Abstract
Mycobacterium tuberculosis (M. tuberculosis) is the causative agent of tuberculosis (TB) that causes millions of death every year. We have sequenced the genome of M. tuberculosis isolated from cerebrospinal fluid (CSF) of a patient diagnosed with tuberculous meningitis (TBM). The isolated strain was referred as M. tuberculosis SB24. Genomic DNA of the M. tuberculosis SB24 was extracted and subjected to whole genome sequencing using PacBio platform. The draft genome size of M. tuberculosis SB24 was determined to be 4,452,489 bp with a G + C content of 65.6%. The whole genome shotgun project has been deposited in NCBI SRA under the accession number SRP076503.
Keywords: Mycobacterium tuberculosis, TBM, CSF, Genome, PacBio
| Specification | |
|---|---|
| Organism/cell line/tissue | Mycobacterium tuberculosis |
| Strain | SB24 |
| Sequencer or array type | PacBio RS II |
| Data format | Raw data |
| Experimental factor | Microbial strain |
| Experimental features | Raw data sequence of Mycobacterium tuberculosis SB24 genome |
| Consent | N/A |
| Sample source location | Kota Kinabalu, Sabah 5.9788°N 116.0753°E |
1. Direct link to deposited data
http://www.ncbi.nlm.nih.gov/bioproject/PRJNA325545
(SAMN05240578).
2. Experimental design, materials and methods
In Malaysia, tuberculosis (TB) remains a major health threat with over 24,000 cases reported annually since 2013 [1]. Among all the states in Malaysia, Sabah has always recorded the highest number of TB cases. Nevertheless, the characteristic of Mycobacterium tuberculosis (M. tuberculosis) strains that are prevailing in Malaysia, mainly in Sabah is poorly understood. Only a limited number of M. tuberculosis genomes in Malaysia have been sequenced [2]. Here, we report the first draft genome sequence of M. tuberculosis, strain SB24 isolated from cerebrospinal fluid (CSF) of a patient diagnosed with tuberculous meningitis (TBM) at a local hospital in Kota Kinabalu, Sabah. The sample was cultured in Middlebrook 7H9 broth supplemented with OADC enrichment (BD) and PANTA antibiotic mixture (BD). Based on in-vitro drug susceptibility testing (DST) using the microscopic observation drug susceptibility (MODS) assay, this isolate was susceptible to both rifampicin (RIF) and isoniazid (INH).
The genomic DNA of M. tuberculosis SB24 was extracted according to the method previously described [3]. Whole genome sequencing (WGS) was performed using the Pacific Biosciences RS II Single-Molecule Real Time (SMRT) sequencing technology. The library was prepared according to the 20-kb Template Preparation Using BluePippin™ Size-Selection System. Library was then sequenced on the PacBio RS II sequencing platform (Pacific Biosciences) using DNA/Polymerase Binding Kit P6 v2 (Pacific Biosciences) and DNA Sequencing Reagent Kit 4.0 (Pacific Biosciences). Titration density was 0.08 nM. Template was loaded into SMRT® cell v3 using a Mag Bead Kit. Sequencing was performed using one SMRT cell and a movie of 360 min was taken.
The sequence data obtained from the M. tuberculosis SB24 genome was assembled by using the Hierarchical Genome Assembly Process 3.0 (HGAP 3.0) in SMRT Portal v2.3.0 and the assembly was improved using Quiver iteratively in the SMRT portal. After quality-filtering and trimming, 87,044 reads were obtained with a mean length of 3543 bp totaling 308,405,967 bp. The complete genome sequence of M. tuberculosis SB24 was determined with a length of 4,452,489 bp and a G + C content of 65.6%. M. tuberculosis SB24 genome size is 40,957 bp and 48,653 bp longer than the reference strains H37Rv (4,411,532 bp) and M. tuberculosis CDC1551 (4,403,836) respectively.
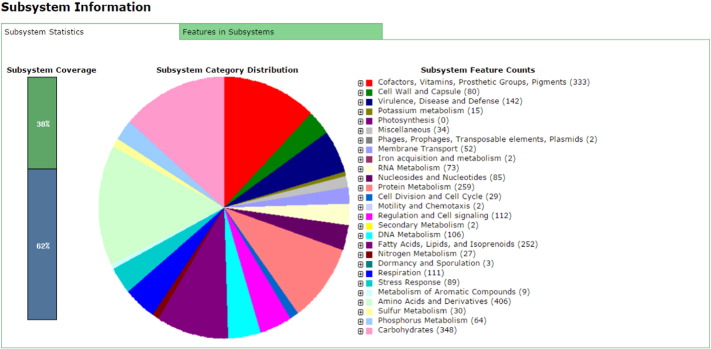
The draft genome of M. tuberculosis SB24 was annotated by using the Rapid Annotation of microbial genomes using Subsystem Technology (RAST) and GeneMarks [4], [5]. The annotation for the M. tuberculosis SB24 genome sequence using RAST showed 398 subsystems, 4586 coding sequences and 48 RNA genes (Fig.1). GeneMarks revealed that the genome of M. tuberculosis SB24 found to encode 4402 proteins.
Fig. 1.
Subsystem distribution of Mycobacterium tuberculosis SB24 strain (based on RAST annotation server).
The genome of M. tuberculosis SB24 encodes a gene for the Arylsulfatase enzyme (EC 3.1.6.1) which is involved in the metabolism of Sphingolipids (KEGG Pathway 00600), which leads to the production of Sulfur, one of the essential nutrients required in the survival of M. tuberculosis. Under sulfate-limiting environments, bacteria usually synthesize proteins, known as sulfate starvation-induced (SSI) proteins to complement their sulfur requirements [6]. Arylsulfatase enzyme is one of the SSI proteins produced and functionally help in the assimilation of sulfur from environmental sources. M. tuberculosis SB24 genome also encodes several polyketide synthase (pks) genes which are the pks1, pks7 and pks10. These genes involved in the synthesis of dimycocerosyl phthiocerol (DIM) which is necessary for the virulence of M. tuberculosis [7], [8].
Screening of antibiotic resistance genes using the curated database, the Antibiotic Resistance Genes Database (ARDB) (http://ardb.cbcb.umd.edu/) [9], M. tuberculosis SB24 was found to be resistant only to aminoglycosides, thus confirming the in-vitro susceptibility of this isolate to RIF and INH.
The closest strains of M. tuberculosis SB24 based on comparison of genome sequences using RAST are M. tuberculosis NCGM2209 (score 472), M. tuberculosis UM 1072388579 (score 417) and M. tuberculosis NA-A0008 (score 413). Based on the nucleotide sequence over the alignable portions using the fast sequence alignment tools BLAST [10], the genomes of M. tuberculosis SB24 was similar to M. tuberculosis HKBS1, M. tuberculosis CCDC5180, M. tuberculosis CCDC5079, M. tuberculosis 18b and M. tuberculosis Beijing-like with 99% similarity. Phylogenetic tree based on the aligned rpoB gene using BLAST showed that M. tuberculosis SB24 strain belongs to the Beijing family.
The availability of this genome sequence would enables the study of comparative analysis and phylogenetic study of this isolate with other M. tuberculosis strains and thus give us important insights into the biology and molecular epidemiology of M. tuberculosis strains that are prevalent in Sabah.
Nucleotide sequence accession number
The whole genome shotgun project has been deposited in NCBI SRA under the accession number SRP076503.
Conflict of interest
None.
Acknowledgments
This study was funded by UMS grant: SBK0163-SKK-2014.
References
- 1.Sman C. Borneopostonline; March 2016. Over 24 000 People Affected by TB Annually.http://www.theborneopost.com/2016/03/25over-24000-people-affected-by-tb-annauly/.25 (accessed 29.03.16) [Google Scholar]
- 2.Kuan C.S., Chan C.L., Yew S.M., Toh Y.F., Khoo J.S., Chong J., Lee K.W., Tan Y.C., Yee W.Y., Ngeow Y.F., Ng K.P. Genome analysis of the first extensively drug-resistant (XDR) Mycobacterium tuberculosis in Malaysia provides insights into the genetics basis of its biology and drug resistance. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0131694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han S.J., Song T., Cho Y.J., Kim J.S., Choi S.Y., Bang H.E., Chun J., Bai G.H., Cho S.N., Shin S.J. Complete genome sequence of Mycobacterium tuberculosis K from a Korean high school outbreak, belonging to the Beijing family. Stand. Genomic Sci. 2015;10:78. doi: 10.1186/s40793-015-0071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A.R., Fia F., Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besemer J., Lomsadze A., Borodovsky M. GeneMarks: a self-training method for prediction of gene starts in microbial genoms. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cregut M., Piutti S., Slezack-Deschaumes S., Benizri E. Compartmentalization and regulation of arylsulfatase activities in Streptomyces sp., Microbacterium sp. and Rhodococcus sp. soil isolates in response to inorganic limitation. Microbiol. Res. 2013;168(1):12–21. doi: 10.1016/j.micres.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhyay S., Nair S., Ghosh S. Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol. Rev. 2011;36(2):463–485. doi: 10.1111/j.1574-6976.2011.00302.x. [DOI] [PubMed] [Google Scholar]
- 8.Sirakova T.D., Fitzmaurice A.M., Kolattukudy P. Regulation of expression of mas and fadD28, two genes involved in production of dimcocerosyl phthiocerol, a virulence factor of Mycobacterium tuberculosis. J. Bacteriol. 2002;184(24):6796–6802. doi: 10.1128/JB.184.24.6796-6802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B., Pop M. ARDB-antibiotic resistance genes database. Nucleic Acids Res. 2009;37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]