Abstract
The 22 γ-Protocadherin (γ-Pcdh) adhesion molecules encoded by the Pcdhg gene cluster play critical roles in nervous system development, including regulation of dendrite arborisation, neuronal survival, and synaptogenesis. Recently, they have been implicated in suppression of tumour cell growth by inhibition of canonical Wnt signalling, though the mechanisms through which this occurs remain unknown. Here, we show differential regulation of Wnt signalling by individual γ-Pcdhs: The C3 isoform uniquely inhibits the pathway, whilst 13 other isoforms upregulate signalling. Focusing on the C3 isoform, we show that its unique variable cytoplasmic domain (VCD) is the critical one for Wnt pathway inhibition. γ-Pcdh-C3, but not other isoforms, physically interacts with Axin1, a key component of the canonical Wnt pathway. The C3 VCD competes with Dishevelled for binding to the DIX domain of Axin1, which stabilizes Axin1 at the membrane and leads to reduced phosphorylation of Wnt co-receptor Lrp6. Finally, we present evidence that Wnt pathway activity can be modulated up (by γ-Pcdh-A1) or down (by γ-Pcdh-C3) in the cerebral cortex in vivo, using conditional transgenic alleles. Together, these data delineate opposing roles for γ-Pcdh isoforms in regulating Wnt signalling and identify Axin1 as a novel protein interactor of the widely-expressed γ-Pcdh-C3 isoform.
Protocadherins (Pcdhs) belong to the cadherin superfamily, a group of cell adhesion molecules that are known to play critical roles in several biological processes, including embryonic morphogenesis, neural circuit formation, angiogenesis, and cancer1,2,3. The clustered Pcdhs, consisting of ~60 proteins encoded by three tandem gene clusters (Pcdha, Pcdhb, and Pcdhg) on human chromosome 5q31 (chromosome 18 in the mouse), represent the largest subgroup within the cadherin superfamily4,5. Within the Pcdha and Pcdhg clusters, large “variable” exons encoding 6 extracellular cadherin (EC) repeats, a transmembrane domain, and a variable cytoplasmic domain (VCD) are expressed from their own promoters and spliced to three short constant exons that encode a C-terminal domain shared by all α- or γ-Pcdhs (β-Pcdhs do not have this shared domain and are thus each encoded by a single exon; Pcdhg cluster schematized in Fig. 1a)4,6,7. Clustered Pcdhs, as a group, are strongly expressed throughout the developing and mature nervous system8,9,10,11,12, with lower levels detected in other organs such as lung and kidney13,14. The expression of individual clustered Pcdh genes is differentially regulated in each cell via promoter methylation and interaction with distant regulatory elements15,16,17,18. Single-cell RT-PCR analysis suggests that each neuron expresses a fraction of the 14 Pcdha, 22 Pcdhb, and 22 Pcdhg genes; most are stochastically and sparsely expressed, though 5 “C” subtype genes (Pcdhac1, Pcdhac2, Pcdhgc3, Pcdhgc4, and Pcdhgc5) appear to be expressed ubiquitously19,20,21. All clustered Pcdhs form cis-multimers that interact in a strictly homophilic manner in trans22,23,24.
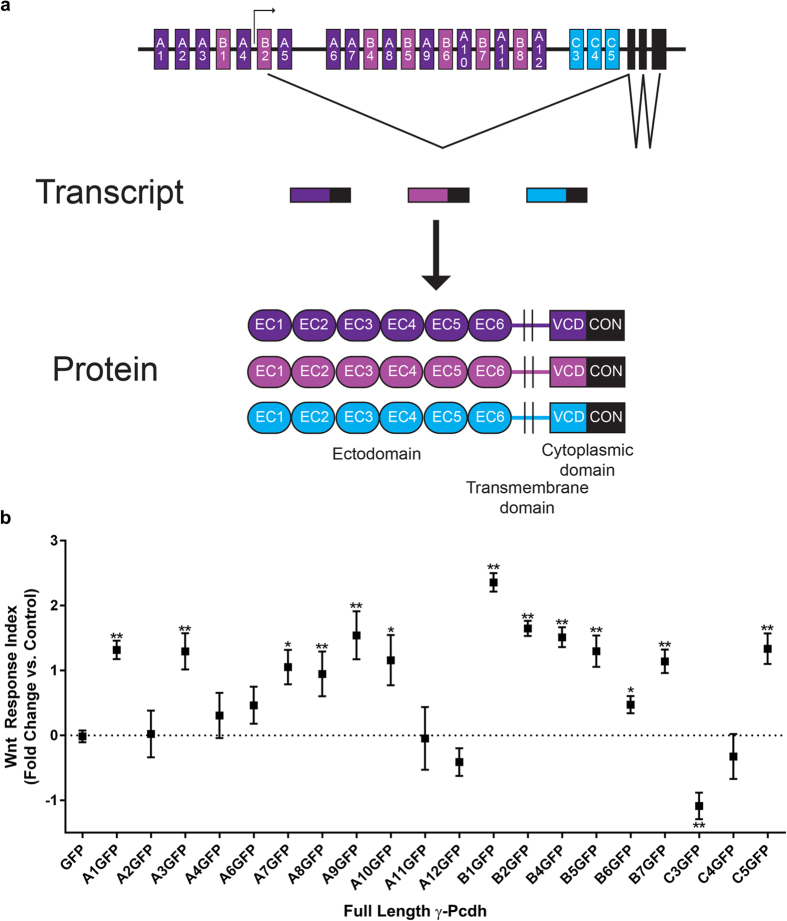
Figure 1. Differential regulation of canonical Wnt signalling by individual γ-Pcdh isoforms.
(a) Schematic of the mouse Pcdhg gene cluster (top), with examples of promoter activation, gene transcription, and splicing. Example transcripts and resultant proteins are schematized below. Each of the 22 isoforms is encoded by 4 exons: 1 variable exon (shades of purple or blue; encodes 6 EC domains, a transmembrane domain, and a VCD) and 3 constant exons (black; encode a shared C-terminal domain). EC: extracellular cadherin repeats; VCD: variable cytoplasmic domain; CON: constant region. (b) TOPFlash assays were performed by co-transfecting plasmids encoding individual γ-Pcdh isoforms with the SUPER-TOPFlash and Renilla luciferase plasmids into HEK293 cells. Cell lysates were assayed for luciferase activity 24 hours after exposure to Wnt3a CM. Results are presented as a “Wnt Response Index”, defined as the fold change in Relative Luciferase Units (RLU; ratio of firefly luciferase to Renilla luciferase [control for transfection efficiency]) following Wnt treatment in cells transfected with individual γ-Pcdh isoforms compared to that in cells expressing only a GFP negative control. Thus, the control (GFP only) Wnt cellular response is the baseline (“0”), and manipulations that increase further that response yield a positive index, while those that decrease that response yield a negative index. Several γ-Pcdh isoforms significantly upregulated Wnt signalling activity, while only γ-Pcdh-C3 significantly downregulated Wnt signalling. Data are presented on a log2 scale of means ± SEM from 6 independent experiments. A two-way ANOVA with Bonferroni post hoc test (to correct for multiple comparisons) was performed to assess statistical significance. *p < 0.05, and **p < 0.01.
The clustered Pcdhs play critical roles in neurodevelopment. Using constitutive and conditional Pcdhg mouse mutants, we demonstrated that the γ-Pcdhs regulate neuronal survival11,25, synaptogenesis26,27, astrocyte-neuron interactions26, dendrite arborisation8,28,29, and axonal patterning30. In retinal starburst amacrine cells, the γ-Pcdhs are also required for normal dendrite self-avoidance31,32, underscoring the importance of this family for the proper formation of dendrite arbours. The relative importance of the γ-Pcdhs is underscored by the fact that mice lacking all 22 isoforms die shortly after birth7, in contrast to the viable and fertile Pcdha cluster nulls33. Nevertheless, the α-Pcdhs have been shown to regulate dendrite arborisation and dendritic spine formation34, as well as axonal targeting33,35,36,37, in a variety of neurons.
Intriguingly, the γ-Pcdhs also have been recently implicated as potential tumour suppressors, via inhibition of canonical Wnt signalling13. A genome-wide analysis of promoter hypermethylation in Wilms’ tumour, a paediatric kidney cancer, identified the clustered Pcdh genes as consistently hypermethylated; many of them, especially those encoding γ-Pcdhs, are indeed transcriptionally silenced in tumour cells. Knockdown of all Pcdhg genes (using an siRNA targeting the constant region) in kidney cell lines led to increased canonical Wnt signalling, while overexpression of individual Pcdhg cDNAs inhibited the Wnt pathway and led to reduced tumour cell growth in soft agar assays13. The ubiquitously-expressed γ-Pcdh-C3 isoform (encoded by Pcdhgc3) was similarly shown to be silenced in colorectal cancer cells, and to reduce growth of these cells when overexpressed via inhibition of a Wnt-mTOR pathway38. Together with other studies identifying aberrant methylation of Pcdhga11 in astrocytomas39, Pcdhb genes in neuroblastoma40, and Pcdhg genes in toxicant-induced malignant cells41, these data suggest that understanding how clustered Pcdhs regulate tumour growth pathways will be of future translational importance. Intriguingly, a non-clustered δ-Pcdh known as PAPC/Pcdh8/Arcadlin can also affect canonical42 and non-canonical43,44 Wnt pathways, suggesting that Pcdhs in general may be of interest to our understanding of Wnt biology.
Here, we have investigated the molecular mechanisms through which γ-Pcdh-C3 inhibits canonical Wnt signalling, a well-known tumourigenic pathway that is also of paramount importance for many steps in neurodevelopment45,46. Binding of Wnt ligands to Frizzled and the Lrp5/6, Ryk, or ROR co-receptors can activate at least three distinct pathways: the canonical (β-catenin/TCF), PCP (planar cell polarity), and Wnt-Ca2 + pathways47,48,49. In the canonical pathway, β-catenin is found in a complex including Axin1, Adenomatous Polyposis Coli (APC) and GSK3β in the absence of Wnt ligand (the OFF state; reviewed in Clevers and Nusse, 2012)50. Axin1 acts as a scaffold for the “destruction complex”: GSK3β phosphorylates β-catenin, which is then targeted for ubiquitin-dependent degradation by the proteasome. In the presence of the Wnt ligand (the ON state), Dishevelled (Dvl) binds at the C-terminus of Frizzled51 and recruits Axin1 to the cytoplasmic tail of Lrp5/6, facilitating the phosphorylation of Lrp5/6 by GSK3β and CK152,53,54,55,56,57. Phospho-Lrp6 is thought to directly inhibit GSK3β activity in the destruction complex, leading to reduced phosphorylation of β-catenin as well as Axin1 itself. Dephosphorylated Axin1 dissociates from the activated receptor complex and also from β-catenin, thus inactivating the destruction complex until Axin1 is again phosphorylated55,58. This inactivation of the destruction complex is thought to allow β-catenin to accumulate in the cytoplasm and to translocate to the nucleus. There, it displaces Groucho/TLE corepressors bound to TCF (T-cell factor)/LEF (Lymphoid enhancer factor) transcription factors, thereby activating Wnt target genes, including many that promote tumour proliferation or regulate brain patterning, dendrite arborisation, and synaptogenesis45,50,59.
Though the γ-Pcdhs have been implicated in the regulation of Wnt signalling13,38, the underlying molecular mechanisms are unknown. Here, we show that γ-Pcdh-C3, but not other γ-Pcdhs, significantly inhibits exogenous activation of canonical Wnt signalling in cultured cells, and identify the C3-specific VCD as the critical site of action. Though it has been reported that the intracellular domain of γ-Pcdhs can be cleaved and trafficked to the nucleus60,61, we show that the C3 VCD acts at the membrane, not the nucleus, to inhibit Wnt signalling. We further show that the C3 VCD physically, and directly, interacts with the DIX domain of Axin1, and present evidence that it competes for this binding site with Dvl1. Binding of Axin1 to the C3 VCD stabilizes Axin1 at the membrane and prevents Lrp6 phosphorylation, suggesting that in this context, membrane localization can actually stabilize the β-catenin destruction complex. Finally, we use a Wnt signalling reporter mouse line and an inducible γ-Pcdh-C3 overexpression allele to show that that increasing C3 levels reduces endogenous Wnt signalling in the mouse cerebral cortex in vivo. Together, these data identify a novel molecular mechanism by which γ-Pcdhs can affect Wnt signalling, and identify Axin1 as the first known intracellular signalling partner for the widely-expressed γ-Pcdh-C3 isoform.
Results
γ-Pcdh isoforms differentially regulate canonical Wnt signalling
We began by asking whether the many γ-Pcdh isoforms (Fig. 1a) uniformly, or differentially, affect the Wnt signalling pathway. To assay Wnt activity, we utilized the well-characterized TOPFlash assay, in which a TCF/LEF-responsive promoter drives firefly luciferase, levels of which can be normalized to constitutively-expressed Renilla luciferase to control for transfection efficiency in HEK293 cells62. To activate canonical Wnt signalling, we applied media conditioned by L cells stably expressing Wnt3a63. Dallosso et al.13,38 previously reported that human γ-Pcdh-A2, -A6, -A12 and -C3 were all capable of inhibiting aberrantly high endogenous TOPFlash signal in Wilms’ tumour cell lines, or the increase seen following Wnt treatment in HEK293 cells. We individually overexpressed 20 γ-Pcdh isoforms in HEK293 cells and found differential activity among γ-Pcdhs. In agreement with Dallosso et al.38, overexpression of the C3 isoform reliably and significantly inhibited Wnt signalling (Fig. 1b), though in contrast to Dallosso et al.13 we found that A2, A6, or A12 overexpression had no significant effect (Fig. 1b). Surprisingly, we also found that overexpression of several other isoforms, including A1, A3, A7-10, B1-7, and C5, all significantly increased the TOPFlash response in HEK293 cells (Fig. 1b). Results are presented throughout as a “Wnt Response Index”, defined as the fold change the Wnt response in cells overexpressing a γ-Pcdh compared to that in cells expressing only a GFP negative control. Example raw TOPFlash and FOPFlash (negative control plasmids in which TCF/LEF sites are mutated) data are presented in Supplementary Figure S1; FOPFlash results were always essentially zero and are not shown for subsequent experiments. In all subsequent analyses, we focus on the C3 isoform because: 1) our results in this case are fully in agreement with those of Dallosso et al.38 in other cell lines; 2) C3 is the only isoform that significantly downregulated Wnt signalling; and 3) C3 is highly and ubiquitously expressed and thus is likely to be of importance in the greatest number of contexts.
The variable cytoplasmic domain of γ-Pcdh-C3 mediates inhibition of Wnt signalling
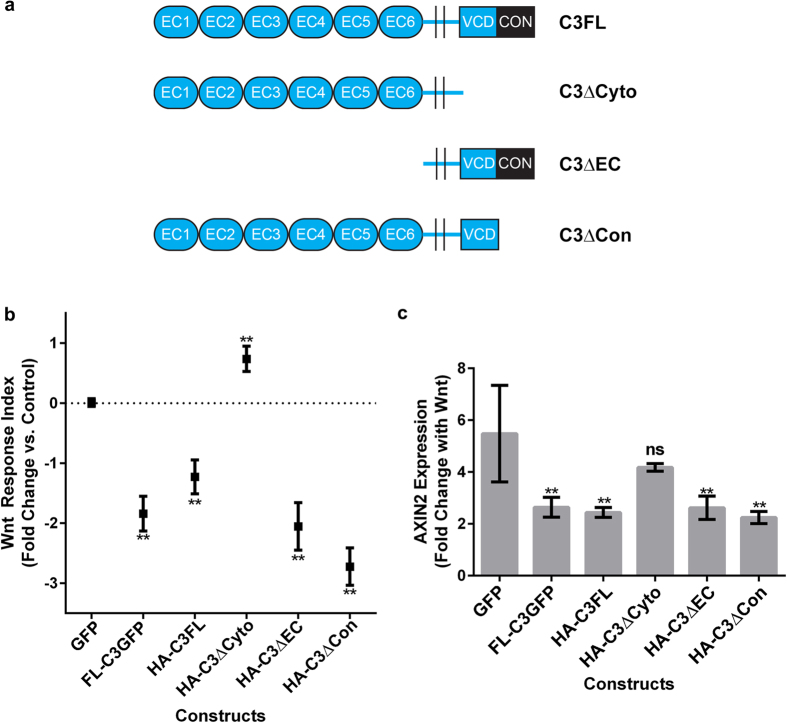
We next asked which domain of the γ-Pcdh-C3 protein was important for its inhibition of Wnt signalling. As noted above, each γ-Pcdh isoform is encoded by four exons: the large variable exon, specific to each isoform, encodes 6 EC repeats (EC2 and 3 determine homophilic binding specificity23;), a transmembrane domain, and the ~90 amino acid VCD; the 3 small constant exons encode a C-terminal domain shared by all γ-Pcdhs (Fig. 1a). We generated and tested the following constructs: γ-Pcdh-C3 ΔEC (lacking all 6 EC repeats); γ-Pcdh-C3 ΔCyto (lacking the entire intracellular domain, including both the VCD and the constant domain); and γ-Pcdh-C3 ΔCon (lacking only the shared constant domain) (Fig. 2a). When these constructs were used in the TOPFlash assay as described above, we found that all except the C3 ΔCyto construct were still able to negatively regulate Wnt signalling activity. Expression of C3 ΔCyto failed to inhibit Wnt activity, and in fact resulted in a small, but statistically significant, increase (Fig. 2b). To confirm these results, we utilized another assay for canonical Wnt signalling, performing quantitative RT-PCR for AXIN2, a Wnt target gene. In control (GFP-transfected) cells exposed to Wnt3a, AXIN2 transcript levels increased more than five-fold (Fig. 2c). Overexpression of C3 full length (FL; either C-terminally GFP-tagged or N-terminally HA-tagged), C3 ΔEC, or C3 ΔCon significantly abrogated this AXIN2 response, while overexpression of C3 ΔCyto did not (Fig. 2c). Together, these data implicate the C3 VCD as the protein domain responsible for inhibiting Wnt signalling.
Figure 2. The variable cytoplasmic domain (VCD) is critical for γ-Pcdh-C3 inhibition of Wnt signalling.
(a) Schematic of constructs used in experiments to determine the domain involved in γ-Pcdh-C3’s regulation of Wnt signalling. All are HA-tagged at the N-terminus. (b) The aforementioned constructs were used in a TOPFlash assay, as previously described. All constructs except HA-C3ΔCyto significantly inhibited Wnt signalling response 24 hours after the addition of Wnt3a CM, implicating the VCD as the key domain. Data are presented on a log2 scale of means ± SEM from 6 independent experiments. A two-way ANOVA with Bonferroni post hoc test (to correct for multiple comparisons) was performed to assess statistical significance. **p < 0.01 (c) Quantitative TaqMan® PCR on AXIN2, a Wnt target gene, was performed 24 hours after exposure to Wnt3a CM in HEK293 cells transfected with the constructs in (a). Data are presented as fold increase in compared to DMEM media change control for cells transfected with each construct. AXIN2 expression increases ~5-fold in the GFP transfected control, 24 hours after addition of Wnt3a CM. This increase is significantly abrogated in cells expressing all γ-Pcdh-C3 constructs except HA-C3ΔCyto, paralleling TOPFlash results in (b). Means ± SEM of AXIN2 expression from 3 independent experiments were graphed. A two-way ANOVA with Bonferroni post hoc test (to correct for multiple comparisons) was performed to assess statistical significance. **p < 0.01.
The variable cytoplasmic domain of γ-Pcdh-C3 acts at the membrane to inhibit Wnt signalling
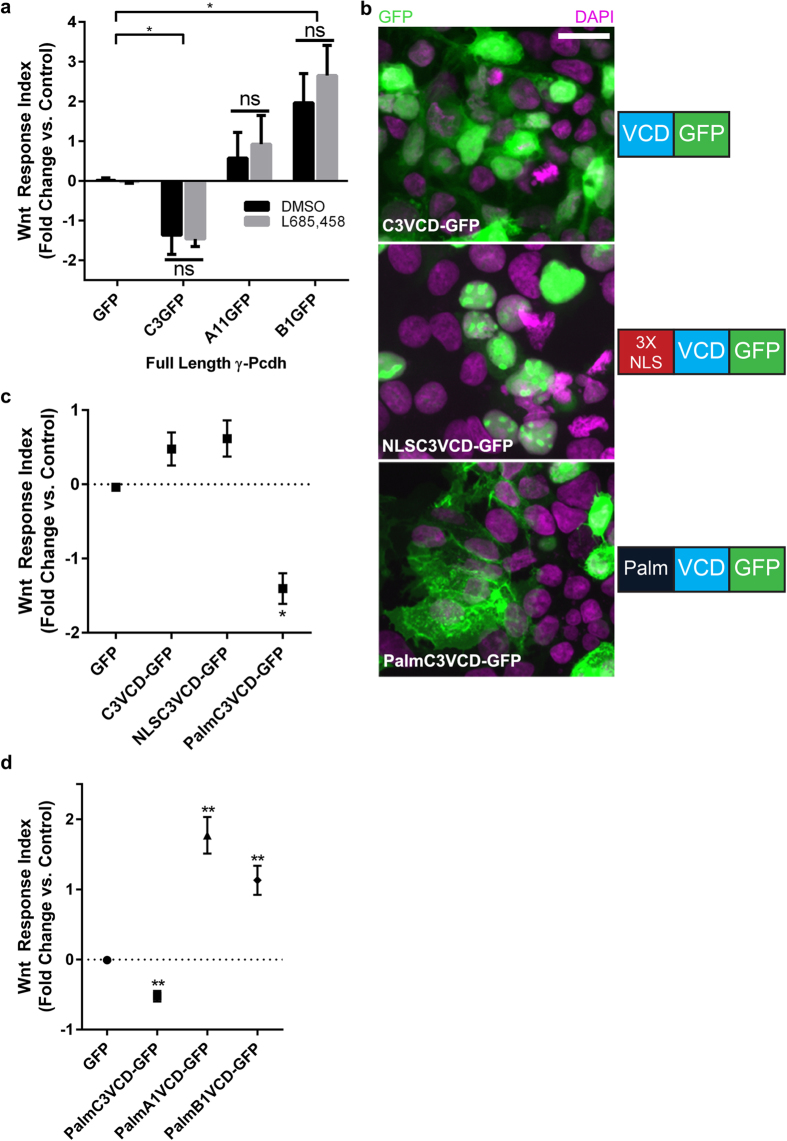
It has been reported that γ-Pcdhs can undergo two cleavage events: ectodomain shedding mediated by matrix metalloproteinases (MMPs) at the junction between the EC repeats and transmembrane domain, and cleavage at the junction between the intracellular and transmembrane domain by γ-secretase60,61. The cleaved intracellular domain (containing both the VCD and the constant domain) can translocate to the nucleus60,61, where we reasoned it might interfere directly with TCF/LEF-dependent Wnt target gene activation. We thus asked whether inhibition of γ-secretase cleavage would abrogate γ-Pcdh-C3’s ability to inhibit Wnt signalling. We treated cells with a specific pharmacological inhibitor of γ-secretase (L685,458), previously shown to block γ-Pcdh intracellular domain cleavage60,61. In the TOPFlash assay, there was no effect of treatment with L685,458 vs. the DMSO vehicle control: C3 significantly inhibited Wnt signalling, A11 had no effect, and B1 significantly potentiated Wnt signalling, as observed previously (Fig. 1b), regardless of inhibitor treatment (Fig. 3a). Western blots for the relevant C-terminal fragment confirmed the efficacy of L685,458 in this assay (Supplementary Figure S2). These observations indicate that γ-Pcdh C-terminal cleavage is not important for effects on Wnt signalling, and argue against a nuclear site of action for the C3 VCD.
Figure 3. The γ-Pcdh-C3 VCD acts at the membrane to inhibit Wnt signalling.
(a) Cells transfected with FL-C3GFP, FL-A11GFP and FL-B1GFP were treated with 10 μM L685,458 (to inhibit γ-secretase), or DMSO (vehicle control) and used in a TOPFlash assay, as previously described. Treatment with L685,458 did not significantly alter individual γ-Pcdh isoforms’ effect on Wnt signalling activity, indicating that C-terminal cleavage is not required. (b) Constructs encoding the isolated VCD of γ-Pcdh-C3 fused to GFP, either unmodified (C3VCD-GFP), tagged with a triplicate nuclear localization signal (NLSC3VCD-GFP) or with a palmitoylation sequence (PalmC3VCD-GFP) were generated. HEK293 cells transfected with these constructs were immunostained for GFP (green) and counterstained with DAPI (purple) to assess subcellular localization and visualize nuclei, respectively. Localization was as predicted: C3VCD-GFP localizes to both cytoplasm and nucleus, NLSC3VCD-GFP is restricted to nuclei, and PalmC3VCD-GFP is concentrated at the cell surface and at cell-cell junctions. Scale bar = 20 μm. (c) These new constructs were used in a TOPFlash assay, as described previously. PalmC3VCD-GFP was the only construct that significantly inhibited Wnt signalling activity; C3VCD-GFP and NLSC3VCD-GFP did not differ significantly from control. (d) Palmitoylated A1VCD-GFP and B1VCD-GFP were made and used in a TOPFlash assay. The VCD alone of these individual γ-Pcdh isoforms are sufficient to significantly upregulate Wnt signalling activity. Data are presented on a log2 scale of means ± SEM from 6 independent experiments. A two-way ANOVA with Bonferroni post hoc test (to correct for multiple comparisons) was performed to assess statistical significance. *p < 0.05 ** p < 0.01.
To confirm this directly, we designed new constructs to investigate the cellular compartment at which the C3 VCD acts to inhibit Wnt signalling. These constructs encoded the isolated C3 VCD fused to GFP, either alone (C3VCD-GFP) or with the addition of a 3X nuclear localization signal (NLSC3VCD-GFP) or a membrane-targeting palmitoylation signal (PalmC3VCD-GFP). Each construct exhibited the expected subcellular localization when expressed in HEK293 cells (Fig. 3b). Of the three constructs, only the membrane-targeted palmitoylated version significantly inhibited Wnt signalling (assayed using TOPFlash) following addition of Wnt3a CM (Fig. 3c). These data indicate that the VCD of γ-Pcdh-C3 acts at the plasma membrane to regulate Wnt signalling, consistent with the normal topology of the full-length protein and with our demonstration that preventing cleavage of the C3 intracellular domain (Fig. 3a) did not disrupt Wnt inhibition. Given these results, we asked whether γ-Pcdh isoforms that upregulate Wnt signalling (Fig. 1b) also do so via their VCDs. We constructed plasmids encoding PalmA1VCD-GFP and PalmB1VCD-GFP and tested them in the TOPFlash assay as above. Indeed, we found that cells expressing either construct exhibited significantly enhanced response to Wnt3a (Fig. 3d), just as cells expressing their full length counterparts did (Fig. 1b).
γ-Pcdh-C3 physically interacts with the DIX domain of Axin1 via its variable cytoplasmic domain
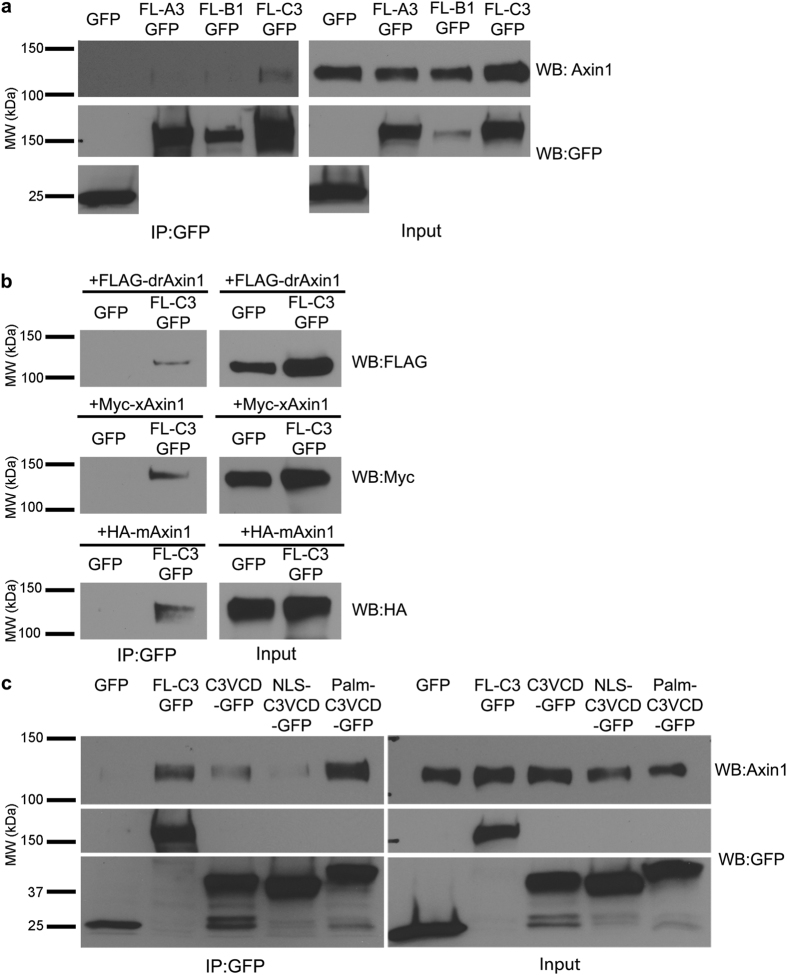
Given that the C3 VCD inhibits Wnt signalling by acting at the plasma membrane, we hypothesized that C3 may physically interact with one of the many components of the Wnt pathway that can localize there during signalling. A previous large-scale proteomics study suggested that Ryk could be a potential target of some γ-Pcdhs, including C364. We transfected HEK293 cells with plasmids encoding FL C3-GFP, or GFP only as a negative control, immunoprecipitated using anti-GFP antibody, and blotted with antibodies to a variety of Wnt pathway proteins including Ryk, Dvls, Frizzleds, Lrp6, Naked, GSK3β, and Axin1. The only endogenous human protein that co-immunoprecipitated with mouse FL C3-GFP was Axin1 (Fig. 4a; we were unable to confirm an interaction with Ryk64 despite multiple attempts). We were unable to co-immunoprecipitate Axin1 reliably with two other γ-Pcdhs tested (A3 or B1), suggesting that this interaction is specific to γ-Pcdh-C3 (Fig. 4a). Using a variety of tagged Axin1 plasmids in a co-transfection immunoprecipitation assay, we found this interaction to be evolutionarily conserved, as mouse FL C3-GFP could pull down FLAG-tagged zebrafish (drAxin1), Xenopus (xAxin1), and mouse (mAxin1) proteins (Fig. 4b) as well as endogenous human Axin1 expressed by HEK293 cells. Given that we found that the VCD of C3 is the critical domain for Wnt pathway inhibition, we next tested whether Axin1 interacted with this domain in particular. We found that the C3 VCD co-immunoprecipitated endogenous Axin1; intriguingly, the PalmC3VCD construct consistently showed a more robust interaction with Axin1 than did the cytoplasmic or nuclear constructs (Fig. 4c). This result suggests that at least some of the C3VCD-Axin1 complex co-immunoprecipitated in this assay formed in the cell, at the membrane, rather than in solution following lysis.
Figure 4. The γ-Pcdh-C3 VCD interacts with Axin1.
(a) Lysates of HEK293 cells transfected with either GFP (control), FL-A3GFP, FL-B1GFP or FL-C3GFP were immunoprecipitated with anti-GFP and blotted for endogenous (human) Axin1. Axin1 co-immunoprecipitated with γ-Pcdh-C3 but not –A3 or –B1. All proteins encoded by the transfected constructs were present in the cell lysates used (right; input) and re-probing of blots with anti-GFP antibody confirmed immunoprecipitation. (b) Lysates from HEK293 cells co-transfected with GFP or FLC3-GFP and either FLAG-tagged zebrafish (Danio rerio) Axin1 (FLAG-drAxin1), Myc-tagged Xenopus Axin1 (Myc-xAxin1) or HA-tagged mouse Axin1 (HA-mAxin1) were immunoprecipitated with anti-GFP and blotted for the tagged Axin1. FL-C3GFP co-immunoprecipitated all three Axin1 orthologues. (c) Co-immunoprecipitation of endogenous HEK293 Axin1 as in (a) was performed using GFP, FLC3-GFP, C3VCD-GFP, NLSC3VCD-GFP and PalmC3VCD-GFP. In all cases, the VCD is sufficient to pull down endogenous Axin1; co-immunoprecipitation was more robust for PalmC3VCD-GFP. Blots shown are representative of at least 3 experiments. MW, molecular weight; kDa, kilodaltons.
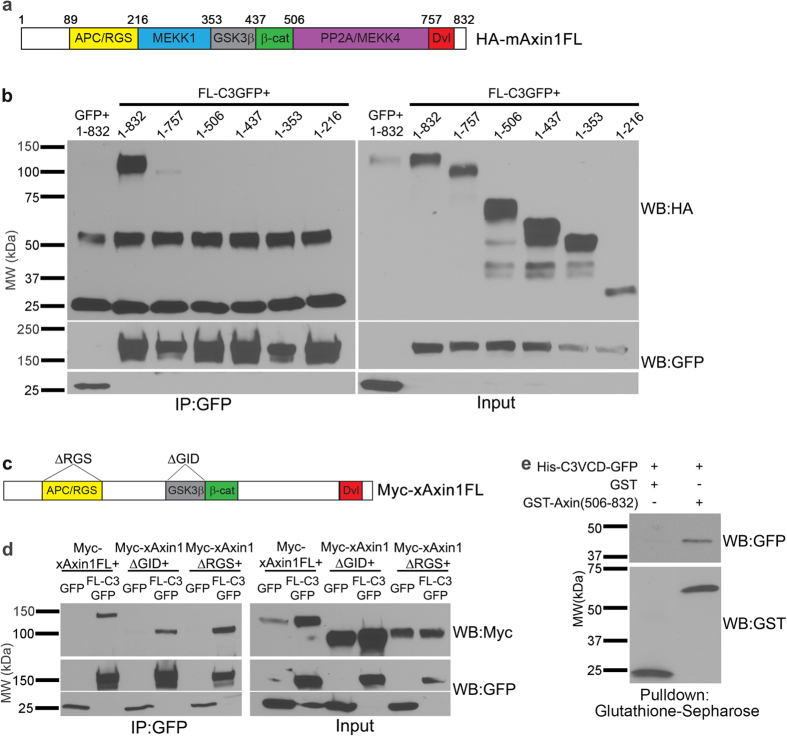
We next sought to identify the domain of Axin1 to which the C3 VCD binds. The protein structure of Axin1 has been well studied, leading to the identification of multiple conserved domains and regions of protein-protein interactions. These include binding regions for APC (the RGS domain), MEKK1, GSK3β, β-catenin, PP2A/MEKK4, and Dvl (DIX domain; Fig. 5a). We utilized constructs65 encoding HA-tagged mAxin1 with progressive 3′ deletions leading to loss of these domains in co-immunoprecipitation assays with FL C3-GFP (Fig. 5b). Full length mAxin1 protein again exhibited robust interaction with γ-Pcdh-C3; this interaction was almost completely abrogated following the loss of amino acids 757-832, and no interaction was observed with further mAxin1 truncation (Fig. 5b). This result indicates that the C3 VCD interacts with mAxin1 via the latter’s C-terminal Dvl-binding DIX domain. However, since nearly all binding was lost with the very first 3′ truncation, we sought to confirm this by utilizing two Myc-tagged xAxin1 constructs with internal deletions of the GSK3β (ΔGID) or APC (ΔRGS) domains (Fig. 5c). As expected, both of these truncated constructs, as well as FL xAxin1, robustly interacted with FL C3-GFP in transfected HEK293 cells (Fig. 5d). Co-immunoprecipitation can reflect either direct binding between two proteins, or their mutual interaction with a complex of proteins. To ascertain whether the interaction between Axin1 and C3 VCD was direct, we generated new constructs for bacterial production of His-tagged C3VCD-GFP and GST-tagged mouse Axin1(506-832; encompassing the PP2A/MEKK4 and DIX domains), and assessed binding in a cell-free assay. Using glutathione-sepharose to pull down GST-fusion proteins, we recovered His-C3VCD-GFP with GST-Axin1(506–832), but not with GST only (Fig. 5e). Together, our data indicate a direct physical interaction between the VCD of γ-Pcdh-C3 and the DIX domain of Axin1.
Figure 5. γ-Pcdh-C3 interacts with the DIX domain of Axin1.
(a) Schematic of specific protein-protein interacting domains previously identified in mouse Axin1. Numbers above the protein correspond with the amino acid at which C-terminal truncations were made to sequentially remove individual domains. (b) Lysates from HEK293 cells co-transfected with GFP or FL-C3GFP and the HA-tagged full-length mouse Axin1 (1-832); or FL-C3GFP with HA-tagged mouse Axin1 with progressive 3′ deletions (containing amino acids 1-757, 1-506, 1-437, 1-353 or 1-216) were immunoprecipitated with anti-GFP and blotted for HA. Interaction of γ-Pcdh-C3 with Axin1 was abrogated with the loss of amino acids 758-832, indicating the importance of the DIX domain. (c) Schematic of Myc-tagged full length Xenopus Axin1 (Myc-xAxin1FL) and Myc-tagged Xenopus Axin1 with the APC/RGS binding domain (Myc-xAxinΔRGS) or the GSK3β binding domain deleted (Myc-xAxin1ΔGID). (d) Lysates from HEK293 cells co-transfected GFP or FL-C3GFP and Myc-xAxin1FL, Myc-xAxinΔRGS, or Myc-xAxin1ΔGID were immunoprecipitated with anti-GFP and blotted for Myc. Interaction between γ-Pcdh-C3 and Axin1 was observed with all three constructs used, confirming that these internal domains are not critical. (e) Purified His-tagged C3VCD-GFP was mixed with either GST alone, or with GST-Axin(506-832) in a direct binding assay. MW, molecular weight; kDa, kilodaltons.
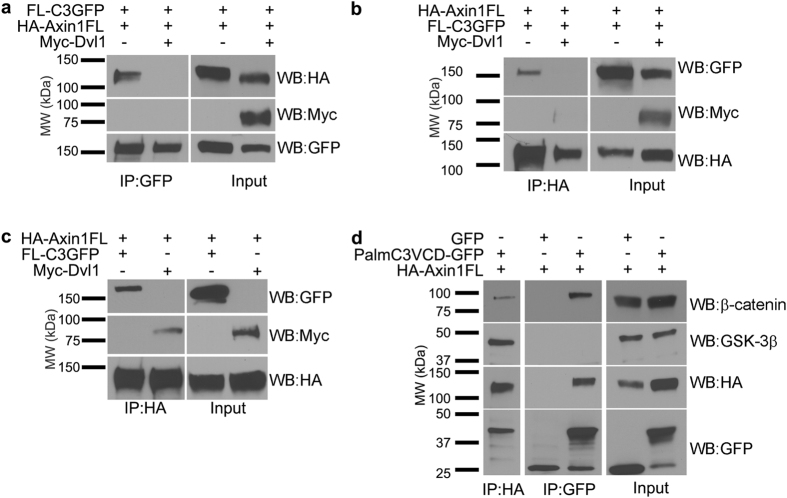
γ-Pcdh-C3 competes with Dvl1 for Axin1 binding and stabilizes Axin1 at the membrane
As noted above, we observed that the Dvl-binding DIX domain of Axin1 was important for its interaction with γ-Pcdh-C3 (Fig. 5b). Thus, we next asked whether Dvl and γ-Pcdh-C3 might compete for Axin1 binding. To test this, HEK293 cells were co-transfected with constructs encoding FL C3-GFP and HA-tagged mAxin1, with or without the addition of a third plasmid encoding Myc-tagged Dvl1. We then immunoprecipitated with antibodies against GFP (for the γ-Pcdh-C3) and blotted for the HA-Axin1, or immunoprecipitated with antibodies against HA (for the Axin1) and blotted for the C3-GFP. These reciprocal immunoprecipitations both show that the interaction between γ-Pcdh-C3 and Axin1 is lost when Dvl1 is overexpressed (Fig. 6a,b). Interestingly, Dvl1 did not co-immunoprecipitate with Axin1 in the presence of γ-Pcdh-C3 (Fig. 6b), although it did, as expected, in its absence (Fig. 6c), indicating that C3 effectively competes with Dvl1 for Axin1 binding. We next asked if γ-Pcdh-C3 interacts with other destruction complex components to regulate Wnt signalling. We overexpressed PalmC3VCD-GFP and full length HA-tagged mAxin1 in HEK293 cells, immunoprecipitated with either anti-HA or anti-GFP, and blotted for endogenous β-catenin or GSK3β. A previous proteomics study66 recovered β-catenin in material pulled down from brain with an antibody recognizing all γ-Pcdhs. We were able to confirm an interaction of the C3VCD with β-catenin in these transfected HEK293 cells (Fig. 6d); note that this likely to be indirect via Axin1, as γ-Pcdhs (unlike classical cadherins) do not exhibit any known catenin binding sites. We also were able to co-immunoprecipitate both β-catenin and GSK3β with Axin1, as expected67. However, we were unable to observe an interaction between γ-Pcdh C3 and GSK3β in this assay (Fig. 6d). This suggests that γ-Pcdh-C3 can interact with Axin1 and β-catenin, but probably not in the context of a complete destruction complex.
Figure 6. γ-Pcdh-C3 competes for Dvl1 binding to Axin1.
(a,b) Lysates from HEK293 cells co-transfected with FL-C3GFP and HA-Axin1FL, with or without Myc-Dvl1 were immunoprecipitated with anti-GFP or anti-HA to pull down γ-Pcdh-C3 or Axin1, respectively. In either case, the interaction between γ-Pcdh-C3 and Axin1 is abrogated in the presence of Dvl1. (c) Lysates from HEK293 cells co-transfected with HA-Axin1FL and FL-C3GFP or Myc-Dvl1 were immunoprecipitated with anti-HA to pull down Axin1, and blotted for GFP to detect γ-Pcdh-C3 or Myc to detect Dvl1. Axin1 can co-immunoprecipitate either γ-Pcdh-C3 or Axin1, as long as both are not present. (d) HEK293 cells co-transfected with HA-Axin1FL and either GFP or PalmC3VCD-GFP were immunoprecipitated with anti-GFP or anti-HA, and blotted for endogenous β-catenin and GSK3β (components of the Wnt destruction complex). Axin1 interacts with both β-catenin and GSK3β, while γ-Pcdh C3 interacts with β-catenin, but apparently not with GSK3β. MW, molecular weight; kDa, kilodaltons.
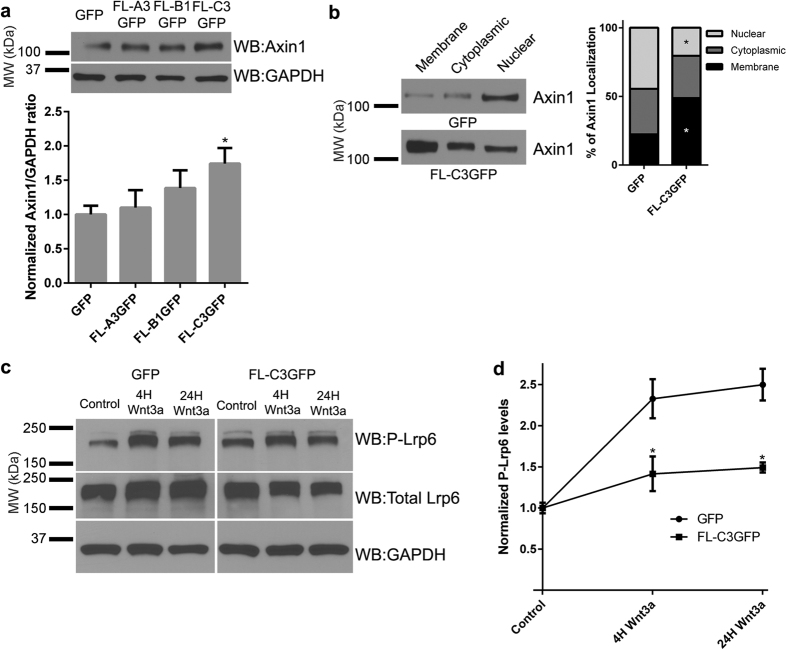
In the course of our immunoprecipitation experiments, we noticed that the overall levels of Axin1 were often increased when γ-Pcdh-C3 was overexpressed in comparison to GFP (e.g., compare GFP to C3 lanes in input blots in Fig. 5b,d). We reasoned that this might indicate that interaction with γ-Pcdh-C3 stabilizes Axin1 at the membrane. To test this, we first quantified total endogenous Axin1 levels in HEK293 cells transfected with GFP only vs. γ-Pcdh-A3, -B1, or -C3. Only the C3 isoform significantly increased total Axin1 levels in this assay (Fig. 7a). Subcellular fractionation also demonstrated that cells overexpressing γ-Pcdh-C3 exhibited a significant shift in Axin1 localization to the membrane compared to cells expressing the GFP-only control (Fig. 7b). Together, these data suggest that Axin1 is stabilized when bound to the C3 VCD at the plasma membrane.
Figure 7. Interaction between γ-Pcdh C3 and Axin1 stabilizes Axin1 at the membrane and prevents phosphorylation of Lrp6.
(a) Representative Western blot showing increased endogenous Axin1 levels in HEK293 cells overexpressing FL-C3GFP in comparison to GFP, FL-A3GFP, or FL-B1GFP. The band intensity of Axin1 was quantified and normalized to that of GAPDH in the same lane. Means ± SEM of 4 independent experiments were graphed. A one-way ANOVA with Bonferroni post hoc test (to correct of multiple comparisons) was used to assess statistical significance. Only γ-Pcdh-C3 significantly increased levels of Axin1. *p < 0.05 (b) Subcellular fractionation of HEK293 cell lysates overexpressing FL-C3GFP shows a significant shift in Axin1 localization to the membrane fraction in comparison cells expressing only the GFP control. The intensity of Axin1 bands from all membrane, cytoplasmic, and nuclear fractions were quantified across six individual experiments and analysed with a two-way ANOVA with Bonferroni post hoc test. *p < 0.05 (c,d) Cells transfected with either GFP or FL-C3GFP were exposed to DMEM (control), or Wnt3a for 4 or 24h before being lysed and immunoblotted for phosphorylated Lrp6 (S1490), total Lrp6 and GAPDH. Levels of phosphorylated Lrp6 (normalized to GAPDH) increase >2-fold in GFP-only transfected cells; this increase is significantly abrogated in the presence of g-Pcdh-C3 expression. The intensity of the phospho-Lrp6 band for each condition was normalized to GAPDH in the same lane, and means of 3 individual experiments ± SEM were graphed. A two-way ANOVA with Bonferroni post hoc test was performed. *p < 0.05 MW, molecular weight; kDa, kilodaltons.
γ-Pcdh-C3 prevents Lrp6 phosphorylation in response to Wnt ligand
The Dvl competition data suggested that γ-Pcdh-C3 might inhibit Wnt signalling at the level of Wnt co-receptor activation, in this case Lrp6 phosphorylation. We addressed this possibility by assaying levels of phospho-Lrp6 (Ser1490) following Wnt3a CM addition in the presence or absence of γ-Pcdh-C3. Indeed, we found that, compared to cells transfected with GFP only, HEK293 cells overexpressing FL C3-GFP had significantly lower induction of Lrp6 phosphorylation at Ser1490 (Fig. 7c,d). Axin1 is required for Lrp6 phosphorylation57; γ-Pcdh-C3 could thus inhibit Lrp6 phosphorylation by sequestering a pool of Axin1, making it less available for Dvl-mediated recruitment to activated co-receptor complexes. This stabilized pool of Axin1 may not be utilized for destruction complex assembly (based on our inability to IP GSK3b with the C3VCD; Fig. 6d); it is possible it’s not solely dedicated to Wnt/beta-catenin regulation but rather shunted to other pathways, such as cytoskeletal regulation.
Evidence that γ-Pcdhs can differentially regulate Wnt signalling in the cerebral cortex in vivo
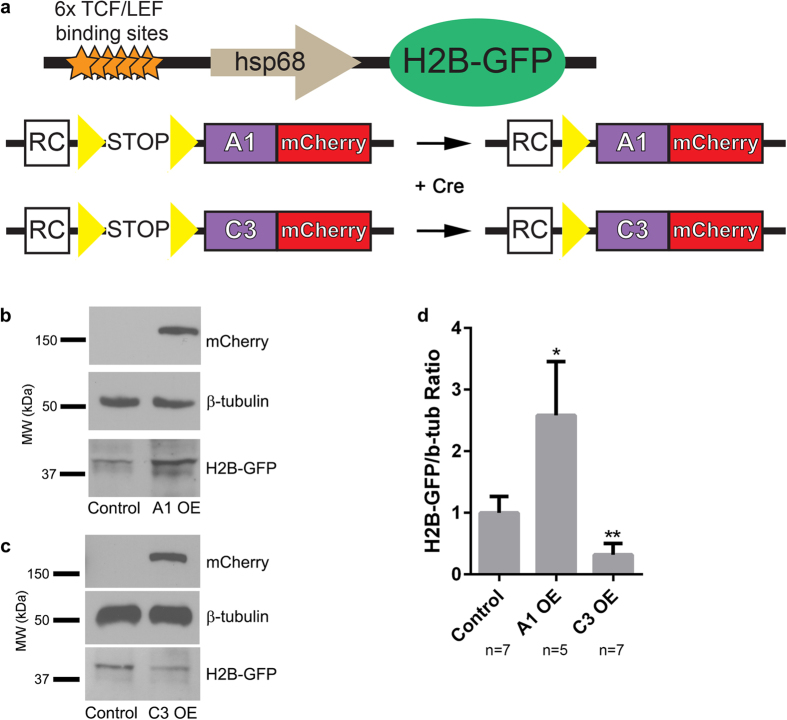
Finally, we asked whether the regulation of Wnt signalling by γ-Pcdhs that we have explored in cultured cells also occurs in the forebrain in vivo, where both Wnt and the γ-Pcdhs have important functions8,45. To answer this question, we crossed a Wnt reporter mouse line, which harbours a transgene containing six TCF/LEF binding sites upstream of a histone 2B (H2B)-GFP fusion68 with mice that overexpress, in a Cre-dependent manner, a cDNA encoding either γ-Pcdh-A1-mCherry (predicted to upregulate Wnt signalling; Fig. 1b) or γ-Pcdh-C3-mCherry (predicted to downregulate Wnt signalling; Figs 1, 2, 3)(refs29,32; Fig. 8a). We activated expression of these γ-Pcdh transgenes in the cerebral cortex by including the Emx1-Cre allele69. We then prepared cortical protein lysates from these compound transgenics and from littermate controls, blotting for GFP to detect the Wnt-responsive H2B-GFP reporter. Consistent with our in vitro data, the cortices of mice overexpressing γ-Pcdh-A1 exhibited a significantly stronger reporter signal (Fig. 8b,d) while those of mice overexpressing γ-Pcdh-C3 exhibited a significantly weaker reporter signal (Fig. 8c,d). This result suggests that, as in HEK293 cells assessed using TOPFlash, distinct γ-Pcdh isoforms can bi-directionally modulate canonical Wnt pathway signalling levels in the cerebral cortex in vivo.
Figure 8. Individual γ-Pcdh isoforms can differentially regulate Wnt signalling in the cerebral cortex in vivo.
(a) Schematic of transgenic mouse alleles utilized. The Wnt reporter allele (top68) has 6 TCF/LEF Wnt-responsive binding sites and an hsp68 minimal promoter driving histone 2B-GFP. The A1-mCherry and C3-mCherry transgenes29,32 are inserted at the ubiquitous Rosa locus. The transgenes are driven by Rosa and CAG elements (RC) only after a floxed stop cassette is excised by Cre. Yellow triangles, loxP sites. (b,c) Cortical lysates from animals overexpressing γ-Pcdh-A1 or –C3 were used in western blots and probed for GFP to visualize changes in H2B-GFP level, reflecting Wnt signalling activity. Representative Western blot shows an increase in H2B-GFP band intensity in A1-overexpressing animals, while C3-overexpressing animals exhibit a decrease in H2B-GFP band intensity. Blots were probed with anti-mCherry antibody to verify expression of exogenous A1 and C3 isoforms, while β-tubulin is used as a loading control. (d) Band intensities of H2B-GFP were normalized to those of β-tubulin in the same lane. Means ± SEM of ≥5 animals were graphed and analysed using one-way ANOVA with Bonferroni post hoc test (to correct for multiple comparisons). *p < 0.05, **p < 0.01. MW, molecular weight; kDa, kilodaltons.
Discussion
In this report, we have demonstrated differential regulation of canonical Wnt signalling by the γ-Pcdhs both in vitro and in the cerebral cortex in vivo. Prior work by Dallosso et al.13,38 indicated that the Pcdhg gene cluster is hypermethylated in both Wilms’ tumour and colorectal cancer, and showed that some γ-Pcdhs could inhibit Wnt signalling and tumour growth. However, few isoforms were tested, and the mechanisms through which γ-Pcdhs might exert an effect on Wnt signalling remained unknown. Using a quantitative TOPFlash assay in HEK293 cells, we tested 20 γ-Pcdh isoforms and found that while the majority of them either have no effect or actually upregulate Wnt signalling, γ-Pcdh-C3, uniquely, significantly inhibits Wnt signalling. Focusing on the C3 isoform, we showed that the VCD of C3 is the domain required for its inhibition of the Wnt pathway, and identified Axin1 as a direct binding partner for this domain. The C3 VCD competes with Dvl for binding to Axin1 via its DIX domain, and this C3-Axin1 interaction stabilizes Axin1 at the membrane and leads to reduced phosphorylation of Lrp6. Finally, we show that overexpression of individual γ-Pcdh isoforms in vivo can up- or down-regulate a canonical Wnt pathway reporter in the cerebral cortex.
Our data may indicate a novel mechanism in which interaction of Axin1 with the VCD of γ-Pcdh-C3 sequesters it away from other Wnt pathway components (Fig. 9). Stabilization of Axin1 at the membrane per se is sufficient to inhibit Wnt activity in human, Drosophila, and Xenopus cells70,71,72, contrary to the standard interpretation of the ON-state of Wnt signalling. Activation of canonical Wnt signalling through binding of the Wnt ligand to its receptors activates Dvl, which in turn recruits Axin1/GSK3β complexes to the membrane, promoting the phosphorylation of Lrp6. Phospho-Lrp6 then acts as a competitive inhibitor of GSK3β activity, preventing the phosphorylation and further degradation of β-catenin, as well as of Axin1, effectively activating Wnt signalling52,73. Because γ-Pcdh-C3 and Dvl interact with the same domain of Axin1, the C-terminal DIX domain as shown here, C3’s competition with Dvl for binding to Axin1 could prevent this downstream activation of Wnt. Consistent with this idea, we show that overexpression of γ-Pcdh-C3 can prevent Lrp6 phosphorylation in response to Wnt. Stabilization and prevention of Axin1 turnover at the membrane by γ-Pcdh-C3 could somehow potentiate or prolong β-catenin degradation and lead to an inhibition of Wnt signalling, since Axin1 is thought to be a limiting component of the destruction complex74. However, at this point, it is unclear whether γ-Pcdh-C3-associated Axin1 can assemble a stable destruction complex, as we were able to co-immunoprecipitate β-catenin, but not GSK3β, with γ-Pcdh-C3. In any case, this mechanism of Wnt inhibition—competition with Dvl for binding to Axin1—has some precedence, as another Axin1-binding protein, Axam, was found to inhibit Wnt signalling by preventing the interaction between Dvl and Axin75. However, Axam binds to residues 507–712 of Axin1 (within the PP2A/MEKK4 binding site), instead of the DIX domain through which γ-Pcdh-C3 interacts, and is a more potent Wnt/β-catenin antagonist. It was subsequently determined that Axam promotes phosphorylation and degradation of β-catenin (thus inhibiting Wnt signalling) as part of its desumoylation activity75. That γ-Pcdh-C3 acts more as a modulator or buffer of Wnt signalling, rather than as a complete inhibitor, is indicated by the fact that a grossly normal cerebral cortex still forms in mice overexpressing this isoform following introduction of Emx1-Cre (Fig. 8 and ref. 29). Given the importance of canonical Wnt signalling to cortical morphogenesis45, complete inhibition would not be compatible with the normal-sized cortex observed in these mice.
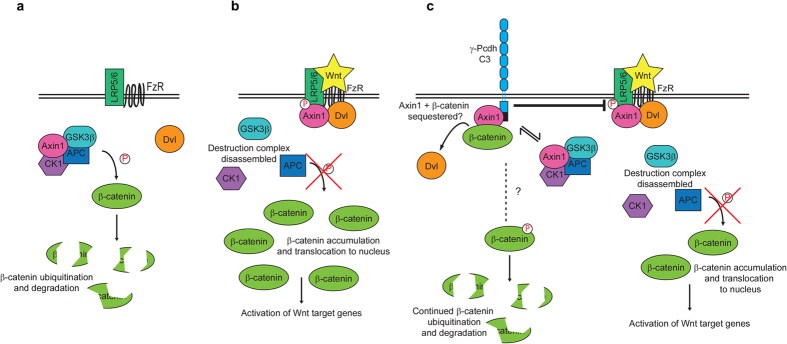
Figure 9. Schematic model of canonical Wnt signalling regulation by γ-Pcdh-C3.
Basic ON- (a) and OFF- (b) states of β-catenin-dependent canonical Wnt signalling. See text for details. (c) A model in which γ-Pcdh-C3 competes with Dvl for binding to Axin1; this interaction stabilizes Axin1 at the membrane, and results in reduced phosphorylation of Lrp6 and decreased Wnt signalling. It remains unclear (dashed lines, question marks) whether γ-Pcdh-C3-bound Axin1 can assemble a functional destruction complex, or precisely how β-catenin is regulated by γ-Pcdh-C3.
The most striking aspect of the clustered Pcdhs is their diversity. Though analyses of mutant and transgenic mice have revealed a multitude of roles for the γ-Pcdhs in neural development11,25,26,27,28,29,31,32,34,76,77,78,79,80, we still know relatively little about how individual isoforms differ in their functions and signalling interactions. By testing 20 γ-Pcdh constructs, we found that multiple A and B subfamily isoforms, as well as γ-Pcdh-C5, can actually upregulate Wnt signalling in HEK293 cells, as measured by the TOPFlash assay, while γ-Pcdh-C3 is the only isoform that consistently downregulates Wnt signalling. Though this is by nature an artificial in vitro assay, we confirmed that overexpression of γ-Pcdh-A1 led to an increase in levels of a Wnt reporter protein in the cerebral cortex in vivo, just as overexpression of γ-Pcdh-C3 led to a decrease in levels of this reporter (Fig. 8). Though we show that γ-Pcdh-C3 inhibits Wnt signalling by binding to, and stabilizing, Axin1 at the membrane, we did not observe an interaction between other γ-Pcdhs and Axin1. This suggests that there are distinct molecular mechanisms by which some γ-Pcdhs isoforms upregulate Wnt signalling. In this regard, it is important to note the differences in regulation of the Pcdhg genes: The A and B subfamily genes are stochastically, monoallelically, and sparsely expressed, while the C isoforms are ubiquitously and biallelically expressed, at least in neurons assayed using single-cell RT-PCR19,21. It may be that the highly and ubiquitously expressed C3 isoform acts as a general “brake” on overall Wnt signalling levels in all cells, while the expression of distinct combinations of other γ-Pcdhs may modulate signalling levels upward. Given that we know nothing about how homophilic trans-interactions or differential cis-interactions22,23 affect the ability of γ-Pcdhs to modulate Wnt signalling, at this point we can only speculate how individual isoforms might collaborate in such a way. The C3 VCD is the site of Axin1 binding and this domain alone is as effective at Wnt inhibition as the FL protein is. This could indicate that Wnt inhibition does not require homophilic trans-interactions between γ-Pcdh-C3 ectodomains, but it is equally possible that such interactions cause a conformational change in the protein that enhances Axin1 binding to the VCD. As our experiments with cells expressing FL γ-Pcdh-C3 were conducted in confluent cultures, homophilic trans-interactions would have been present. It would be interesting in the future to utilize FL γ-Pcdh-C3 constructs harbouring mutations in EC2/3 predicted (on the basis of recent structural data22) to disrupt homophilic interaction to address this possibility.
In addition to its role in Wnt signalling, Axin1 has also been found to interact with synaptic scaffolding molecule (S-SCAM), cyclin-dependent kinase 5 (Cdk5) and Ca2 + /calmodulin-dependent protein kinase II (CaMKII) to play multiple roles in dendrites and dendritic spines65,81,82,83. Dishevelled is also known to regulate Rho GTPases and the JNK pathway to affect dendrite morphogenesis84. Loss of γ-Pcdhs in the cerebral cortex in vivo28, or in hippocampal neurons in vitro34, leads to reduced dendrite arborisation. We recently manipulated γ-Pcdh repertoire in the cortex in vivo and found that homophilic interactions between neurons, and between neurons and astrocytes, promote dendrite complexity29. Consistent with these data, knockout of CTCF, a major regulator of the Pcdh gene clusters, led to reduced cortical and hippocampal neuron dendrite arborisation85. The γ-Pcdhs are also required for normal dendrite self-avoidance in retinal starburst amacrine cells (SAC) and cerebellar Purkinje cells31,32. The regulation of Wnt signalling by binding and stabilizing Axin1 at the membrane could thus be a novel mechanism through which γ-Pcdh-C3 regulates dendrite development.
In this respect, it will be important to utilize novel mouse models to further delineate the regulation of Wnt signalling by γ-Pcdhs in vivo. For example, what is the net effect on Wnt signalling in the Pcdhg null mouse lacking all 22 γ-Pcdhs? Given how many γ-Pcdhs were found to upregulate Wnt in vitro in this study, one might expect a net decrease in Wnt activity in a null mutant; however, the sole inhibitory γ-Pcdh, C3, is more ubiquitously and more highly expressed than any other isoform, so the opposite result is possible as well. Mice with a constitutive null mutation die at birth11, which limits the analyses that can be performed, but a conditional mutant allele25,26,28 could be used to generate tissue-specific knockouts. We are also in the process of generating new lines of mice that either lack one (e.g., a C3 single-isoform mutant) or many (e.g., 8 or 12 of the 22) of the functional variable exons. Given that Wnt pathways in general84,86 and Axin1 in particular65,83 can regulate dendrite arborisation and synaptogenesis, it will be important to determine the extent to which the γ-Pcdhs exert their many functions in neural development25,26,28,30,76,79 through Wnt pathway modulation. Similarly, as the Pcdhg genes are hypermethylated in a number of cancers13,38,39,41 and may act as tumour suppressors, it will be important in the future to develop in vivo models of tumour growth and metastasis in mice lacking or overexpressing varying complements of the 22 γ-Pcdh isoforms.
Methods
Constructs
Full length γ-Pcdh-GFP constructs were reported in Lobas et al.79, and full length and truncated HA-tagged γ-Pcdh constructs were reported in Schreiner and Weiner23. For VCD constructs, the C3 (or A1 or B1) VCD domain, with or without a 3X nuclear localization signal from SV40 or a palmitoylation signal, were generated by PCR from existing plasmids and cloned into pcDNA3.1 ( + ) with a C-terminal GFP within the ApaI restriction site of the vector. Mouse Axin1 expression constructs were the kind gift of Dr. Nancy Ip. For protein production, mouse Axin(506-832) was amplified from HA-mAxin1 FL and inserted into pGEX-6P-1 (GE Healthcare; GST tagged). The VCD of γ-Pcdh C3 with GFP was amplified from the VCD only constructs and was cloned into pET-24A (Novagen; His-tagged).
Cell culture
HEK-293 (ATCC® CRL-1573™) cells were maintained in DMEM with 10% fetal bovine serum and penicillin/streptomycin at 37 °C with 5% CO2. Wnt3a conditioned media was harvested from LWnt3A (ATCC® CRL2647™) cells as previously described63.
Protein Lysis
Cells were lysed in Mild Lysis Buffer (50 mM Tris-HCl, pH7.4, 150 mM NaCl, 25 mM NaF, 1% TritonX-100). Cell lysates were vortexed for 20 seconds and incubated on ice for 10 minutes and centrifuged at 8000 X g for 5 minutes to pellet cellular debris. The supernatant was collected and used for western blotting and immunoprecipitation.
Immunoprecipitation
Cell lysates were incubated with 25 μl ThermoFisher Agarose A/G beads and 5 μl of the targeted antibody (antibody concentration of 1 mg/ml) overnight at 4 °C with rotation. Cell lysates were then centrifuged briefly at 2,000 X g for 3 minutes to harvest bead/antibody/protein complex. The beads were washed several times using Mild Lysis Buffer. The protein complexes were eluted by boiling samples for 5 minutes after adding 50 μl of BioRad 2 X Laemmli Buffer.
Western Blot
Samples were loaded on to a 7.5% Stain-Free TGX Mini Gel and run at 155 V for 58 minutes. The TURBO Mini TGX transfer program on the TransBlot Turbo Transfer System (BioRad) was used to transfer proteins from the gel onto a nitrocellulose membrane. Membranes were blocked for an hour with 5% non-fat milk in TBS-Tween (0.1%) and probed with the primary antibody (usually a 1:1000 dilution in 5% BSA TBS-Tween solution) at 4 °C with rotation. Following 3 5-minute washes in TBST, membranes were probed with host-specific HRP secondary (1:5000 for mouse or rat, 1:1000 for rabbit in 5% non-fat milk in TBS-Tween) for an hour with shaking at RT. Membranes were then washed 5 times with TBST after which the membranes were incubated with SuperSignal West Pico or Femto ECL reagents (ThermoFisher) for signal detection. X-ray films were exposed for a range of times and developed. Films were scanned and analysed in FIJI using the Gel Analyzer plugin.
Antibodies
The following antibodies were used: mouse anti-GST (Sigma), rat anti-mCherry (Invitrogen), rabbit anti-Axin1 (C76H11) (Cell Signaling Technologies; CST), rabbit anti-Lrp6 (C47E12) (CST), rabbit anti-phospho-Lrp6 (Ser1490) (CST), mouse anti-Myc (9E10) (DSHB), rabbit anti-Myc (71D10) (CST), mouse anti-β-tubulin (Sigma), mouse anti-GAPDH (Abcam), rat anti-HA (Roche), rabbit anti-GFP (Life Technologies), mouse anti-GFP (Roche), chicken anti-GFP (Life Technologies), rabbit anti-FLAG tag (CST).
Luciferase Assay
Cells were plated to be approximately 75–80% confluent at time of transfection. Transfections for luciferase assays were performed according to manufacturer’s recommendation of 3:1 FuGeneHD to DNA ratio for HEK293 cells, using 0.1 μg of M72 (SuperTOPFlash), 0.1 ng of Renilla luciferase and 0.4 μg construct of interest (GFP or individual γ-Pcdh isoforms) per well of a 24-well plate. Other transfections were done following FugeneHD’s protocol database. Cells were incubated with the transfection mixture for 24 hours, after which media were changed to either DMEM (control) or Wnt3a conditioned media (CM). Cells were exposed to Wnt3a CM or DMEM for 24 H before being lysed for Dual Luciferase Reporter Assay (Promega). Cells were lysed using 100 μl 1 x Passive Lysis Buffer on a rocker for 15 minutes. Twenty μl of cell lysates were added to Dual Luciferase Reagent and mixed well through pipetting. Firefly luciferase activity was measured using a Turner Biosystems 20/20 n for 10 seconds, and 100 μl of Stop & Glo Reagent was added. Tubes were vortexed briefly before assaying for Renilla luciferase activity for 10 seconds again. The ratio of the two is referred to as the Relative Luciferase Units. In most cases, data are presented on a log2 scale as a “Wnt Response Index”, which is the fold increase or decrease in the Wnt response seen in cells expressing a γ-Pcdh relative to those expressing a control (GFP only) plasmid.
Real-Time Quantitative PCR
HEK293 cells plated in 6-well plates were transfected as described above. Cells were incubated with the transfection mixture for 24 hours, after which media were changed to either DMEM (control) or Wnt3a conditioned media (CM). Cells were exposed to Wnt3a CM or DMEM for 24 H before RNA isolation using TRIzol reagent (ThermoFisher) according to manufacturer’s protocol. Two μg of total RNA was used for cDNA synthesis using SuperScript II Reverse Transcriptase (ThermoFisher). The cDNA produced was diluted 1:10 for use in TaqMan® Real Time PCR Assays (ThermoFisher) using a Roche LightCycler 480. The relative expression of genes was analysed using the comparative CT method with the formula: relative expression = 2−ΔΔCT (ΔΔCT = ΔCT (Wnt3a) − ΔCT (DMEM), in which ΔCT = CT(AXIN2) − CT(GAPDH)). The relative expression of the DMEM control was set to 1.
Subcellular Fractionation
Samples were harvested and processed 48 hours following transfection with appropriate constructs, according to the Subcellular Protein Fractionation Kit for Cultured Cells’ (ThermoFisher Scientific, 78840) manual.
Expression of Recombinant Proteins from Bacteria
LB-medium containing the appropriate antibiotics (100 μg/ml ampicillin, 10 μg/ml kanamycin) was inoculated with 10 ml overnight culture per 500 ml medium and shaken at 37 °C until OD600nm between 0.6–0.8 was reached. Expression of protein was induced by addition of IPTG to 200 μM or 500 μM final concentration for pGEX or pET vectors respectively. The culture was further grown at 30 °C for 3 hours. Cells were pelleted by centrifugation at 4000 X g at 4 °C for 20 minutes. Cell pellets were immediately used for protein purification.
Purification of His6-tagged Proteins
The purification of His6-tagged proteins was performed using Ni Sepharose 6 Fast Flow (GE Healthcare) The cell pellet was resuspended in 5 ml per 500 ml culture of His binding buffer (50 mM Na2H2PO4, pH 7.4, 300 mM NaCl, 5% glycerol, 10 mM imidazole, 0.1% β-mercaptoethanol, 0.1% Triton-X100) complemented with 1 mg/ml lysozyme and 1 tablet of cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail. Cell homogenate was incubated on ice for 30 minutes before sonication. Cell lysate was cleared by centrifugation at 5,000 X g for 20 minutes at 4 °C. The supernatant was incubated with 200 μl of 50% Ni Sepharose 6 Fast Flow agarose slurry for an hour at 4 °C with inversion. The agarose was washed 3 times with 5 ml of binding buffer and bound proteins was eluted using 750 μl binding buffer with 250 mM imidazole. Eluted proteins were concentrated and buffer exchanged using an Amicon centrifugal filter unit (Millipore) to PBS before being used in direct binding assays.
Purification of GST-fusion Proteins
The purification of GST-fusion proteins was performed using Glutathione SepharoseTM 4B (GE Healthcare) The cell pellet was resuspended in 5 ml per 500 ml culture of GST binding buffer (1 X PBS, pH 7.4, 5 mM DTT, 5% glycerol, 0.025% Triton-X100, 10 mM EDTA) complemented with 1 mg/ml lysozyme and 1 tablet of cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail. Cell homogenate was incubated on ice for 30 minutes before sonication. Cell lysate was cleared by centrifugation at 5,000 X g for 20 minutes at 4 °C. The supernatant was incubated with 200 μl of 50% Glutathione SepharoseTM 4B agarose slurry for an hour at 4 °C with inversion. The agarose was washed 3 times with 5 ml of binding buffer and bound proteins was eluted using 750 μl GST elution buffer (100 mM Tris-HCl, pH8.0, 20 mM reduced glutathione, 100 mM NaCl, 0.025% Triton-X100). Eluted proteins were concentrated and buffer exchanged using an Amicon centrifugal filter unit (Millipore) to PBS before being used in direct binding assays.
Direct Binding Assay
One μg of His-C3VCD-GFP was mixed with 0.5 μg GST or GST-tagged Axin(506-832) in PBS supplemented with 1 mg/ml BSA and 0.2% NP-40 for 2 hours at 4 °C with inversion. 30 μl of 50% Glutathione SepharoseTM 4B agarose slurry was added to the protein mixture and incubated for an additional hour. The agarose was washed 3 times with PBS supplemented with 0.2% Tween-20 and proteins were eluted using 25 μl GST elution buffer. 2X BioRad Laemmli buffer was added to the eluted proteins and boiled for 5 minutes before use in Western Blot.
Animals
All animal procedures were approved by the University of Iowa Institutional Animal Care and Use Committee and conformed to all NIH guidelines on the use of animals. The A1-mCherry and C3-mCherry transgenic lines, in which a single γ-Pcdh tagged at the C-terminus with mCherry is expressed from the Rosa locus following Cre excision of a STOP cassette, have been described previously29,32 and were the kind gift of Julie Lefebvre and Joshua Sanes (Harvard University). The Emx1-Cre (ref. 69; JAX stock #005628) and TCF/Lef:H2B-GFP lines (ref. 68; JAX stock #013752) were obtained from The Jackson Laboratory (Bar Harbor, ME). For all experiments, at least three animals were analysed per genotype.
Statistical Analyses
Comparisons between constructs, genotypes and treatments were performed by one- or two-way ANOVA using the recommended Bonferroni post-hoc test (corrected for multiple comparisons) in GraphPad Prism. When only two conditions were being compared, a two-way t-test was utilized. Asterisks in figures denote the following significance levels: *p < 0.05; **p < 0.01; ***p < 0.001.
Additional Information
How to cite this article: Mah, K. M. et al. The γ-Protocadherin-C3 isoform inhibits canonical Wnt signalling by binding to and stabilizing Axin1 at the membrane. Sci. Rep. 6, 31665; doi: 10.1038/srep31665 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Diane Slusarski and members of the Weiner laboratory for helpful discussions; Drs. Nancy Ip and Yu Chen for providing published mouse Axin1 constructs; Drs. Joshua Sanes and Julie Lefebvre for providing the single γ-Pcdh isoform transgenic lines; Leah Fuller for mouse colony management, and Joshua James for preliminary work not included here. This work was funded by NIH R01 NS055272 awarded to J.A.W.
Footnotes
Author Contributions K.M., D.W.H. and J.A.W. conceived the experiments; K.M. performed the experiments; and K.M., D.W.H. and J.A.W. wrote the manuscript.
References
- Gumbiner B. M. Regulation of cadherin-mediated adhesion in morphogenesis. Nature reviews Molecular cell biology 6, 622–634 (2005). [DOI] [PubMed] [Google Scholar]
- Sotomayor M., Gaudet R. & Corey D. P. Sorting out a promiscuous superfamily: towards cadherin connectomics. Trends Cell Biol 24, 524–536, doi: 10.1016/j.tcb.2014.03.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nature Reviews Neuroscience 8, 11–20 (2007). [DOI] [PubMed] [Google Scholar]
- Wu Q. & Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790 (1999). [DOI] [PubMed] [Google Scholar]
- Yagi T. Molecular codes for neuronal individuality and cell assembly in the brain. Frontiers in molecular neuroscience 5, 45, doi: 10.3389/fnmol.2012.00045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B. et al. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Molecular cell 10, 21–33 (2002). [DOI] [PubMed] [Google Scholar]
- Wang X., Su H. & Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes & development 16, 1890–1905, doi: 10.1101/gad.1004802 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeler A. B., Molumby M. J. & Weiner J. A. Protocadherins branch out: multiple roles in dendrite development. Cell adhesion & migration 9, 214–226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmura N. et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20, 1137–1151 (1998). [DOI] [PubMed] [Google Scholar]
- Phillips G. R. et al. γ-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. The Journal of neuroscience 23, 5096–5104 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Gamma protocadherins are required for survival of spinal interneurons. Neuron 36, 843–854 (2002). [DOI] [PubMed] [Google Scholar]
- Zou C., Huang W., Ying G. & Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience 144, 579–603 (2007). [DOI] [PubMed] [Google Scholar]
- Dallosso A. R. et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS genetics 5, e1000745, doi: 10.1371/journal.pgen.1000745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M. et al. Differential expression of individual gamma-protocadherins during mouse brain development. Molecular and cellular neurosciences 29, 603–616, doi: 10.1016/j.mcn.2005.05.001 (2005). [DOI] [PubMed] [Google Scholar]
- Kehayova P., Monahan K., Chen W. & Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proceedings of the National Academy of Sciences of the United States of America 108, 17195–17200, doi: 10.1073/pnas.1114357108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda S. et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron 82, 94–108, doi: 10.1016/j.neuron.2014.02.005 (2014). [DOI] [PubMed] [Google Scholar]
- Yokota S. et al. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. The Journal of biological chemistry 286, 31885–31895, doi: 10.1074/jbc.M111.245605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S., Tasic B. & Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proceedings of the National Academy of Sciences of the United States of America 103, 19719–19724, doi: 10.1073/pnas.0609445104 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi S. et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nature genetics 37, 171–176 (2005). [DOI] [PubMed] [Google Scholar]
- Hirano K. et al. Single-neuron diversity generated by Protocadherin-beta cluster in mouse central and peripheral nervous systems. Frontiers in molecular neuroscience 5, 1–13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko R. et al. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. The Journal of biological chemistry 281, 30551–30560, doi: 10.1074/jbc.M605677200 (2006). [DOI] [PubMed] [Google Scholar]
- Rubinstein R. et al. Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell 163, 629–642, doi: 10.1016/j.cell.2015.09.026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner D. & Weiner J. A. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proceedings of the National Academy of Sciences of the United States of America 107, 14893–14898, doi: 10.1073/pnas.1004526107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu C. A. et al. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell 158, 1045–1059, doi: 10.1016/j.cell.2014.07.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T., Wang X., Gray P. A. & Weiner J. A. A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-gamma gene cluster. Development 135, 4153–4164, doi: 10.1242/dev.026807 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett A. M. & Weiner J. A. Control of CNS synapse development by {gamma}-protocadherin-mediated astrocyte-neuron contact. The Journal of neuroscience: the official journal of the Society for Neuroscience 29, 11723–11731, doi: 10.1523/JNEUROSCI.2818-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. A., Wang X., Tapia J. C. & Sanes J. R. Gamma protocadherins are required for synaptic development in the spinal cord. Proceedings of the National Academy of Sciences of the United States of America 102, 8–14, doi: 10.1073/pnas.0407931101 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett A. M., Schreiner D., Lobas M. A. & Weiner J. A. gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron 74, 269–276, doi: 10.1016/j.neuron.2012.01.028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molumby Michael J., Keeler, Austin B. & Weiner Joshua A. Homophilic Protocadherin Cell-Cell Interactions Promote Dendrite Complexity. Cell Reports, doi: 10.1016/j.celrep.2016.03.093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T. & Weiner J. A. Direct and Indirect Regulation of Spinal Cord Ia Afferent Terminal Formation by the gamma-Protocadherins. Frontiers in molecular neuroscience 4, 54, doi: 10.3389/fnmol.2011.00054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostadinov D. & Sanes J. R. Protocadherin-dependent dendritic self-avoidance regulates neural connectivity and circuit function. eLife 4, e08964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre J. L., Kostadinov D., Chen W. V., Maniatis T. & Sanes J. R. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517–521, doi: 10.1038/nature11305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S. et al. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Molecular and cellular neurosciences 38, 66–79, doi: 10.1016/j.mcn.2008.01.016 (2008). [DOI] [PubMed] [Google Scholar]
- Suo L., Lu H., Ying G., Capecchi M. R. & Wu Q. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. Journal of molecular cell biology 4, 362–376, doi: 10.1093/jmcb/mjs034 (2012). [DOI] [PubMed] [Google Scholar]
- Katori S. et al. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. The Journal of neuroscience: the official journal of the Society for Neuroscience 29, 9137–9147, doi: 10.1523/JNEUROSCI.5478-08.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro R. et al. Impaired clustered protocadherin-alpha leads to aggregated retinogeniculate terminals and impaired visual acuity in mice. Journal of neurochemistry 133, 66–72, doi: 10.1111/jnc.13053 (2015). [DOI] [PubMed] [Google Scholar]
- Yamashita H. et al. Restoration of contralateral representation in the mouse somatosensory cortex after crossing nerve transfer. PloS one 7, e35676 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallosso A. R. et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene 31, 4409–4419, doi: 10.1038/onc.2011.609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waha A. et al. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia 7, 193–199, doi: 10.1593/neo.04490 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banelli B. et al. A pyrosequencing assay for the quantitative methylation analysis of the PCDHB gene cluster, the major factor in neuroblastoma methylator phenotype. Laboratory investigation; a journal of technical methods and pathology 92, 458–465, doi: 10.1038/labinvest.2011.169 (2012). [DOI] [PubMed] [Google Scholar]
- Severson P. L., Tokar E. J., Vrba L., Waalkes M. P. & Futscher B. W. Agglomerates of aberrant DNA methylation are associated with toxicant-induced malignant transformation. Epigenetics 7, 1238–1248, doi: 10.4161/epi.22163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzmann A., Wang Y., Weber D. & Steinbeisser H. Xenopus paraxial protocadherin inhibits Wnt/β-catenin signalling via casein kinase 2β. EMBO reports 13, 129–134, doi: 10.1038/embor.2011.240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B., Köhler A., Schambony A. & Wedlich D. PAPC and the Wnt5a/Ror2 pathway control the invagination of the otic placode in Xenopus. BMC developmental biology 11, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft B., Berger C. D., Wallkamm V., Steinbeisser H. & Wedlich D. Wnt-11 and Fz7 reduce cell adhesion in convergent extension by sequestration of PAPC and C-cadherin. The Journal of cell biology 198, 695–709, doi: 10.1083/jcb.201110076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Uy S. J. & Pleasure S. J. Wnt signaling and forebrain development. Cold Spring Harbor perspectives in biology 4, a008094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inestrosa N. C. & Varela-Nallar L. Wnt signalling in neuronal differentiation and development. Cell and tissue research 359, 215–223, doi: 10.1007/s00441-014-1996-4 (2015). [DOI] [PubMed] [Google Scholar]
- Kohn A. D. & Moon R. T. Wnt and calcium signaling: β-catenin-independent pathways. Cell calcium 38, 439–446 (2005). [DOI] [PubMed] [Google Scholar]
- Niehrs C. The complex world of WNT receptor signalling. Nature reviews. Molecular cell biology 13, 767–779, doi: 10.1038/nrm3470 (2012). [DOI] [PubMed] [Google Scholar]
- Slusarski D. C., Corces V. G. & Moon R. T. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature 390, 410–413 (1997). [DOI] [PubMed] [Google Scholar]
- Clevers H. & Nusse R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- Wong H.-C. et al. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Molecular cell 12, 1251–1260 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilić J. et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316, 1619–1622 (2007). [DOI] [PubMed] [Google Scholar]
- Davidson G. et al. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature 438, 867–872, doi: 10.1038/nature04170 (2005). [DOI] [PubMed] [Google Scholar]
- He X., Semenov M., Tamai K. & Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development 131, 1663–1677, doi: 10.1242/dev.01117 (2004). [DOI] [PubMed] [Google Scholar]
- Kim S.-E. et al. Wnt Stabilization of β-Catenin Reveals Principles for Morphogen Receptor-Scaffold Assemblies. Science 340, 867–870, doi: 10.1126/science.1232389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K. et al. A mechanism for Wnt coreceptor activation. Molecular cell 13, 149–156 (2004). [DOI] [PubMed] [Google Scholar]
- Zeng X. et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438, 873–877, doi: 10.1038/nature04185 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Wang S. & Li L. New insights into the regulation of Axin function in canonical Wnt signaling pathway. Protein & cell 5, 186–193, doi: 10.1007/s13238-014-0019-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K. M. & Waterman M. L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harbor perspectives in biology 4, a007906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas I. G., Frank M., Veron N. & Kemler R. Presenilin-dependent processing and nuclear function of gamma-protocadherins. The Journal of biological chemistry 280, 9313–9319, doi: 10.1074/jbc.M412909200 (2005). [DOI] [PubMed] [Google Scholar]
- Hambsch B., Grinevich V., Seeburg P. H. & Schwarz M. K. {gamma}-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. The Journal of biological chemistry 280, 15888–15897, doi: 10.1074/jbc.M414359200 (2005). [DOI] [PubMed] [Google Scholar]
- DasGupta R., Kaykas A., Moon R. T. & Perrimon N. Functional Genomic Analysis of the Wnt-Wingless Signaling Pathway. Science 308, 826–833, doi: 10.1126/science.1109374 (2005). [DOI] [PubMed] [Google Scholar]
- Willert K. H. Isolation and application of bioactive Wnt proteins. Wnt Signaling: Pathway Methods and Mammalian Models, 17–29 (2008). [DOI] [PubMed] [Google Scholar]
- Berndt J. D. et al. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/β-catenin signaling. The Journal of cell biology 194, 737–750, doi: 10.1083/jcb.201107021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. Axin Regulates Dendritic Spine Morphogenesis through Cdc42-Dependent Signaling. PLoS One 10, e0133115, doi: 10.1371/journal.pone.0133115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.-H., Lin C., Meng S. & Wang X. Proteomics analysis reveals overlapping functions of clustered protocadherins. Molecular & Cellular Proteomics 9, 71–83 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. et al. Axin, an inhibitor of the Wnt signalling pathway, interacts with β-catenin, GSK-3β and APC and reduces the β-catenin level. Genes to Cells 3, 395–403, doi: 10.1046/j.1365-2443.1998.00198.x (1998). [DOI] [PubMed] [Google Scholar]
- Ferrer-Vaquer A. et al. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC developmental biology 10, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. The Journal of neuroscience 22, 6309–6314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620, doi: 10.1038/nature08356 (2009). [DOI] [PubMed] [Google Scholar]
- Schneider P. N., Slusarski D. C. & Houston D. W. Differential role of Axin RGS domain function in Wnt signaling during anteroposterior patterning and maternal axis formation. PloS one 7, e44096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolwinski N. S. Membrane bound axin is sufficient for Wingless signaling in Drosophila embryos. Genetics 181, 1169–1173, doi: 10.1534/genetics.108.098236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Symes K., Seldin D. C. & Dominguez I. Threonine 393 of β‐catenin regulates interaction with Axin. Journal of cellular biochemistry 108, 52–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Salic A., Kruger R., Heinrich R. & Kirschner M. W. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS biology 1, E10, doi: 10.1371/journal.pbio.0000010 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya T. et al. Inhibition of Wnt signaling pathway by a novel axin-binding protein. The Journal of biological chemistry 275, 37030–37037, doi: 10.1074/jbc.M005984200 (2000). [DOI] [PubMed] [Google Scholar]
- Keeler A. B., Schreiner D. & Weiner J. A. Protein Kinase C Phosphorylation of a gamma-Protocadherin C-terminal Lipid Binding Domain Regulates Focal Adhesion Kinase Inhibition and Dendrite Arborization. The Journal of biological chemistry 290, 20674–20686, doi: 10.1074/jbc.M115.642306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledderose J., Dieter S. & Schwarz M. K. Maturation of postnatally generated olfactory bulb granule cells depends on functional [ggr]-protocadherin expression. Scientific reports 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre J. L., Zhang Y., Meister M., Wang X. & Sanes J. R. gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development 135, 4141–4151, doi: 10.1242/dev.027912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobas M. A. et al. Molecular heterogeneity in the choroid plexus epithelium: the 22-member gamma-protocadherin family is differentially expressed, apically localized, and implicated in CSF regulation. Journal of neurochemistry 120, 913–927, doi: 10.1111/j.1471-4159.2011.07587.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H. et al. Gamma-protocadherins regulate the functional integrity of hypothalamic feeding circuitry in mice. Developmental biology 339, 38–50, doi: 10.1016/j.ydbio.2009.12.010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W.-Q. et al. Cdk5-mediated phosphorylation of Axin directs axon formation during cerebral cortex development. The Journal of neuroscience 31, 13613–13624 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S. et al. Synaptic scaffolding molecule interacts with axin. Journal of neurochemistry 90, 332–339 (2004). [DOI] [PubMed] [Google Scholar]
- Ye T., Fu A. K. & Ip N. Y. Emerging roles of Axin in cerebral cortical development. Frontiers in cellular neuroscience 9, 217, doi: 10.3389/fncel.2015.00217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso S. B., Sussman D., Wynshaw-Boris A. & Salinas P. C. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nature neuroscience 8, 34–42, doi: 10.1038/nn1374 (2005). [DOI] [PubMed] [Google Scholar]
- Hirayama T. & Yagi T. Clustered protocadherins and neuronal diversity. Progress in molecular biology and translational science 116, 145–167, doi: 10.1016/B978-0-12-394311-8.00007-8 (2013). [DOI] [PubMed] [Google Scholar]
- Krylova O. et al. WNT-3, expressed by motoneurons, regulates terminal arborization of neurotrophin-3-responsive spinal sensory neurons. Neuron 35, 1043–1056 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.