Abstract
Myelodysplastic syndrome (MDS) is characterized by ineffective hematopoiesis with resultant cytopenias. Increased apoptosis and aberrantly functioning progenitors are thought to contribute to this phenotype. As is the case for other malignancies, overcoming apoptosis is believed to be important in progression toward acute myeloid leukemia (AML). Using the NUP98-HOXD13 (NHD13) transgenic mouse model of MDS, we previously reported that overexpression of the anti-apoptotic protein BCL2, blocked apoptosis and improved cytopenias, paradoxically, delaying leukemic progression. To further understand this surprising result, we examined the role of p53 and its pro-apoptotic effectors, PUMA and NOXA in NHD13 mice. The absence of p53 or PUMA but not NOXA reduced apoptosis and expanded the numbers of MDS-repopulating cells. Despite a similar effect on apoptosis and cell numbers, the absence of p53 and PUMA had diametrically opposed effects on progression to AML: absence of p53 accelerated leukemic progression, while absence of PUMA significantly delayed progression. This may be explained in part by differences in cellular responses to DNA damage. The absence of p53 led to higher levels of γ-H2AX (indicative of persistent DNA lesions) while PUMA-deficient NHD13 progenitors resolved DNA lesions in a manner comparable to wild-type cells. These results suggest that targeting PUMA may improve the cytopenias of MDS without a detrimental effect on leukemic progression thus warranting further investigation.
Myelodysplastic syndromes (MDS) are a genetically and clonally heterogeneous group of myeloid malignancies, likely arising from the hematopoietic stem cell.1, 2, 3 Despite their genetic heterogeneity, they share common phenotypic features including low peripheral blood counts (cytopenias) in spite of a hypercellular bone marrow (ineffective hematopoiesis), dysplasia and a variable propensity for transformation to acute myeloid leukemia (AML).4 Programmed cell death or apoptosis of white blood cells is a common feature, and is thought to contribute to the cytopenias particularly in early stages of the disease. The basis of ineffective hematopoiesis in MDS may also lie in the dysfunction of hematopoietic progenitors and their inability to support adequate bone marrow function. Apoptosis can be triggered by extrinsic factors, such as FAS ligand or TNF-α, or by cell-intrinsic factors such as ribosomal stress or increased reactive oxygen species. A cell extrinsic mechanism for apoptosis in MDS has been favored for many years;5 however, recent evidence suggests a cell-intrinsic trigger, especially in del(5q) MDS for which there is evidence for activation of the tumor suppressor p53 by ribosomal dysfunction.6, 7
More aggressive stages of MDS have less apoptosis than earlier stages,8 supporting the paradigm that acquired resistance to apoptosis of malignant progenitors is an important step in cancer progression.9 Among hematological malignancies, there is strong experimental data to support the tumor suppressor function of apoptosis in lymphoid malignancies10, 11, 12 but there is a paucity of data addressing this paradigm in myeloid malignancies including MDS. Genetic events that could inhibit apoptosis include overexpression of BCL2 or loss of p53, although a genetic or epigenetic mechanism for increasing BCL2 expression has not been identified and loss of p53 function is seen in only 10–15% of MDS.13 Furthermore, loss of p53 affects many cellular processes in addition to apoptosis, including cell-cycle arrest, cell senescence and DNA repair.14
To address the mechanism and relevance of apoptosis in MDS, we have used the NUP98-HOXD13 (NHD13) transgenic mouse in which the t(2;11)(q31;p15) fusion protein is expressed in hematopoietic stem and progenitor cells under the control of the hematopoietic specific promoter, Vav.15 Although this translocation is rare in human MDS, these mice develop a number of key features characteristic of human disease. From 3 months of age onwards, NHD13 transgenic mice develop cytopenias (macrocytic anemia and thrombocytopenia) and show evidence of dysplasia in their bone marrow. Approximately one-third of mice progress to AML within 10–14 months and this is frequently associated with acquisition of Ras or Cbl mutations.16 NHD13 mice also show evidence of increased apoptosis of bone marrow stem and progenitor cells.17 In this model, we have demonstrated that apoptosis is driven by cell-intrinsic factors because it is effectively blocked by enforced expression of BCL2.17 Challenging the paradigm that overcoming apoptosis is an important step toward malignant transformation, we have found that blocking apoptosis by BCL2 in this mouse model actually prevents transformation to AML. However, several caveats need to be considered for the effects seen with transgenic expression of BCL2. First, expression of the BCL2 transgene is much higher (10-fold) than that seen for BCL-2 in human MDS. Additionally, prevention of disease progression contrasts with the absence of p53 in the NHD13 mouse model, which is thought to prevent apoptosis (although not directly examined) yet was reported to promote disease progression.18 To address these caveats, we have used knockout mice of p53 and its transcriptional pro-apoptotic targets Noxa and Puma to clarify the relationship between apoptosis and progression of MDS to AML.
Results
Defining apoptosis of the MDS-repopulating cells
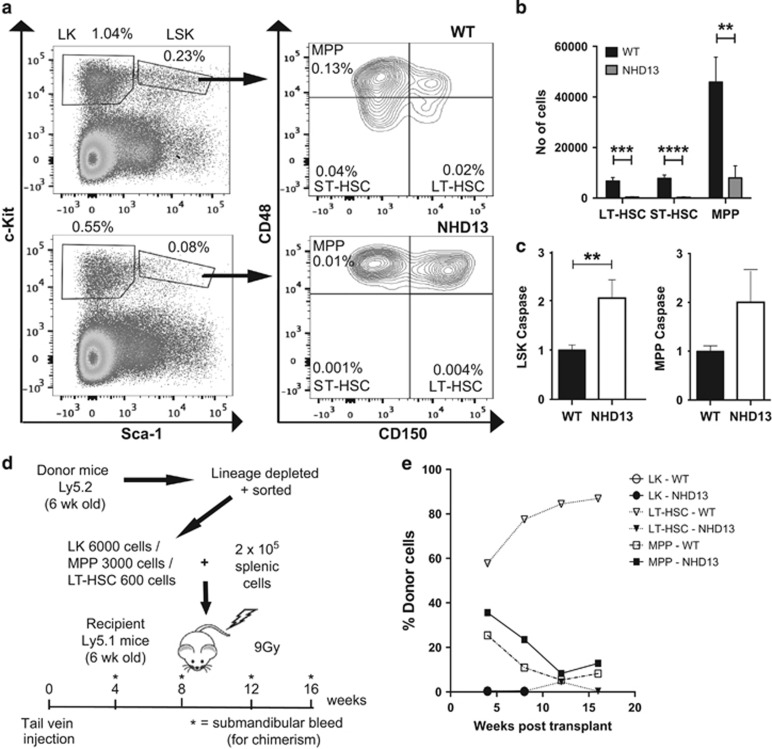
Using the NUP98-HOXD13 (NHD13) transgenic mouse model of MDS, we demonstrated previously that enforced expression of BCL2 could rescue oncogenic stress-induced apoptosis of hematopoietic stem and myeloid progenitors and found this prevented progression to AML.17 To define the hematopoietic subsets affected by apoptosis in NHD13 mice, we measured the numbers and percentages of cells undergoing apoptosis among defined hematopoietic stem and progenitor cells by flow cytometry. As previously reported,18, 19 the proportions of LineagenegSca-1+c-Kit+ (LSK) stem/multi-potent progenitor cells and LineagenegSca-1−c-Kit+ (LK) progenitor cells were reduced approximately three-fold and two-fold, respectively (Figure 1a). Using the Signaling Lymphocyte Activation Molecules (SLAM) CD48 and CD150, we found that long-term (LT)-HSCs (LSK,CD150+CD48−) and short-term (ST)-HSCs (LSK, CD150−CD48−) were more markedly reduced (>10-fold) in proportion and absolute numbers than multi-potent progenitors, MPP (LSKCD150−CD48+) (Figures 1a and b). Apoptosis, as measured by intracellular staining for activated caspase-3/7, was increased two-fold in total LSK cells and the MPP sub-population from NHD13 mice (Figure 1c). In the more mature LK population, we found approximately two-fold reduced numbers of granulocyte-macrophage progenitors (GMP), burst-forming units-erythroid (BFU-E) and colony-forming units-erythroid (CFU-E) (Supplementary Figure S1A and B). Unlike the more immature stem cell populations, we did not observe increased intracellular activated caspase-3/7 in the GMP and erythroid sub-populations (Supplementary Figure S1C). Therefore, expression of the NHD13 oncogene led to reduced numbers and increased apoptosis of the immature LSK population, with the greatest effect seen in LT-HSC and ST-HSC fractions.
Figure 1.
Stem/progenitor cell characterization in NHD13 mice. (a) Flow-cytometric analysis of LK and LSK cell populations gated on lineage-negative cells; and SLAM populations as a subset of LSK cells. Cell populations examined include LT-HSCs (CD150+CD48−), ST-HSCs (CD150−CD48−) and MPPs (CD150−CD48+). Numbers depict percentages of total nucleated cells. Plots shown are representative of three individual experiments. (b) Absolute quantification of individual SLAM cell populations (per femur and tibia of each mouse) (n=9 mice per genotype). (c) Apoptosis analysis of LSK and MPP cell populations from WT (n=14) and NHD13 (n=15) mice. Results were normalized to caspase-3/7 activation levels in WT cells. (d) Schema for chimeric transplant experiment. Four recipient mice were used for each population of cells. Total number of recipient mice=24. (e) Chimerism results for transplanted mice. Results are derived from analyses of monthly bleeds for 4 months in total (except for LK cell transplants) and represent percentage of peripheral blood cells that were donor-derived. **P<0.01; ***P<0.001; ****P<0.0001
Arising from cells with self-renewal capacity, leukemic transformation is a multi-step process in which oncogenic lesions are sequentially acquired. Activating mutations in Nras and Kras are typical of AML in the NHD13 model.16 The presence of self-renewing cells capable of generating MDS was demonstrated by transplantation of NHD13 bone marrow cells.19 However, the phenotype of these cells was not defined beyond the absence of lineage markers; a population that includes both HSCs and committed progenitors. Therefore, we transplanted into lethally irradiated recipients, immature HSC subsets including LT-HSCs or MPPs and more terminally differentiated and mature LK cells to define which cell is most relevant for our studies on the role of apoptosis in leukemic transformation (Figure 1d). Excluding aberrant self-renewal of committed progenitors was important in the setting of the NHD13 transgene because it is known to induce an aberrant Hox gene signature in committed progenitors similar to MLL fusion proteins, which induce self-renewal of GMPs (a subset of the LK population).20 LT-HSCs from NHD13 mice had significantly reduced repopulating activity up to 16 weeks post transplantation compared with wild-type LT-HSCs. In contrast, MPPs from NHD13 mice had detectable long-term repopulating ability, although in our hands wild-type MPPs also had some activity up to 16 weeks. Importantly, the LK fraction that contains GMP had no demonstrable repopulating ability (Figure 1e). Thus, the phenotype of the predominant long-term repopulating cell in mice expressing the NHD13 transgene resides within the immature LSK population and most closely resembles that of the MPP cells. As the chimeric mice were not analyzed beyond 16 weeks, we cannot definitively exclude the contribution of LT-HSC to longer-term reconstitution beyond 16 weeks.
Apoptosis of LSK cells and multi-potent progenitors is p53-dependent
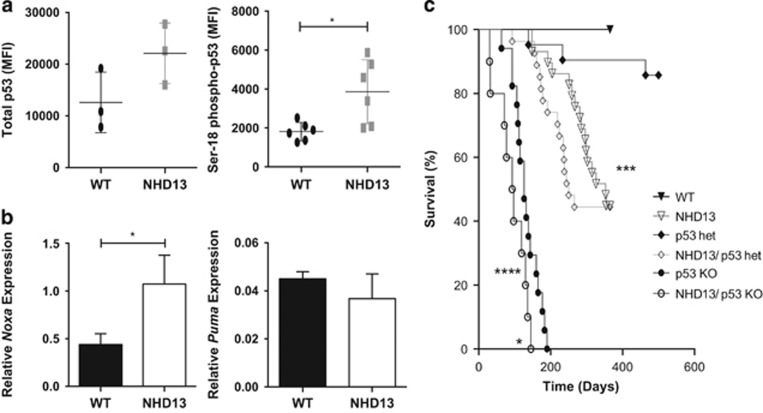
Oncogenic stress is a well-recognized activator of the tumor suppressor p53.21 Although uncommon, loss of p53 portends a poor prognosis in human MDS.13 Consistent with this, it has been reported that loss of p53 accelerated leukemic transformation in NHD13 mice although the impact of p53 loss on apoptosis of hematopoietic cells was not directly examined.18 Examination of MPP (the predominant LSK sub-population) for activation of p53 revealed a two-fold increase in the total and the Ser18 phosphorylated form of p53 (Figure 2a). Gene expression profiling of the LSK population (which comprises 90% MPP) revealed increased expression of Noxa, a direct transcriptional target of p53, but not the other major pro-apoptotic BH3-only target Puma (Figure 2b). Other classic p53 target genes were not elevated in cells expressing the NHD13 transgene (Supplementary Figure S2).
Figure 2.
Expression of p53 and its transcriptional targets; and impact of loss of p53 on NHD13 mouse lifespan. (a) Total p53 and phosphorylated (i.e., activated) Ser18-p53 levels as determined by flow-cytometric analysis in WT and NHD13 MPP cells. Results are expressed as mean fluorescent intensity (MFI); (n=3 per genotype for total p53; and n=6 for phospho-p53). (b) Quantification of Noxa and Puma mRNA levels relative to actin and HPRT (n=3 mice per genotype); error bars represent standard deviation. (c) Kaplan–Meier analysis of animal survival for WT (n=16); NHD13 (n=29); p53−/− (n=17); NHD13/p53+/− (n=27) and NHD13/p53−/− (n=10) mice; *P<0.05; ***P<0.001; ****P<0.0001
We generated NHD13 mice on a p53-deficient background to definitively assess the role of p53 on the apoptotic phenotype. Similar to the previous study,18 we observed a shortened lifespan in NHD13 mice lacking p53, with a median survival of 95 days compared with 126 days in p53-deficient mice alone (P<0.01) and 353 days in NHD13 mice alone (P<0.0001) (Figure 2c). NHD13 mice on a p53 heterozygous (p53+/−) background also had a shortened survival (median 250 days) compared with p53 heterozygous mice alone (median survival undefined, P=0.03). The absence of p53 had no obvious effect on types of leukemia, with examples of transformation to both acute myeloid leukemia and T-cell lymphoblastic leukemia observed in the NHD13;p53−/− cohort (Supplementary Figure S3). Thus, consistent with a previous report,18 the absence of p53 accelerates leukemic progression.
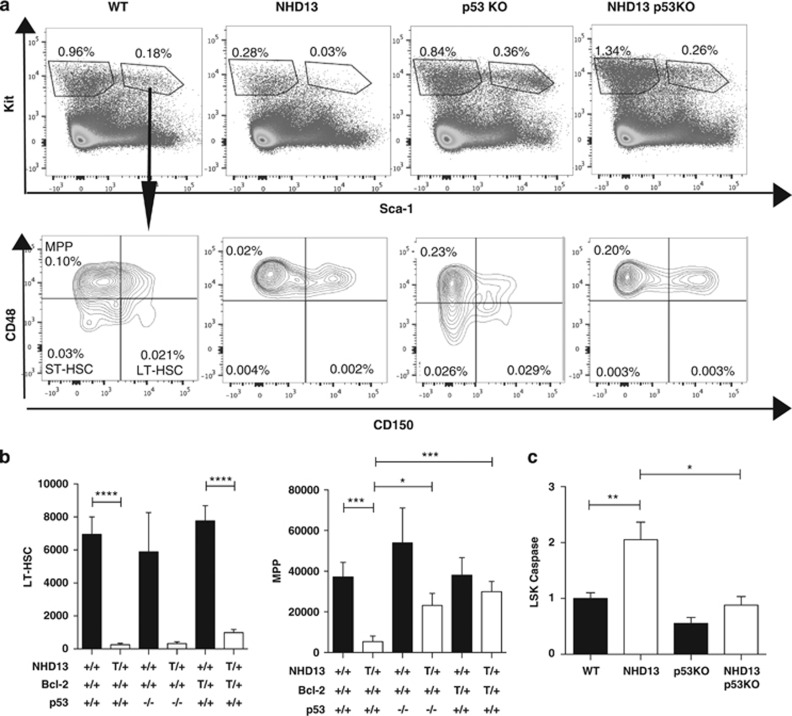
Given the rapid development of leukemia in NHD13;p53−/− mice, we examined apoptosis in mice younger than 6 weeks of age to reduce the chances of analyzing leukemic samples. The absence of p53 rescued the proportion and absolute numbers of LSK, although this was exclusive in the MPP sub-population (Figures 3a and b). This was somewhat different from the previous report where absence of p53 rescued LT-HSC numbers.18 Nevertheless, in our hands, this selective rescue of MPPs but not LT-HSCs was not specific for loss of p53 alone because enforced expression of BCL-2 had a similar effect (Figure 3b). Importantly, the rescue of MPPs by the absence of p53 was associated with reduced apoptosis (Figure 3c). Due to the shortened lifespan of the NHD13;p53−/− mice, we were unable to determine whether rescue of apoptosis improved the cytopenias that arise in older NHD13 mice.
Figure 3.
Phenotype of NHD13 mice deficient for p53. (a) Flow-cytometric analysis of LSK and SLAM populations. Numbers represent percentages of total nucleated cells. Plots represent one replicate of four individual experiments. (b) Absolute numbers of LT-HSCs and MPPs in bone marrow per mouse leg (femur plus tibia)—quantified using flow cytometry for WT (n=14); NHD13 (n=16); p53−/− (n=4) and NHD13;p53−/− (n=4) mice. Results are compared with data from NHD13;Bcl-2 transgenic mice (n=5 for BCL2 and NHD13;BCL2 transgenic mice). (c) Graph of flow-cytometric analysis of activated Caspase-3/7 positivity in LSK cells of WT (n=14); NHD13 (n=15); p53−/− (n=6) and NHD13;p53−/− (n=7) from four individual experiments. Results are normalized to the levels observed in WT cells; +/+, wild-type; T/+, transgenic; −/−, knockout; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001
Apoptosis of erythroid but not myeloid progenitors is p53 dependent
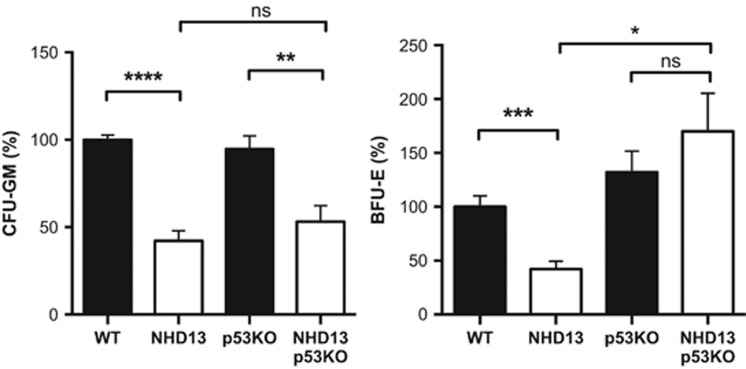
We also examined the role of p53 in the more mature LK progenitor fraction, in which overcoming apoptosis by BCL-2 overexpression could abrogate loss of both myeloid and erythroid progenitors in NHD13 mice.17 Analysis of NHD13 mice lacking p53 revealed a potent rescue of erythroid (BFU-E) but not myeloid progenitor colony formation (CFU-GM) (Figure 4). Phenotypic analysis by flow cytometry was also consistent with the differential rescue of the erythroid progenitor subset (pre-CFU-E), although the analysis of GMP was indeterminate because control p53−/− mice had reduced GMP numbers (Supplementary Figure S4A and B). p53-dependent apoptosis has been specifically implicated in the anemia of 5q-MDS, in which haplo-insufficiency for RPS14 causes ribosomal dysfunction and p53 activation.6, 7 We therefore sought to determine whether a genetic signature of ribosomal dysfunction might underlie the differential rescue of erythroid precursor numbers through loss of p53 in our NHD13 mice. However, we found that expression of RPS14 was normal and genes increased by loss of RPS14 (ref. 22) were paradoxically reduced in NHD13 LK cells (Supplementary Figure S5A). Further interrogation of the 108 ribosomal proteins detected by RNA-seq revealed significantly reduced expression of ribosomal protein S4-like gene (Rps4l) although the functional relevance of this was not investigated further (Supplementary Figure S5B).
Figure 4.
Differential impact of loss of p53 on erythroid versus myeloid progenitors. Progenitor assays enumerating colony-forming units granulocyte-macrophage (CFU-GM) and burst-forming units-erythroid (BFU-E) for WT (n=10); NHD13 (n=10); p53−/− (n=11) and NHD13;p53−/− (n=14) mice. Each sample was processed in triplicate. Number of colonies was normalized to those seen in wild-type mice; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001
NOXA is not required for apoptosis of NHD13 stem and progenitor cells
Noxa was the BH3-only p53 target gene significantly upregulated in NHD13 HSCs, MPPs and LK cells (Figure 2b). The NOXA protein can inhibit the anti-apoptotic BCL-2 family member MCL-1, which has been shown to be important for the survival of normal and MLL-transformed HSCs.23, 24 To address the role of elevated Noxa mRNA in MDS, we generated NHD13 mice on a NOXA-deficient background. The absence of NOXA had no discernible impact on the extent of apoptosis (Supplementary Figure S6A) or in vitro colony growth (Supplementary Figure S6B). These findings suggest that NOXA is not the mediator of p53-dependent apoptosis in NHD13 stem/progenitor cells. In support of this, we noted that Noxa mRNA levels in NHD13 mice deficient for p53 remained elevated, suggesting that Noxa upregulation is p53 independent (Supplementary Figure S6C). Given the lack of any demonstrable role for NOXA in apoptosis, we did not age mice to examine the impact of NOXA deficiency on cytopenias or leukemic transformation.
PUMA mediates apoptosis of NHD13 stem and progenitor cells
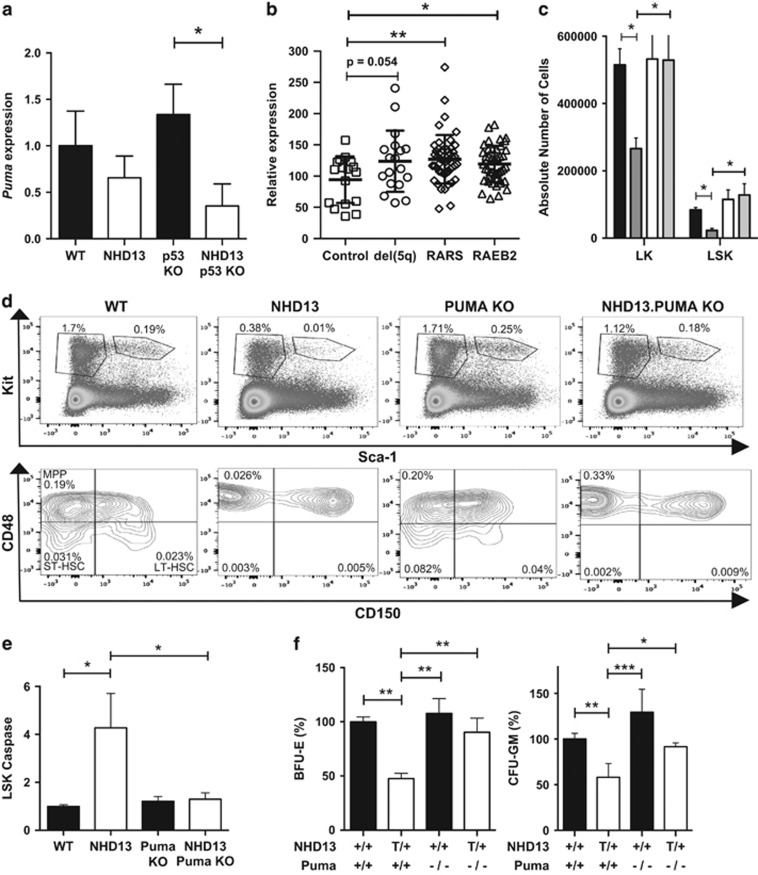
Although expression of Puma mRNA was normal in NHD13 stem and progenitor cells (Figure 2a), baseline Puma mRNA levels were significantly reduced in NHD13;p53−/− mice suggestive of an underlying p53-mediated basis of activation (Figure 5a). PUMA is the major mediator of p53-dependent apoptosis and, unlike NOXA, is capable of inhibiting all pro-survival BCL-2 family members.25 Puma is also significantly elevated in CD34+ stem/progenitor cells isolated from humans with MDS (Figure 5b). Therefore, to determine whether PUMA is the mediator of apoptosis downstream of p53 in NHD13 stem and progenitor cells, we generated NHD13 mice on a PUMA-deficient background. Analysis of bone marrow from 12-week-old NHD13;Puma−/− mice revealed a rescue of relative and absolute numbers of LSK and LK cells (Figures 5d and e). Of the LSK population, MPP cell numbers but not LT- or ST-HSCs were rescued (Figure 5d and Supplementary Figure S7). This is identical to the pattern seen in the absence of p53 or overexpression of BCL-2 (Figure 3b). The rescue of MPP cell numbers was associated with a substantial reduction in cells undergoing apoptosis (Figure 5e). Unlike the absence of p53, the absence of PUMA rescued both erythroid and myeloid progenitor cell growth in vitro (Figure 5f).
Figure 5.
Puma expression in human MDS and phenotype of NHD13;Puma−/− mice. (a) Puma mRNA expression levels for WT, NHD13, p53 KO and NHD13; p53 KO mice (n=3). Results are expressed relative to actin and HPRT. (b) Puma gene expression in human MDS—data have been extracted from Pellagatti et al.44 and separate MDS into low-risk del(5q) and RARS and higher risk RAEB2. (c) Absolute quantification of LK and LSK subsets per mouse leg based on flow-cytometric analysis (n=9 per genotype). (d) Results from flow-cytometric analysis showing relative quantification of LK, LSK and SLAM subsets. Results are expressed as percentages of total nucleated cells. Plots shown are a single representation of three individual experiments. (e) Apoptosis of LSK stem cells as measured by staining for activated caspase-3/7, with data expressed relative to levels seen in WT mice (n=11 per genotype; 4 independent experiments). (f) In vitro colony assays—BFU-E (n=5 per genotype) and GMP (n=4 per genotype). Data have been normalized to wild-type numbers; *P<0.05; **P<0.01; ***P<0.001
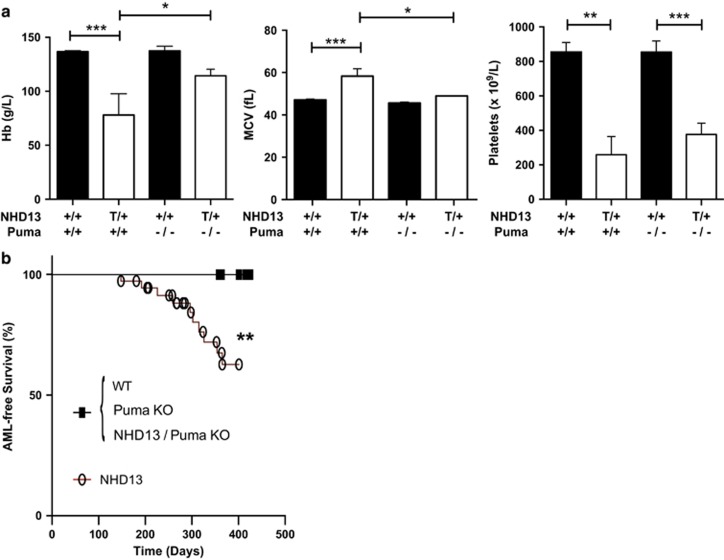
Germline deletion of Puma has no detrimental effect on long-term survival of mice and (at least on its own) does not lead to spontaneous hematopoietic tumors,26 thus allowing an examination of the role of PUMA in later stages of MDS. Consistent with an important role for apoptosis in cytopenias, the absence of PUMA was able to rescue the macrocytic anemia in older NHD13 mice (Figure 6a). However, similar to the effects of BCL-2 overexpression,17 thrombocytopenia was not corrected by loss of PUMA (Figure 6a), suggesting that thrombocytopenia is not attributable to increased apoptosis or that the apoptosis of this cell population occurs independent of PUMA. Finally, cohorts of mice were aged to determine the effect of loss of PUMA on the development of acute leukemia. Remarkably, NHD13 mice lacking PUMA never developed AML (Figure 6b). Of the eight NHD13;Puma−/− mice that died, five were available for analysis and all were diagnosed with T-ALL based on the findings of a thymic lymphoma and flow-cytometric analysis. Median survival of NHD13 mice with or without PUMA was comparable (301 days versus 327 days; P=0.15) (Supplementary Figure S8). Thus, PUMA is required for apoptosis of hematopoietic stem cells and progression of disease to AML.
Figure 6.
Impact of loss of PUMA on peripheral blood counts and survival of NHD13 mice. (a) Peripheral blood analysis of Hb, MCV and platelet count in 8-month-old mice (n>6 per genotype). (b) Kaplan–Meier curve analysis for AML-free survival; +/+, wild-type; T/+, transgenic; −/−, knockout; *P<0.05; **P<0.01; ***P<0.001
Absence of p53 and PUMA have disparate effects on cellular responses to DNA damage
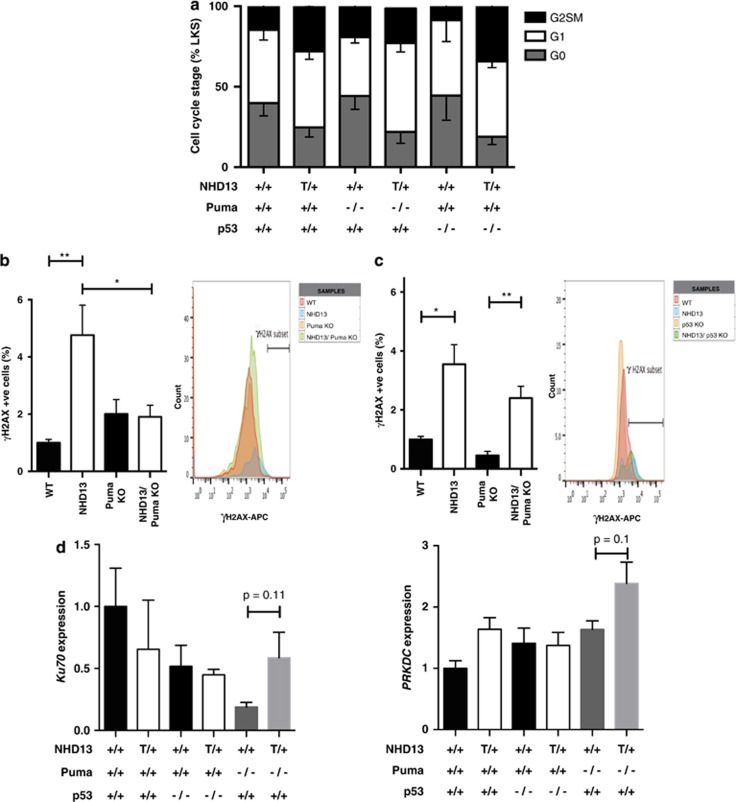
The rescue from apoptosis and prevention of myeloid transformation in the absence of PUMA phenocopies the impact of BCL-2 overexpression.17 In contrast, loss of p53, which also prevents apoptosis of hematopoietic stem/progenitor cells in NHD13 mice, accelerated myeloid transformation (Figure 2c). This dichotomous effect is consistent with recent experimental data, suggesting that the tumor suppressor activity of p53 may be independent of, or at least not solely reliant on its apoptosis-inducing function.14, 27 To further understand different functions of p53 and PUMA in MDS stem and progenitor cells, we examined cell cycling and DNA damage responses in NHD13 mice lacking p53 or PUMA. As previously reported,17 the NHD13 LSK population had reduced percentages of cells in quiescence compared with wild-type mice (Figure 7a), which may be explained by reduced levels of Cdkn1a. However, unlike BCL-2 overexpression, the absence of p53 or PUMA did not significantly increase the proportion of quiescent cells in NHD13 mice (Figure 7a). Measurement of DNA damage using staining for phospho-γ-H2AX was most revealing. As previously reported, NHD13 mice had 2- to 4-fold increase in γ-H2AX+ LSK stem/progenitor cells compared with wild-type controls. This increase was abrogated by loss of PUMA (Figure 7b). In contrast, the absence of p53 had no effect on the proportion of γ-H2AX-positive LSK cells in NHD13 mice, with a persistent increase above baseline (p53−/− LSK cells) (Figure 7c). Examination of mRNA expression levels of DNA damage response genes involved in non-homologous end-joining (NHEJ), which is particularly relevant in the repair of HSCs, revealed a trend for higher expression of genes such as Ku70 and PRKDC (Figure 7d) and Ku80 (not shown) in NHD13 LSK cells deficient for Puma—suggesting a possible mechanistic basis for reduced γ-H2AX levels in these cells compared with those deficient for p53.
Figure 7.
The differing roles of p53 and PUMA in progression of MDS to AML. (a) Cell-cycle analysis of LSK cells based on DAPI and Ki67 staining. Data show percentage of total cells in various stages of cell cycle. (b) DNA damage analysis with data normalized to the levels seen in wild-type cells for Puma-deficient mice. γH2AX staining is expressed as a percentage of activated caspase-3/7 negative, G0 (non-cycling) cells (n=9 per genotype); Histogram to the right shows a single graphical representation of the data. (c) DNA damage analysis with data normalized to the levels seen in wild-type cells for WT (n=9), NHD13 (n=8), p53 KO (n=4) and NHD13;p53 KO (n=5) mice; Histogram to the right shows a single representation of the data. (d) mRNA expression of genes involved in the NHEJ DNA damage response. Results have been normalized to HPRT and actin levels (n=3 per genotype); T/+, transgenic; *P<0.05; **P<0.01
Discussion
We have used knockout mice lacking the tumor suppressor p53 and its pro-apoptotic transcriptional target genes Noxa and Puma to examine the importance of oncogene-induced apoptosis in myelodysplasia and progression to AML. We show that p53 and its BH3-only target gene Puma are essential for apoptosis of the MDS-repopulating cell population in mice expressing the NUP98-HOXD13 fusion protein. Although we acknowledge the NUP98-HOXD13 fusion is rare in human MDS and there are a number of limitations of this model, the resultant phenotype of the mice recapitulates many of the features of human disease and is thus useful as a tool to study apoptosis and myeloid transformation.
Although the absence of p53 and PUMA led to comparable effects on apoptosis, the absence of p53 and PUMA had opposite effects on progression to AML; absence of p53 accelerated myeloid transformation, while absence of PUMA prevented the development of AML, permitting ongoing development of thymic lymphoma. To our knowledge, this is the first study examining a potential functional role for PUMA in MDS.
Mutations in p53 are found in ~10% of human MDS and are associated with poor prognosis, even in patients with the otherwise low-risk del(5q) syndrome.13, 28 As recently reported,18 the absence of p53 accelerated transformation to AML in the NHD13 model of MDS. The rescue of stem/progenitor cells from apoptosis might explain accelerated disease progression by expanding the pool of self-renewing MDS-repopulating cells, which could acquire the necessary additional mutations to drive AML. However, the observation, that loss of its pro-apoptotic transcriptional target Puma can also block apoptosis and expand the pool of self-renewing MDS-repopulating cells but not accelerate leukemic progression in the same mouse model, suggests that loss of apoptosis alone does not explain the more aggressive disease caused by absence of p53. Increased γ-H2AX staining in NHD13; p53−/− LSKs (Figure 7b) supports recent evidence that the tumor suppressor activity of p53 is mediated (at least in part) by efficient DNA damage repair.14 Indeed, activation of DNA damage response genes appears more prominent where p53 is intact (i.e., NHD13; Puma−/−) in comparison with cases where p53 is lost (Figure 7d). Loss of p53 might also enhance progression to AML by promoting aberrant self-renewal29 or a switch to oxidative metabolism causing DNA damage.27
In response to DNA damage (e.g., γ-irradiation) and cytokine withdrawal, PUMA appears to be more important than NOXA for apoptosis of normal hematopoietic stem and progenitor cells.30, 31, 32, 33, 34 Here, we provide the first evidence of a similar predominant role for PUMA in oncogene-induced apoptosis of hematopoietic stem and progenitor cells in vivo. Interestingly, the absence of PUMA but not loss of p53 rescued myeloid progenitors, suggesting that the mechanism of apoptosis is not a simple linear pathway from p53 to PUMA. Notably, p53-independent activation of PUMA has already been reported: for example, glucocorticoid-induced killing of lymphoid cells is substantially reduced by loss of PUMA but loss of p53 has no impact.35, 36 Pifithrin-α, an inhibitor of p53, has therapeutic potential for improving erythropoiesis in del(5q) MDS, but the obvious concern is leukemic progression.7 Our results suggest that blocking PUMA may be a safer strategy for improving cytopenias and may even delay leukemic progression. This may be relevant in human MDS where Puma levels are increased above those of normal controls (Figure 5a) and certainly warrants further consideration.
This study further challenges the ‘Hanahan and Weinberg' paradigm that overcoming apoptosis is an important hallmark of cancer.9 Previously, it has been shown that the absence of PUMA (or overexpression of BCL-2) could greatly delay or even prevent development of γ-radiation-induced thymic lymphoma, a malignancy that is, like MDS, also derived from bone marrow HSCs.31, 37 We previously showed that overexpression of BCL-2 with consequent blocking of apoptosis could have a similar effect in the presence of an established MDS-initiating cell.16 The effects seen with loss of PUMA extend our previous work by identifying the BH3-only protein that is essential for initiating apoptosis in this setting.
It is notable that both BCL-2 overexpression10 and loss of PUMA12 accelerate MYC-induced lymphoma development. So, why then does loss of PUMA prevent progression of MDS to AML? A likely explanation is that, similar to what was seen in γ-radiation-induced thymic lymphoma development,31, 37 loss of PUMA maintains survival of leukocytes (and their progenitors) under conditions of stress. This likely obviates the need for mobilization and increased division of stem/progenitor cells preventing the acquisition of oncogenic mutations that may be associated with persistent replication. Our results also suggest better mobilization of a DNA damage response (Figure 7d) may be of significance in this case.
Although the use of genetically modified mice is a powerful method for understanding oncogenesis, it is not without its limitations. It will thus be important to determine whether similar effects are seen in human MDS and in additional mouse models that resemble the genetic and cellular heterogeneity of acquired human MDS. Furthermore, it will be important to understand how preventing apoptosis might act to delay leukemic progression in MDS. In summary, we provide experimental evidence that preventing apoptosis by loss of PUMA may present a means for improving blood counts and delaying disease progression in MDS.
Materials and Methods
Mice
NHD13 mice were generated as previously described.15 BCL2 transgenic mice and Noxa (Noxa−/−) and Puma (Puma−/−) knockout mice were provided by Professor J Adams (Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia).26, 38, 39 Mice were maintained on a C57BL/6 background. Animal experiments were approved by the Alfred Medical Research and Education Precinct (AMREP) ethics committee and all experimental procedures were conducted in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (2004). Bone marrow was harvested from femora and tibiae into mouse-tonicity phosphate-buffered saline (PBS, in-house) containing 2% fetal bovine serum (FBS, Sigma, Castlehill, NSW, Australia). Mice were 3 months of age at time of organ harvest unless specified and both males and females were used for experimentation without randomization. Peripheral blood was collected into EDTA (Sarstedt, Newton, NC, USA) acid-coated tubes for analysis on the Hemavet Multispecies Hematology Analyzer (Drew Scientific, Dallas, TX, USA).
Flow cytometry
Lineage-negative, c-KIT+, SCA-1+ (LK(S)) progenitors, SLAM subsets as well as erythroid and myeloid progenitors were stained with antibodies from Becton Dickinson (North Ryde, NSW, Australia). These included: CD150 (TC15-12F12.2) as a phycoerythrin (PE) conjugate, SCA-1 (Dy) as a tandem PE and Cy7 conjugate (PE-Cy7); CD105 (MJ7/18) as a Pacific Blue conjugate (PB); CD16/32 (2.4G2) as a perdinin chlorophyll protein complex (PerCP) conjugate; c-KIT (2B8) as an allophycocyanin (APC) conjugate; CD48 (HM48-1) as a tandem APC and Cy7 conjugate (APC-Cy7) and biotinylated B220 (RA3-6B2), Mac-1 (M1/70), Gr-1 (RB6-8C5), Ter119 and CD3 (145-2C11). For secondary antibody staining, streptavidin (SAv) brilliant violet-605 was used (BioLegend, San Diego, CA, USA). For acute leukemia profiling, stains were used to analyze lineage markers (CD3 PE, CD19 APC-Cy7 (1D3), B220 as a tandem PE and Cy5 conjugate (PE-Cy5), Ter119 as a fluorescein isothiocyanate conjugate (FITC), Gr-1 PE-Cy7, MAC-1 APC) and T-cell markers (CD4 APC (RM4-5), CD8 PB (53-6.7), Thy1.2 FITC (53-2.1), TCR-β PE (H57-597), CD44 APC-Cy7 (IM7) and CD25 PE-Cy7 (PC61)). Total p53 was stained using Mouse mAb #2524 (1C12, Cell Signaling, Danvers, MA, USA). Phospho-p53 (Ser15) #9284 (Cell Signaling) was used to detect phosphorylation at the murine equivalent (p53-Ser18). Apoptosis was measured using Annexin-V (51-6874, BD Pharmingen) or activated Caspase-3/7 (Vybrant assay kit, Molecular Probes; 35118, Mulgrave, VIC, Australia). Caspase-3/7 staining was undertaken for 1 h at 37 °C following the manufacturer's guidelines. Cell sorting was undertaken on the BD FACS Aria II. For cell cycle and DNA damage analysis, cells were fixed with 4% paraformaldehyde for 10 min at 4 °C after initial surface staining, permeabilized using BD Perm/Wash buffer for 30 min at room temperature and stained for Ki67 FITC (B56, BD Pharmingen) at a 1/10 dilution overnight or γH2AX AF647 (20E3, Cell Signaling) for 2 h at room temperature in the dark. Before analysis, cells were stained with a 1-μM solution of 4',6-diamidino-2-phenylindole (DAPI) for 15 min at 4 °C.
Bone marrow transplantation
After harvesting, whole bone marrow from donor mice (6 weeks) was lineage depleted using BioMag Goat Anti-Rat IgG secondary antibody suspension (Qiagen 310107, Valencia, CA, USA) after lineage marker staining. The schema used for transplantation is summarized in Figure 1e. Mice were monitored regularly and bled monthly for hematopoietic chimerism analysis (Ly5.1 versus Ly5.2). Apart from LK Ly5.2 donor cells that did not show evidence of engraftment, mice were bled for 16 weeks.
Quantitative RT-PCR analysis
Total RNA was extracted from FACS-sorted cells after resuspension in TRIzol (Invitrogen). cDNA was transcribed from RNA using Roche Transcriptor First-Strand cDNA Synthesis Kit. PCR was performed using the Promega GoTaq mastermix on a LightCycler480 (Roche) and analyzed using the Roche LightCycler 480 software. Primer pairs are listed in Supplementary Table 1. Cycling conditions included denaturation (95 °C, 60 s) followed by 95 °C for 10 s, 55 °C for 10 s and 72 °C for 30 s for a total of 50 cycles.
RNA-seq analysis
Sequence reads were aligned against the mouse genome (mm10) using Bowtie 2 (ref. 40) and read counts quantified using HTSeq.41 Differential expression analysis was performed using Voom/Limma42 and visualization performed using Degust (http://victorian-bioinformatics-consortium.github.io/degust/). Genes were sorted by log-fold change, and GSEA was used to test for rank-based enrichment of particular gene sets.43 The RNA-seq data from this publication have been submitted to the GEO data set database http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=gjmticimxxwrvij&acc=GSE66264 and assigned the identifier: accession number GSE66264.
Hematopoietic progenitor cell assays
Agar and methylcellulose colony assays (n=3) were performed to assess for granulocyte-macrophage (CFU-GM) colony growth and burst-forming units-erythroid (BFU-E), respectively. Whole BM cells were plated at a density of 5 × 104 and 1 × 105 respectively per 35-mm dish. Agar cultures were generated combining 0.6% bacto agar with Dulbecco's Modified Eagle's Medium (mod-DME) and FBS supplemented with recombinant human SCF (Amgen, Kew, VIC, Australia), mIL-3 (PeproTech) and mIL-6 (PeproTech). Methylcellulose cultures were generated by combining commercial methylcellulose (MethoCult M3134 Stem Cell Technologies), Iscove's Modified Dulbecco's Medium (IMDM) and FBS supplemented with rh-SCF, mIL-3 and human erythropoietin. Plates were incubated at 37 °C in 10% CO2 for 7 days.
Statistical analysis
An unpaired Student's t-test was used to determine significance of data unless otherwise stated. Animal survival studies were plotted on Kaplan–Meier curves and analyzed using the Mantel–Cox log rank test. All error bars represent standard error of the mean (S.E.M.) unless otherwise indicated.
Acknowledgments
We thank Dr J Adams for providing mouse strains; S Beeraka for technical assistance; N Borg and K Spark for animal husbandry and the AMREP Flow Cytometry Core facility for its assistance with flow analysis and cell sorting. This study was supported by project grants from the Australian National Health and Medical Research Council (NHMRC) (628367) and Cancer Council Victoria awarded to CIS and DJC; a Viertel Senior Medical Research Fellowship awarded to DJC; a Leukaemia Foundation Clinical PhD Scholarship awarded to AAG; project grants from the NHMRC (1016701), Leukemia and Lymphoma society (7001-13), Cancer Council Victoria, an NHMRC SPRF fellowship (1020363) and the estate of Anthony (Toni) Redstone OAM awarded to AS.
Author contributions
AAG and DJC wrote the manuscript; AAG, DJC and CIS designed the research; AAG, CIS, LMF, CST and JS performed the research; DRP and FR performed and analyzed bioinformatics data; and AW and AS provided intellectual input, valuable reagents and edited the manuscript.
Glossary
- AML
acute myeloid leukemia
- BFU-E
burst-forming units-erythroid
- CFU-E
colony-forming units-erythroid
- CFU-GM
colony-forming units- granulocyte/macrophage
- GMP
granulocyte-macrophage progenitors
- LK cells
lineage neg Sca-1-c-Kit+
- LSK cells
lineage neg Sca-1+c-Kit+
- LT-HSCs
long-term hematopoietic stem cells
- MDS
myelodysplastic syndromes
- MLL
mixed-lineage leukemia
- MPPs
multipotent progenitors
- NHD13
Nup98-HoxD13
- SLAM
Signaling Lymphocyte Activation Molecules
- ST-HSCs
short-term hematopoietic stem cells
- TNF
tumor necrosis factor
- WT
wild-type
CIS receives royalties from the National Institutes of Health Technology Transfer office for the invention of the NUP98-HOXD13 mouse.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by A Ashkenazi
Supplementary Material
References
- Bravo GM, Lee E, Merchan B, Kantarjian HM, Garcia-Manero G. Integrating genetics and epigenetics in myelodysplastic syndromes: advances in pathogenesis and disease evolution. Br J Haematol 2014; 166: 646–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang WW, Pluvinage JV, Price EA, Sridhar K, Arber DA, Greenberg PL et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci USA 2013; 110: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woll PS, Kjällquist U, Chowdhury O, Doolittle H, Wedge DC, Thongjuea S et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell 2014; 25: 794–808. [DOI] [PubMed] [Google Scholar]
- Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer 2012; 12: 849–859. [DOI] [PubMed] [Google Scholar]
- Kerbauy D, Deeg H. Apoptosis and antiapoptotic mechanisms in the progression of myelodysplastic syndrome. Exp Hematol 2007; 35: 1739–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med 2010; 16: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 2011; 117: 2567–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Mufti GJ, Rasool F, Mijovic A, Devereux S, Pagliuca A. The role of apoptosis, proliferation, and the Bcl-2-related proteins in the myelodysplastic syndromes and acute myeloid leukemia secondary to MDS. Blood 2000; 96: 3932–3938. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 1990; 348: 331–333. [DOI] [PubMed] [Google Scholar]
- Vandenberg CJ, Waring P, Strasser A, Cory S. Plasmacytomagenesis in Eμ-v-abl transgenic mice is accelerated when apoptosis is restrained. Blood 2014; 124: 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ 2009; 16: 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekararaj AG, Smith AE, Mian SA, Mohamedali AM, Krishnamurthy P, Lea NC et al. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br J Haematol 2013; 160: 660–672. [DOI] [PubMed] [Google Scholar]
- Valente LJ, Gray DHD, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep 2013; 3: 1339–1345. [DOI] [PubMed] [Google Scholar]
- Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 2005; 106: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slape C, Liu LY, Beachy S, Aplan PD. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood 2008; 112: 2017–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slape CI, Saw J, Jowett JBM, Aplan PD, Strasser A, Jane SM et al. Inhibition of apoptosis by BCL2 prevents leukemic transformation of a murine myelodysplastic syndrome. Blood 2012; 120: 2475–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Menendez S, Schlegelberger B, Bae N, Aplan PD, Göhring G et al. Loss of p53 accelerates the complications of myelodysplastic syndrome in a NUP98-HOXD13-driven mouse model. Blood 2012; 120: 3089–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YJ, Choi CW, Slape C, Fry T, Aplan PD. Transplantation of a myelodysplastic syndrome by a long-term repopulating hematopoietic cell. Proc Natl Acad Sci USA 2008; 105: 14088–14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006; 442: 818–822. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell 2009; 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 2008; 451: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser SP, Lee EF, Trounson E, Bouillet P, Wei A, Fairlie WD et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev 2012; 26: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 2003; 426: 671–676. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17: 393–403. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 2003; 4: 321–328. [DOI] [PubMed] [Google Scholar]
- Timofeev O, Schlereth K, Wanzel M, Braun A, Nieswandt B, Pagenstecher A et al. p53 DNA binding cooperativity is essential for apoptosis and tumor suppression in vivo. Cell Rep 2013; 3: 1512–1525. [DOI] [PubMed] [Google Scholar]
- Jädersten M, Saft L, Smith A, Kulasekararaj A, Pomplun S, Göhring G et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol 2011; 29: 1971–1979. [DOI] [PubMed] [Google Scholar]
- Akala OO, Park I-K, Qian D, Pihalja M, Becker MW, Clarke MF. Long-term haematopoietic reconstitution by Trp53-/-p16Ink4a-/-p19Arf-/- multipotent progenitors. Nature 2008; 453: 228–232. [DOI] [PubMed] [Google Scholar]
- Jabbour AM, Daunt CP, Green BD, Vogel S, Gordon L, Lee RS et al. Myeloid progenitor cells lacking p53 exhibit delayed up-regulation of Puma and prolonged survival after cytokine deprivation. Blood 2010; 115: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Vandenberg CJ, Delbridge ARD, Wu L, Scott CL, Adams JM et al. Apoptosis-promoted tumorigenesis: gamma-irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev 2010; 24: 1608–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Sun Y, Zhang Z, Feng W, Gao Y, Cai Z et al. Deletion of proapoptotic Puma selectively protects hematopoietic stem and progenitor cells against high-dose radiation. Blood 2010; 115: 4707–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W-S, Heinrichs S, Xu D, Garrison SP, Zambetti GP, ADAMS JM et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 2005; 123: 641–653. [DOI] [PubMed] [Google Scholar]
- Yu H, Shen H, Yuan Y, XuFeng R, Hu X, Garrison SP et al. Deletion of Puma protects hematopoietic stem cells and confers long-term survival in response to high-dose gamma-irradiation. Blood 2010; 115: 3472–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038. [DOI] [PubMed] [Google Scholar]
- Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 2005; 106: 4131–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labi V, Erlacher M, Krumschnabel G, Manzl C, Tzankov A, Pinon J et al. Apoptosis of leukocytes triggered by acute DNA damage promotes lymphoma formation. Genes Dev 2010; 24: 1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvy S, Metcalf D, Print CG, Bath ML, Harris AW, Adams JM. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci USA 1999; 96: 14943–14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploner C, Kofler R, Villunger A. Noxa: at the tip of the balance between life and death. Oncogene 2008; 27: S84–S92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellagatti A, Cazzola M, Giagounidis A, Perry J, Malcovati L, Porta Della MG et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 2010; 24: 756–764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.