska A.. 2013. Mitochondrial protein import: Mia40 facilitates Tim22 translocation into the inner membrane of mitochondria. Mol. Biol. Cell. 24:543–554. 10.1091/mbc.E12-09-0649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L., Sokol A.M., Chojnacka M., and Chacinska A.. 2016. The presence of disulfide bonds reveals an evolutionarily conserved mechanism involved in mitochondrial protein translocase assembly. Sci. Rep. 6:27484 10.1038/srep27484 [DOI] [PMC free article] [PubMed] [Google Scholar]