Abstract
The maintenance of genomic integrity during early embryonic development is important in order to ensure the proper development of the embryo. Studies from cultured cells have demonstrated that cyclin-dependent kinase 12 (Cdk12) is a multifunctional protein that maintains genomic stability and the pluripotency of embryonic stem cells. Perturbation of its functions is also known to be associated with pathogenesis and drug resistance in human cancers. However, the biological significance of Cdk12 in vivo is unclear. Here we bred mice that are deficient in Cdk12 and demonstrated that Cdk12 depletion leads to embryonic lethality shortly after implantation. We also used an in vitro culture system of blastocysts to examine the molecular mechanisms associated with the embryonic lethality of Cdk12-deficient embryos. Cdk12−/− blastocysts fail to undergo outgrowth of the inner cell mass because of an increase in the apoptosis of these cells. Spontaneous DNA damage was revealed by an increase in 53BP1 foci among cells cultured from Cdk12−/− embryos. Furthermore, the expression levels of various DNA damage response genes, namely Atr, Brca1, Fanci and Fancd2, are reduced in Cdk12−/− embryos. These findings indicate that Cdk12 is important for the correct expression of some DNA damage response genes and indirectly has an influence on the efficiency of DNA repair. Our report also highlights that DNA breaks occurring during DNA replication are frequent in mouse embryonic cells and repair of such damage is critical to the successful development of mouse embryos.
In mammals, the first cell lineage specification is the formation of the trophectoderm (TE) and inner cell mass (ICM) during the blastocyst stage.1 The TE forms tissues that make up the placenta, while the ICM gives rise to the embryo proper. Murine ICM cells proliferate rapidly between embryonic day 3.5 (E3.5) and E6.5 and then differentiate into three germ layers. As ICM cells give rise to the cells that make up an entire mammalian organism, the maintenance of genomic integrity within these cells is critical. Results from investigations using embryonic stem (ES) cells that are derived from the ICM, suggest two mechanisms help to support the genome integrity of ICM cells. Firstly, ES cells are sensitive to DNA damage and readily undergo apoptosis or differentiation in order to remove damaged cells from the pluripotent pool.2, 3 Secondly, ES cells possess a potent set of DNA repair machinery. Repair of double-strand breaks (DSBs), the most lethal form of DNA damage,4 is rapidly induced in ES cells. Homologous recombination-mediated repair (HRR), which is the most accurate way to repair DSB,5 is the predominate process involved in repairing DSB in ES cells.6, 7 The above studies underscore the importance of the DNA damage response (DDR) being set in motion efficiently following DNA damage within an ES cell. Knockout of the genes involved in the DDR leads to early embryonic lethality, which provides strong evidence of the critical roles played by DDR genes during early development. For example, a deficiency in Rad51 leads to pre-implantational lethality,8 while disruption of Rad50, Atr and Brca1 reduces cell proliferation and causes abnormal embryonic development at the peri-implantation stage.9, 10, 11 Therefore, investigation of the various proteins and other molecules involved in regulating DDR-related gene expression is crucial to obtaining a proper understanding of early embryonic development.
Cyclin-dependent kinase (Cdk) 12 is a protein kinase that belongs to the Cdk family.12 By associating with cyclin K or cyclin L, Cdk12 executes diverse functions. For example, by interacting with cyclin L, Cdk12 regulates RNA splicing.12 Furthermore, through an interaction with cyclin K, Cdk12 phosphorylates the C-terminal domain of RNA polymerase II, which facilitates elongation during transcription.13 Finally, silencing of Cdk12 results in the decreased expression of various DDR genes, such as Brca1 and Fanci, which induces spontaneous DNA damage.14
In mouse ES cells, Cdk12 is required for self-renewal because knockdown of Cdk12 leads to a reduced expression of a number of self-renewal related genes as well as an increased expression of a range of differentiation markers.15 It has been speculated that DDR genes are downstream effectors of Cdk12 during the maintenance of pluripotency among ES cells because genomic stability is linked to mouse ES cell differentiation.15, 16 In human cancers, perturbation and/or mutation of Cdk12 has been implicated in pathogenesis and drug resistance. Cdk12 can be co-amplified with ERBB2 or may be fused with ERBB2 in breast cancer17 and gastric cancer.18 Furthermore, Cdk12 has been identified as a candidate tumor suppressor gene in ovarian cancer.19, 20 Those Cdk12 mutations associated with ovarian cancer show low levels of kinase activity, which leads to a downregulation of the DDR genes; this was found to sensitize cells to DNA damage drugs in HGS-OvCa patient samples.21, 22
The expression of Cdk12 protein is abundant in mouse ES cells and it then decreases as cell differentiation begins.15 Cdk12 mRNA has been detected in whole embryos at E6.5 and is at a high level in the primitive streak at E7.5.23 Based on the expression pattern and biological functions of Cdk12, it is expected that Cdk12 has an important role in maintaining genomic stability during early embryonic development. To investigate the role of Cdk12 during embryonic development, Cdk12 knockout mice were generated. Cdk12 knockout in mice was found to be fatal during the peri-implantation stage. In addition, Cdk12−/− blastocysts fail to undergo ICM outgrowth in in vitro culture because of an increased level of apoptosis. The expression of DDR genes is lowered and there is an increase in the number of DSBs that can be detected in Cdk12−/− embryonic cells. These findings identify Cdk12 as a critical factor that is involved in maintaining genomic stability during the early embryonic development in mice.
Results
Generation of Cdk12 knockout mice
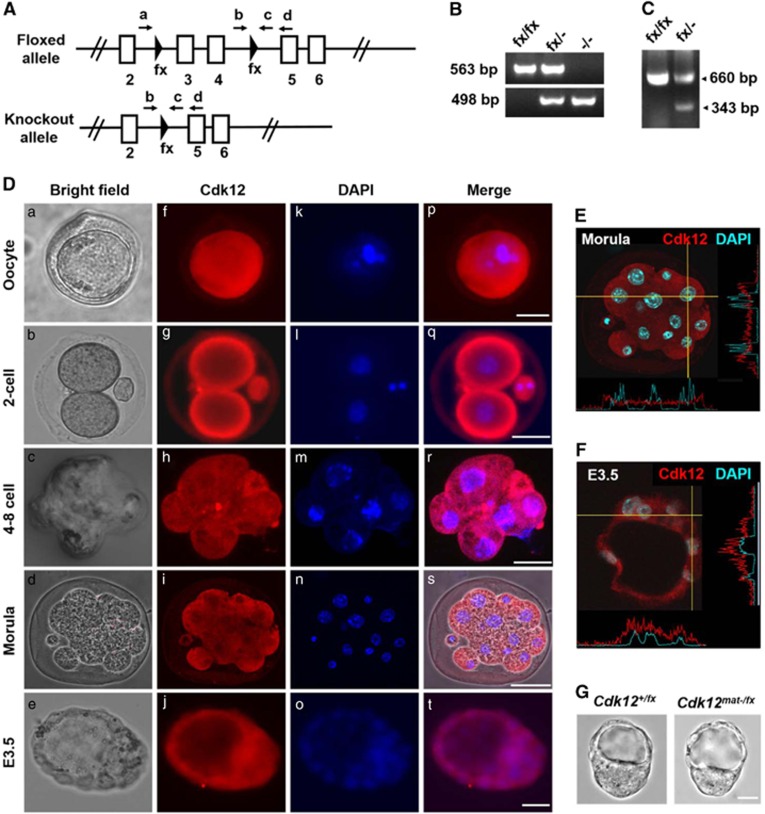
To investigate biological role of Cdk12 in mouse embryonic development, Cdk12 knockout mice were generated from Cdk12fx/fx mice by mating them with Protamin-Cre transgenic mice expressing the Cre recombinase during spermatogenesis.23, 24 Exons 3 and 4 of Cdk12 were deleted by Cre-mediated recombination (Figure 1A) and the knockout allele is predicted to give rise to a prematurely terminated Cdk12 protein lacking the kinase domain. Germline transmission of the knockout allele and the presence of truncated mRNA were verified by nested-PCR and RT-PCR (Figures 1B and C). The Cdk12fx/− mice are viable and fertile. However, when Cdk12fx/− mice are crossed with each other, they do not give rise to viable Cdk12−/− neonates. No postnatal Cdk12−/− mice were found among 108 live-born mice from the intercross (Table 1), which suggests that a homozygous deficiency in Cdk12 leads to embryonic lethality. To detect the exact stage at which Cdk12−/− mice exhibit developmental failure, embryos were collected at various gestation time points and it was found that there were no surviving Cdk12−/− embryos at E14.5, E7.5 or E6.5. Furthermore, the number of empty decidua was found to increase after E6.5 and to match the expected number of Cdk12−/− embryos by Mendel's law (Table 1). Upon examination of E3.5 embryos, we found that the number of Cdk12−/− embryos present were in agreement with Mendel's ratio (Table 1), and that the morphology of the Cdk12−/− embryos was indistinguishable from that of Cdk12fx/−and Cdk12fx/fx embryos (Figure 2Aa–c). These findings indicate that Cdk12 deficiency seems to cause embryonic lethality immediately after implantation.
Figure 1.
Generation of Cdk12 knockout mice. (A) A schematic diagram of Cdk12 floxed and knockout alleles is shown. Arrows represent the locations of the genotyping primers. (B) E3.5 blastocysts were collected after the Cdk12fx/− intercross and genotyped by nested-PCR. The bands representing floxed and knockout alleles are 563 and 498 bp, respectively. (C) mRNA was extracted from Cdk12fx/fx and Cdk12fx/−E14.5 embryos and transcripts derived from the floxed and knockout Cdk12 allele were detected by RT-PCR. The bands representing the normal transcript and the knockout transcript are 660 and 343 bp, respectively. (D) Oocyte and embryos were examined using Cdk12 antibody. Nuclei are indicated by DAPI staining (E and F) Intensity profiles of Cdk12 expression in the cytoplasm and the nucleus along the indicated lines of interest were obtained by confocal sectioned images that reveal the cellular localization of Cdk12 protein at morula stage and in E3.5 embryos. (G) Photographs of E3.5 embryos derived from mating between Zp3-Cre+/0; Cdk12fx/+ female mice with Cdk12fx/fx male mice. Scale bar, 20 μm
Table 1. Genotypes of offspring from the Cdk12 fx/– intercrosses.
| Stage | Cdk12fx/fx (expected: 25%) | Cdk12fx/– (expected: 50%) | Cdk12–/– (expected: 25%) | Empty decidua |
|---|---|---|---|---|
| E3.5 | 28 (29.1%) | 46 (47.9%) | 22 (22.9%) | — |
| E6.5 | 6 (27.3%) | 16 (72.7%) | 0 | 6 |
| E7.5 | 12 (27.3%) | 32 (72.7%) | 0 | 24 |
| E14.5 | 5 (31.3%) | 11 (68.8%) | 0 | 0 |
| Postnatal | 43 (39.8%) | 65 (60.2%) | 0 | — |
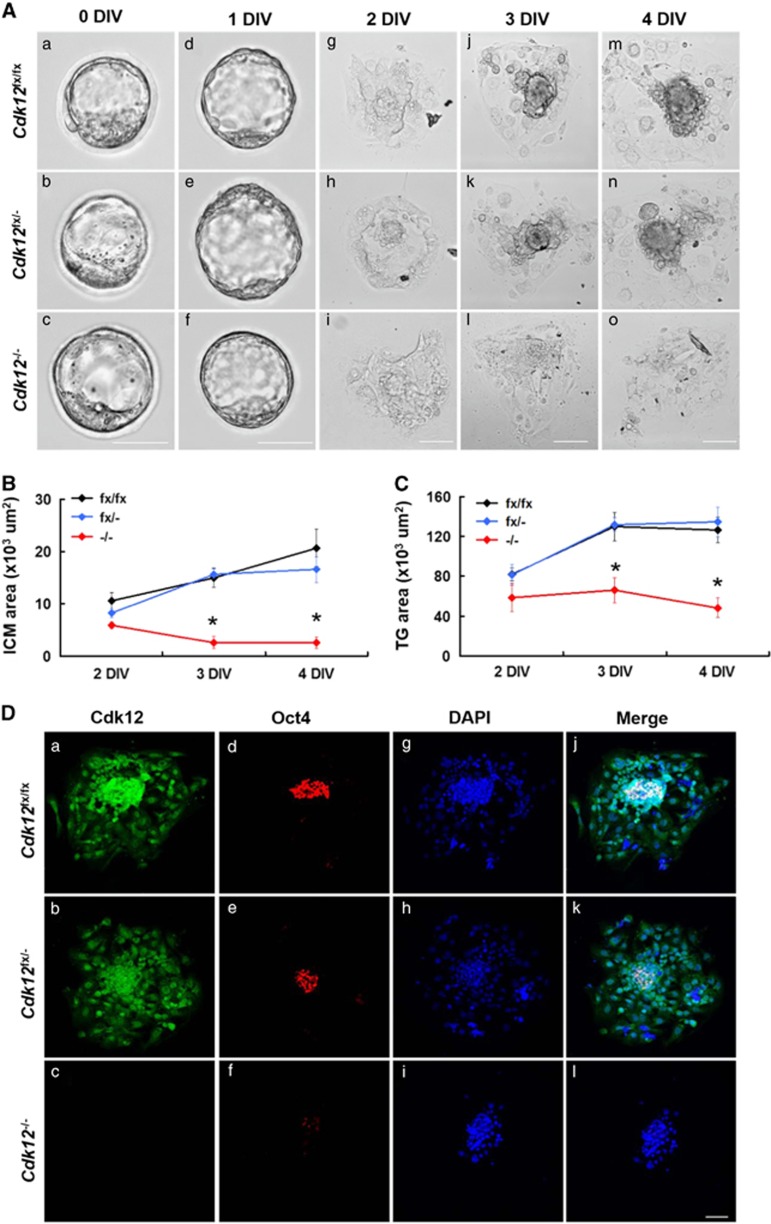
Figure 2.
Cdk12 deficiency results in impaired outgrowth of the blastocyst. (A) Blastocysts were collected by Cdk12fx/− intercrossing and cultured in vitro for 4 days. At the indicated day, embryos were photographed under bright-field conditions. The surface areas of the ICM (B) and TG (C) outgrowth at daily intervals are showed as mean±S.E.M. *P<0.05, by one-way ANOVA. Cdk12fx/fx, n=10; Cdk12fx/−, n=21; Cdk12−/−, n=8. (D) E3.5 embryos were cultured in vitro for 4 days and then immunostained with anti-Cdk12 and anti-Oct4 antibodies. Nuclei are marked by DAPI staining. Scale bar, 50 μm
Expression and cellular location of the Cdk12 protein in pre-implantation embryos
Our recent study has demonstrates that Cdk12 mRNA is detected ubiquitously from E6.5 to E9.5 by in situ hybridization.23 Since we have discovered that Cdk12 is required for the embryonic development before E6.5, we next examined the expression pattern of Cdk12 protein during the early embryonic stages; this was done by immunofluorescence staining. Cdk12 is detected in oocytes and expression continues from the two-cell stage to the blastocyst stage at E3.5 (Figure 1Df–j). At the blastocyst stage, Cdk12 can be detected in both ICM and TE cells (Figure 1Dj). Cdk12 protein is evenly distributed across the cytoplasm and nucleus from the two-cell stage to the morula stage (Figure 1E). Strikingly, a greater amount of Cdk12 protein is present in the nucleus at E3.5 stage than at morula and earlier stages (Figure 1F). As Cdk12 functions in the nucleus,12, 13 these results indicate that Cdk12, while present in the earlier stages of embryo development, may only become fully activated after E3.5. However, because Cdk12 can be detected in oocytes (Figure 1Df), we tested whether a maternal source of Cdk12 protein is able to support embryo development before E3.5. To investigate this possibility, Zp3-Cre transgenic mice, which exhibit Cre activity during oogenesis,25 were used to generate Cdk12 knockout oocytes. The morphology of the maternally Cdk12-null (Cdk12mat-/fx) embryos is indistinguishable from Cdk12+/fx embryos at E3.5 (Figure 1G), and Cdk12mat-/fxpups are viable (Table 2). These results show that maternal Cdk12 protein is not essential for mouse embryonic development before E3.5.
Table 2. Genotypes of the offspring from Zp3-Cre +/0 ; Cdk12 fx/+ × Cdk12 fx/fx crosses.
| Stage | Cdk12mat-/fx (expected: 50%) | Cdk12+/fx (expected: 50%) |
|---|---|---|
| E3.5 | 2 (50%) | 2 (50%) |
| Postnatal | 24 (45.3%) | 29 (54.7%) |
Cultured Cdk12 −/− blastocysts exhibit impaired ICM and TG cell outgrowth
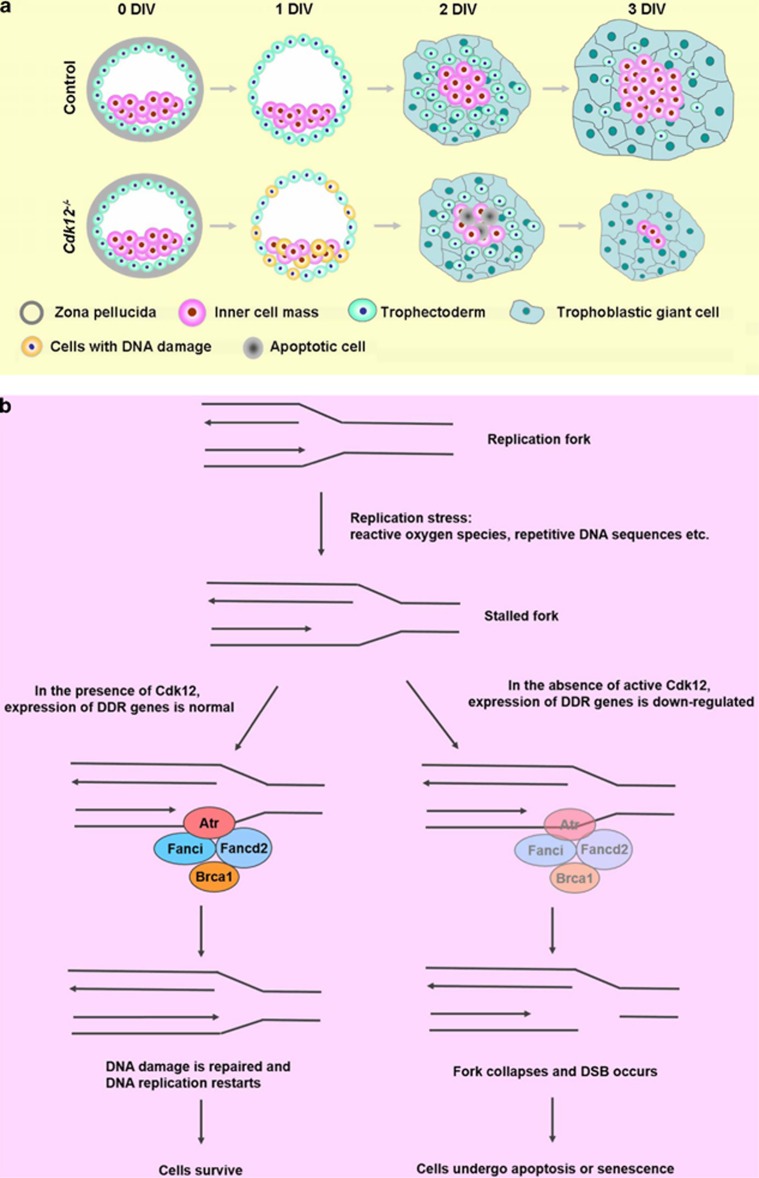
Based on the fact that Cdk12−/− embryos exhibit peri-implantation lethality, it is difficult to study the molecular mechanism underlying the lethality in vivo. As a result of this problem, we used the in vitro culture of blastocysts as an alternative approach. E3.5 blastocysts from intercrosses of Cdk12fx/− mice were isolated and cultured for 4 days in vitro (DIV). Both Cdk12fx/fx and Cdk12fx/−embryos break out from the zona pellucida and possess an enlarged blastocyst cavity at 1 DIV (Figure 2Ad and e). They then attach to the culture plate and initiate outgrowth at 2 DIV (Figure 2Ag and h). The ICM cells expand as a round-shape aggregate on top of the trophoblast giant (TG) cell monolayer at 3 DIV (Figure 2Aj and k). ICM and TG cells keep expanding during 4 DIV (Figure 2Am and n). It was found that the morphology of the Cdk12−/− embryos is similar to that of the Cdk12fx/fx and Cdk12fx/− embryos at 0 and 1 DIV (Figure 2Ac and f). Furthermore, the attachment of Cdk12−/− embryos to the culture plate, and the outgrowth areas from ICM and TG cells are also similar to those with Cdk12fx/fx and Cdk12fx/− embryos during 2 DIV (Figures 2Ai, B and C). However, Cdk12-deficient ICM cells exhibit impaired expansion and the monolayer area of TG cells does not increase during 3 and 4 DIV (Figures 2Al and o, B and C). We further characterized remaining cells at 4 DIV and Cdk12 expression during outgrowth of Cdk12−/− embryos was investigated by immunostaining. In Cdk12fx/fx and Cdk12fx/− embryos, Cdk12 staining is distributed generally across both ICM and TG cells with a stronger signal in the nucleus than in the cytoplasm (Figure 2Da and b). ICM cells were detected using Oct4 expression (Figure 2Dd and e). When the Cdk12−/− embryos were examined, no Cdk12 staining signal could be detected, and very few cells display any significant Oct4 staining (Figure 2Df). When taken together, these findings indicate that Cdk12 deficiency leads to impaired ICM and TG cell outgrowth.
Cdk12 deficiency leads to apoptosis within the ICM
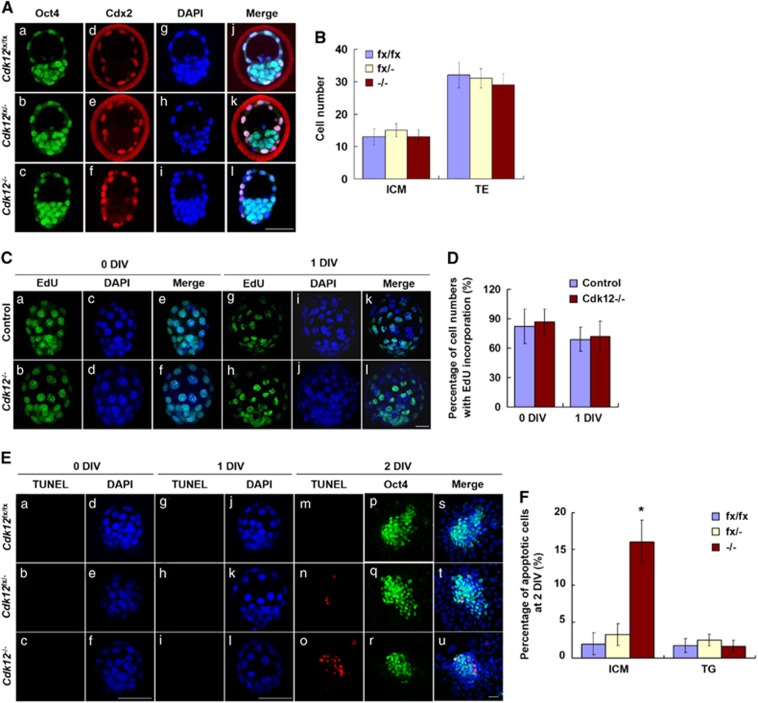
To analyze the underlying mechanisms associated with the failed outgrowth of Cdk12−/− embryos, we examined whether Cdk12 is required for lineage specification, cell proliferation and/or the prevention of apoptosis. E3.5 blastocysts were collected and then probed with Oct4 and Cdx2 antibodies to identify ICM and TE lineage cells, respectively. Oct4 is present in all blastomeres and Cdx2 is restricted to the outside blastomeres at E3.5.26 The number of ICM (Oct4+/Cdx2−) and TE cells (Cdx2+) were counted (Figure 3Aj–l) and no difference was found between the cell numbers of the ICM lineage compared with the TE lineage when Cdk12fx/fx, Cdk12fx/− and Cdk12−/− embryos were compared (Figur 3B). This indicates that Cdk12 is not required for specifying that the ICM and TE lineages at E3.5. We further examined whether there are proliferation defects in Cdk12−/− embryos or perhaps whether there is an abnormal apoptosis occurring in Cdk12−/− embryos. Cell proliferation was measured using a 45-min EdU incorporation assay and apoptotic cells were detected by TUNEL assay. The ratio of EdU positive cells in control and Cdk12−/− embryos is similar at 0 and 1 DIV (Figures 3C and D). No TUNEL positive cells are observed in Cdk12fx/fx, Cdk12fx/− or Cdk12−/− embryos at 0 DIV and 1 DIV (Figure 3Ea–c and g–i). At 2 DIV, however, there were numerous TUNEL positive cells present among the ICM cells of Cdk12−/− embryos (Figures 3Eo and F). The ratio of TUNEL positive cells among the TG cells of Cdk12fx/fx, Cdk12fx/− and Cdk12−/− embryos were found to be similar at 2 DIV (Figure 3F). These findings indicate that Cdk12 deficiency induces apoptosis within the ICM at 2 DIV. To further verify the importance of Cdk12 to maintaining ICM viability, Sox2-Cre transgenic mice that express Cre specifically in the ICM from E3.5 onwards were crossed with Cdk12fx/fx mice.27 No Sox2-Cre+/0; Cdk12−/− embryos can be detected when embryos were examined from E6.5 to E13.5, although numerous empty decidua could be observed (Table 3). These findings indicate that Cdk12 is required to be expressed in the ICM by E6.5 to allow cell survival.
Figure 3.
Cdk12 deficiency induces apoptosis of ICM cells. (A) E3.5 embryos were collected and immunostained with anti-Oct4 and anti-Cdx2 antibodies. Nuclei were indicated by DAPI staining. Individual confocal sections of whole-mount embryos are shown. (B) The cell numbers of ICM and TE lineages in E3.5 embryos were quantified. Cdk12fx/fx, n=5; Cdk12fx/−, n=8; Cdk12−/−, n=4. (C) Cell proliferation of 0 and 1 DIV embryos was measured using a 45-min EdU incorporation assay. (D) Quantitative results of (C). At 0 DIV, control, n=8; Cdk12−/−, n=3. At 1 DIV, control, n=11; Cdk12−/−, n=3. (E) Apoptotic cells were detected by TUNEL assay at 0, 1 and 2 DIV. (F) Quantitative results of (E) at 2 DIV. Cdk12fx/fx, n=3; Cdk12fx/−, n=5; Cdk12−/−, n=3. Results are shown as mean±S.E.M. *P<0.05, by one-way ANOVA. Scale bar, 20 μm
Table 3. Genotypes of the offspring from Sox2-Cre+/0; Cdk12+/– × Cdk12fx/fx crosses.
| Stage | Cdk12+/fx (expected: 25%) | Cdk12−/fx (expected: 25%) | Sox2-Cre+/o; Cdk12+/− (expected: 25%) | Sox2-Cre+/o; Cdk12−/− (expected: 25%) | Empty decidua |
|---|---|---|---|---|---|
| E6.5 | 3 (27.3%) | 3 (27.3%) | 5 (45.5%) | 0 | 5 |
| E7.5 | 13 (30.2%) | 14 (32.6%) | 16 (37.2%) | 0 | 13 |
| E8.5 | 5 (29.4%) | 7 (41.2%) | 5 (29.4%) | 0 | 9 |
| E9.5 | 14 (36.8%) | 13 (34.2%) | 11 (28.9%) | 0 | 11 |
| E10.5 | 4 (36.4%) | 4 (36.4%) | 3 (27.3%) | 0 | 6 |
| E13.5 | 3 (37.5%) | 2 (25%) | 3 (37.5%) | 0 | 0 |
| P0 | 4 (36.4%) | 3 (27.3%) | 4 (36.4%) | 0 | — |
Cdk12 deficiency decreases the expression of DDR genes
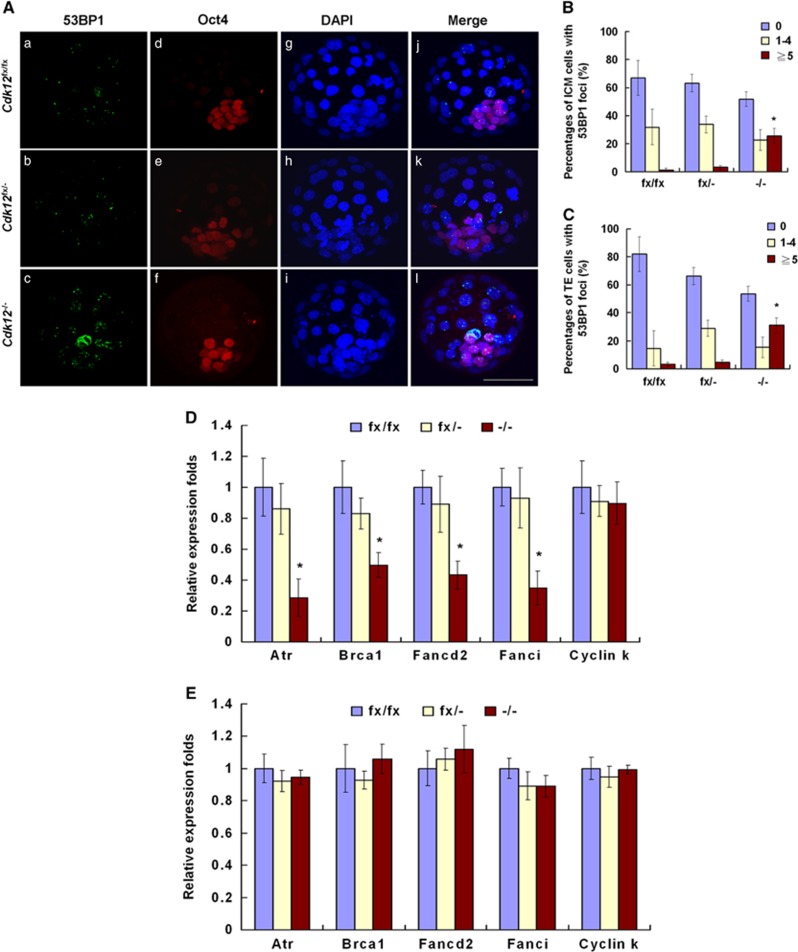
As Cdk12 is involved in the maintenance of genomic stability via regulation of expression of DDR genes,14 we investigated whether apoptosis in Cdk12-deficient embryos is a result of improper expression of DDR genes in response to DNA damage. As increase in apoptotic cells can be detected in Cdk12−/− embryos at 2 DIV (Figure 3Eo), it is expected that Cdk12 deficiency-mediated cellular changes ought to be able to be detected at 1 DIV. DSBs are the most lethal form of DNA damage,4 and 53BP1 foci can be used as a marker for the presence of DSBs.28 It was found that there were few cells with five or more 53BP1 foci per nucleus among ICM cells (1.32%±1.32% and 3.19%±1.48% in Cdk12fx/fx and Cdk12fx/− embryos, respectively) and TE cells (3.41%±3.41% and 4.70%±2.04% in Cdk12fx/fx and Cdk12fx/− embryos, respectively) of Cdk12fx/fx and Cdk12fx/− embryos at 1 DIV (Figures 4Aa and b, B and C). Among Cdk12−/− embryos, cells with five or more 53BP1 foci per nucleus showed a significantly increased trend of 25.50%±5.52% and 31.04%±9.45% among ICM and TE cells, respectively (Figures 4Ac, B and C). In accordance with increased number of DSBs, it was possible to detect lower levels of expression of various DDR genes, namely Atr, Brca1, Fancd2 and Fanci, in Cdk12−/− embryos at 1 IDV (Figure 4D). The mRNA level of Cyclin K shows a similar pattern of expression across all embryos regardless of their genotype. We also examined the expression of DDR genes at the morula stage, a time point where Cdk12 is not functional. The mRNA level of Atr, Brca1, Fancd2, Fanci and Cyclin K shows a similar pattern of expression across all embryos regardless of their genotype (Figure 4E). These findings show that Cdk12 deficiency reduces the expression of DDR genes, resulting in a failure to repair DNA damage in these cells.
Figure 4.
Cdk12 deficiency leads to an increase in the number of DSBs and reduces the expression of DDR genes. (A) Blastocysts were cultured for 1 day and then immunostained with anti-53BP1 and anti-Oct4 antibodies. Nuclei were marked by DAPI staining. The percentage of cell numbers with 53BP1 foci in ICM cells (B) and TE cells (C) were quantified. ICM and TE cells were defined as Oct4+ and Oct4−, respectively. Cdk12fx/fx, n=4; Cdk12fx/−, n=6; Cdk12−/−, n=5. Relative mRNA levels of the indicated genes were determined by real-time PCR at 1 DIV ((D), Cdk12fx/fx, n=4; Cdk12fx/−, n=8; Cdk12−/−, n=5) and morula stage ((E), Cdk12fx/fx, n=6; Cdk12fx/−, n=11; Cdk12−/−, n=4). Results are shown as mean±S.E.M. *P<0.05, by one-way ANOVA
Discussion
In this study, we have demonstrated the importance of Cdk12 protein to mouse embryonic development. Depletion of its activity in knockout mice or Sox2-Cre-mediated conditional knockout mice delivers no embryos that survived beyond E6.5 (Tables 1 and 3). However, E3.5 embryos were found to be morphologically normal, which points toward a requirement for Cdk12 activity that starts after E3.5. The fact that depletion of maternal-derived Cdk12 allows the development of normal pups further supports this view (Table 2). An interesting correlation is that the presence of Cdk12 protein in the nucleus becomes more apparent after E3.5 (Figure 1), which is a prerequisite for exerting its known functions regarding transcription and splicing. Using cultured embryos, we discovered that the expression levels of four DDR genes were reduced to below 50% of that found in Cdk12fx/fx or Cdk12fx/− embryos when Cdk12 was absent (Figure 4D). Reduced expression of these genes in combination will lead to a greater number of DSBs in the genome of the mutant ICM and TE cells, which would subsequently trigger apoptosis of the ICM cells and senescence of the TE cells (Figures 2A and 3E).
Cdk12 deficiency leads to an arrest of embryonic development right after implantation based on the fact that no Cdk12−/− embryos can be identified at E6.5 and that empty decidua increase after E6.5. This observation is further supported by our blastocyst cultured in vitro results. At the suspension stage, namely 0 and 1 DIV, the morphology, cell lineage specification and rates of cell proliferation and apoptosis are similar for all embryos, Cdk12−/−, Cdk12fx/fx and Cdk12fx/−. After embryo attachment at 2 DIV, ICM cells of Cdk12−/− embryos are not able to outgrow because of the increase in apoptosis of these cells (Figure 5a). Although at 2 DIV the number of apoptotic cells is significantly increased only among the ICM cells of Cdk12−/− embryos and this does not occur among the TG cells of Cdk12−/− embryos, a greater number of DNA DSBs can be detected in both ICM cells as well as TE cells of Cdk12−/− embryos at 1 DIV. The discrepancy can be explained by a differential response to DNA damage between ICM and TE cells. At 2 DIV, Cdk12−/− ICM cells undergo apoptosis due to accumulated DNA damage; however, TE cells are induced to differentiate into TG cells which are able to evade DNA damage-induced apoptosis and rather undergo senescence via an elevation of p21 expression.29, 30
Figure 5.
A schematic diagram demonstrates that Cdk12 is required to sustain the viability of the ICM and TE through the maintenance of genome integrity. (a) In the absence of active Cdk12, blastocysts failed to outgrow because of an increase in DNA DSBs in ICM and TE cells. (b) Cdk12 has a crucial role in the regulation of expression of DDR genes that preserve the replication fork structure and repair lesions. In the absence of Cdk12, the fork collapses and as a consequence of this DSBs occur
During early embryonic development, cell cycle progression is rapid and DNA replication is prominent. The integrity of the genome is thus constantly at risk due to the mistakes intrinsic to DNA replication.31 Highly efficient DNA repair is mediated by homologous recombination and this is crucial for the proper development of the organism during this period. For example, Atr−/− and Brca1−/− embryos display growth retardation after implantation, and in vitro culture of their blastocysts show impaired ICM outgrowth.10, 11 Our findings that there are many more DSBs in cultured embryos after loss of Cdk12 activity after just 1 DIV (Figure 4A) and that no Cdk12−/− embryos survive beyond E6.5 further supports the importance of DNA repair during early embryonic development. In addition, the similarity between the phenotypes of mutant embryos and cultured blastocysts of Cdk12−/−, Atr−/− and Brca1−/− mice10, 11 is consistent with Cdk12 affecting the expression of Atr and Brca1. Although mRNAs encoding Atr, Brca1, Fanci and Fancd2 are still present in Cdk12−/− embryos, our results show a parallel and significant decrease in the expression of these four genes when Cdk12-deficient embryos are investigated and this decrease is likely to be sufficient to disrupt DNA damage repair (Figure 5b).
Interestingly, we found that Cdk12−/− embryos develop normally up to the blastocyst stage. We originally considered whether maternal Cdk12 protein may allow normal development before E3.5, as Cdk12 protein can be detected in oocytes and fertilized eggs. Thus, it was a surprise when we found that maternal-null blastocysts are morphologically normal and maternal-null mouse embryos are viable. It is likely that there are other proteins regulating expression of the DDR genes and maintaining efficient DNA damage repair before E3.5. Alternatively, the expression of maternal DDR gene products is able to repair DNA damage and thus supports embryonic development until E3.5. It has been shown that embryos deficient of important DDR genes still develop normally up to peri-implantation stage,8, 9, 10, 11 and maternal Pms2 and Bcas2 proteins are able to maintain integrity of genome before implantation.32, 33
We noted that there is conspicuous nuclear localization of Cdk12 at E3.5, but not at the morula stage, which suggests that Cdk12 may not be active in the cell nucleus with respect to transcription and splicing until E3.5. Measurement of DDR genes' transcripts supports this line of reasoning as expression of DDR genes was decreased in Cdk12−/− embryos compared with that in Cdk12fx/fx and Cdk12fx/−embryos at 1 DIV but not at morula stage (Figure 4D and E). One molecule that may facilitate the translocation of Cdk12 from cytoplasm to the nucleus is cyclin K, as the activation of Cdk12 depends on an association with Cyclin K, and Cyclin K knockout mice exhibit similar embryonic lethality phenotype to that of Cdk12−/− mice.14 Exploring cyclin K expression and its cellular localization during morula and blastocyst stages will be the first step in exploring this possibility in the future.
Dai et al.15 previously reported that the knockdown of Cdk12 in mouse ES cells results in a loss of expression of the pluripotent makers, Oct4 and Sox2, and instead allows the expression of differentiation markers. Here, we have found that cultured ICM cells that are deficient in Cdk12 still express Oct4 even though they are undergoing apoptosis (Figure 3E). This discrepancy may reflect intrinsic differences between ICM cells and cultured ES cells. Alternatively, Cdk13, a protein with many properties identical to Cdk12,14, 15, 23, 34 may be able to compensate for the lack of Cdk12 and bring about regulation of the expression of the relevant self-renewal markers and differentiation markers.
Our results reveal that a deficiency in Cdk12 increases genomic instability and decreases the expression of DDR genes in vivo. These findings shed a new light on the important role that Cdk12 plays in embryonic development and possibly also carcinogenesis. In support of our hypothesis, recent studies have demonstrated that expression of Cdk12 is perturbed in several cancers, including breast cancer,35 gastric cancer18 and ovarian cancer.21, 22, 36 Thus our conditional knockout mouse model may also be useful as a means of clarifying the role of Cdk12 in cancer.
Materials and methods
Mice, genotyping, RT-PCR and real-time PCR
Adult C57BL/6J mice were obtained from the National Laboratory Animal Center (Taipei, Taiwan). All mice were handled according to the University guidelines and all experiments were approved by the National Yang-Ming University Animal Care and Use Committee. For the timed pregnancies, mice were set up in the late afternoon and when plugs were detected next morning, this stage of the mouse embryos was designated as E0.5. Generation of Cdk12fx/fx mice has be described previously.23 To generate the maternal deleted embryos, Zp3-Cre+/0; Cdk12fx/+ female mice were mated with Cdk12fx/fx male mice; one-half of their progeny were maternal Cdk12 null.25 Genotyping of the postnatal mice was performed by PCR using DNA extracted from their tails. Blastocysts were genotyped by nested-PCR. After the embryos were isolated and outgrowth assays or immunostaining performed, the blastocysts were dissolved in DNA lysis buffer containing 50 mM Tris-HCl, pH 8.0, 0.5% Triton X-100 and 200 μg/ml proteinase K. The embryos were digested at 55 °C for 2 h and then the proteinase K was inactivated at 95 °C for 5 min.
Total RNA from E14.5 embryos was extracted using the RNeasy Plus Mini kit (Qiagen, Valencia, CA, USA). The RNA was used for cDNA synthesis with the SuperScript reverse transcriptase III (Invitrogen, Carlsbad, CA, USA). Total RNA of blastocysts and morula was purified and reverse transcribed to cDNA using a TaqMan Gene Expression Cells-to-CT Kit (Ambion, Austin, TX, USA). cDNA was pre-amplified with a specific set of primers using TaqMan PreAmp Master Mix kit (Applied Biosystems, Foster, CA, USA) and used for real-time PCR on an ABI StepOne real-time PCR machine (Applied Biosystems). Samples were assayed in triplicate and normalized against TATA-box binding protein. The sequences of the primers used for the PCR and real-time PCR reactions are provided in Table 4.
Table 4. Primers used for PCR, RT-PCR and real-time PCR.
| Primer | Sequence | Probe (Roche) |
|---|---|---|
| Genotyping | ||
| a | 5′-CTTCCTGCCTCCTCTTCATCATCAGGTATTG-3′ (NT165773.2)a | |
| b | 5′-GTTCAGACAGTGTCAGACCACCTGGAGAAGC-3′ (NT165773.2)a | |
| c | 5′-CCTCTGACCTCCCAATGTGTGCATGACAC-3′ (NT165773.2)a | |
| d | 5′-GTCCAGCCGAACCTTCTTCAGAGCTACTAG-3′ (NT165773.2)a | |
| Transcription detection | ||
| Cdk12 | (F) 5′-CAGCTACTACCCCTCCACCTCAGACAC-3′ (NM_026952.2)a | |
| (R) 5′-GTCCAGCCGAACCTTCTTCAGAGCTACTAG-3′ (NM_026952.2)a | ||
| Real-time PCR | ||
| Atr | (F) 5′-CCCATGCGAATCATGACC-3′ | 104 |
| (R) 5′-ACATCATCGAAGCCTGCAA-3′ | ||
| Brca1 | (F) 5′-ACGAGTCAGGGATGGGAAG-3′ | 46 |
| (R) 5′-CCTGAGAGGGGGAGTTTCTC-3′ | ||
| Cyclin K | (F) 5′-TCCCAAACTTGAGGCTACTCAC-3′ | 67 |
| (R) 5′-GAGGCTTCCGGTCTGGAG-3′ | ||
| Fancd2 | (F) 5′-AACGAAGGGAAGCAACTGG-3′ | 29 |
| (R) 5′-CTAGCCGACGTCTTTTGGAA-3′ | ||
| Fanci | (F) 5′-CACCCTAACAGCATTCGTCA-3′ | 6 |
| (R) 5′-CGACAGCTTCACCAGTTTTTC-3′ | ||
| Tbp | (F) 5′-GGCGGTTTGGCTAGGTTT-3′ | 104 |
| (R) 5′-GGGTTATCTTCACACACCATGA-3′ | ||
Accession number of the reference sequence
Blastocyst isolation and in vitro blastocyst culture
Blastocysts were flushed out from the uterus of pregnant mice at E3.5. To culture the blastocysts, the embryos were transferred to 0.2% gelatin-coated tissue culture plates with mouse ES medium (DMEM, 0.1 mM nonessential amino acids, 0.1 mM sodium pyruvate, 2 mM GlutaMAX-I (all from Gibco, Billings, MT, USA), 15% ES cell qualified FBS (Hyclone, South Logan, Utah, USA), 0.1 mM β-mercaptoethanol (Sigma, St Louis, MI, USA), and 1000 unit/ml leukemia inhibitory factor (Chemicon, Billerica, MA, USA)). Embryos were cultured in the presence of 5% CO2 at 37 °C and photographed daily using a microscope (ZEISS Axio Observer A1, Jena, Germany).
Immunostaining, DNA synthesis assay and apoptosis assay
Embryos at the indicated stage were fixed in 4% paraformaldehyde in PBS at room temperature for 20 min. The fixed embryos were washed with PBS and then permeabilized with 0.2% Triton X-100 in PBS at room temperature for 20 min. Blocking was performed with PBS containing 5% goat serum and 0.2% Triton X-100 at room temperature for 1 h. Immunostaining was carried out overnight at 4 °C in the presence of a primary antibody. The primary antibodies used in this study were rabbit anti-Cdk12 antibody (1:500),12 mouse anti-Cdx2 antibody (Cdx2-88, 1:500, BioGenex, San Ramon, CA, USA), rabbit anti-Oct4 antibody (H-134, 1:500, Santa Cruz, CA, USA) and rabbit anti-53BP1 antibody (1:500, Santa Cruz). After washing with PBS three times, embryos were incubated with appropriate secondary antibodies conjugated with TRITC, Alexa 488 or Alexa 630 at room temperature for 1 h. The DNA was then stained with 1 μg/ml DAPI for 30 min. DNA synthesis was measured by EdU incorporation. Embryos were labeled with 10 μM EdU in mouse ES medium for 45 min and then labeling was detected using a Click-iT EdU Imaging kit (Invitrogen) according to the manufacturer's instructions. The apoptosis assay was performed by TUNEL staining using an In Situ Cell Death Detection Kit (Roche, Indianapolis, IN, USA). Images were taken every 3 μm throughout the embryo by confocal microscopy (Olympus FV10i, Tokyo, Japan and ZEISS LSM700).
Acknowledgments
We thank Y-H Wong, J-Y Yu and J-Y Su for their discussion and critical reading of the manuscript. We thank the technical services provided by TMMC of NCFPB, MOST, Taiwan. This research was supported by Ministry of Science and Technology (NSC101-2320-B-010-064, MOST104-2320-B-010-022-MY3) and the Ministry of Education (Aim for the Top University Plan) to MJF.
Author contributions
H-C Juan, Y Lin, and M-J Fann conceived and designed the experiments. H-C Juan, Y Lin and H-R Chen conducted experiments. H-C Juan and M-J Fann wrote the manuscript.
Glossary
- Cdk12
cyclin-dependent kinase 12
- Cdk13
cyclin-dependent kinase 13
- DDR
DNA damage response
- DIV
days in vitro
- DSB
double-strand breaks
- ES cells
embryonic stem cells
- HRR
homologous recombination-mediated repair
- ICM
inner cell mass
- TE
trophectoderm
- TG
trophoblast giant
The authors declare no conflict of interest.
Footnotes
Edited by H Ichijo
References
- Papaioannou VE. Lineage analysis of inner cell mass and trophectoderm using microsurgically reconstituted mouse blastocysts. J Embryol Exp Morphol 1982; 68: 199–209. [PubMed] [Google Scholar]
- de Waard H, de Wit J, Gorgels TG, van den Aardweg G, Andressoo JO, Vermeij M et al. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2003; 2: 13–25. [DOI] [PubMed] [Google Scholar]
- Van Sloun PP, Jansen JG, Weeda G, Mullenders LH, van Zeeland AA, Lohman PH et al. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Res 1999; 27: 3276–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001; 27: 247–254. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 2008; 18: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Richardson C. Multipotent hematopoietic cells susceptible to alternative double-strand break repair pathways that promote genome rearrangements. Genes Dev 2007; 21: 1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 2004; 23: 3872–3882. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA 1996; 93: 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci USA 1999; 96: 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev 1996; 10: 1835–1843. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 2000; 14: 397–402. [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Wang YC, Fann MJ. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol Cell Biol 2006; 26: 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SW, Kuzyk MA, Moradian A, Ichu TA, Chang VC, Tien JF et al. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol Cell Biol 2012; 32: 4691–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z et al. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 2011; 25: 2158–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Lei T, Zhao C, Zhong J, Tang YZ, Chen B et al. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J Biol Chem 2012; 287: 25344–25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7: 165–171. [DOI] [PubMed] [Google Scholar]
- Sircoulomb F, Bekhouche I, Finetti P, Adelaide J, Ben Hamida A, Bonansea J et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer 2010; 10: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang ZJ, Ong CK, Cutcutache I, Yu W, Zhang SL, Huang D et al. Genetic and structural variation in the gastric cancer kinome revealed through targeted deep sequencing. Cancer Res 2011; 71: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012; 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem 2014; 289: 9247–9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekumi KM, Paculova H, Lenasi T, Pospichalova V, Bosken CA, Rybarikova J et al. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic Acids Res 2015; 43: 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HR, Lin GT, Huang CK, Fann MJ. Cdk12 and Cdk13 regulate axonal elongation through a common signaling pathway that modulates Cdk5 expression. Exp Neurol 2014; 261: 10–21. [DOI] [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA 1997; 94: 14602–14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol 1997; 7: 148–151. [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development 2007; 134: 4219–4231. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 2003; 17: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 2000; 151: 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAuley A, Cross JC, Werb Z. Reprogramming the cell cycle for endoreduplication in rodent trophoblast cells. Mol Biol Cell 1998; 9: 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev 2008; 22: 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett LJ, Plastaras JP. Endogenous DNA damage and mutation. Trends Genet 2001; 17: 214–221. [DOI] [PubMed] [Google Scholar]
- Larson JS, Stringer SL, Stringer JR. Impact of mismatch repair deficiency on genomic stability in the maternal germline and during early embryonic development. Mutat Res 2004; 556: 45–53. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wang F, Xiang Y, Zhang X, Zhao Z, Gao Z et al. Maternal BCAS2 protects genomic integrity in mouse early embryonic development. Development 2015; 142: 3943–3953. [DOI] [PubMed] [Google Scholar]
- Chen HH, Wong YH, Geneviere AM, Fann MJ. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem Biophys Res Commun 2007; 354: 735–740. [DOI] [PubMed] [Google Scholar]
- Natrajan R, Wilkerson PM, Marchio C, Piscuoglio S, Ng CK, Wai P et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol 2014; 232: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res 2014; 74: 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]