Abstract
Phagocytic cells of the immune system must constantly survey for, recognize, and efficiently clear the billions of cellular corpses that arise as a result of development, stress, infection, or normal homeostasis. This process, termed efferocytosis, is critical for the prevention of autoimmune and inflammatory disorders, and persistence of dead cells in tissue is characteristic of many human autoimmune diseases, notably systemic lupus erythematosus. The most notable characteristic of the efferocytosis of apoptotic cells is its ‘immunologically silent' response. Although the mechanisms by which phagocytes facilitate engulfment of dead cells has been a well-studied area, the pathways that coordinate to process the ingested corpse and direct the subsequent immune response is an area of growing interest. The recently described pathway of LC3 (microtubule-associated protein 1A/1B-light chain 3)-associated phagocytosis (LAP) has shed some light on this issue. LAP is triggered when an extracellular particle, such as a dead cell, engages an extracellular receptor during phagocytosis, induces the translocation of autophagy machinery, and ultimately LC3 to the cargo-containing phagosome, termed the LAPosome. In this review, we will examine efferocytosis and the impact of LAP on efferocytosis, allowing us to reimagine the impact of the autophagy machinery on innate host defense mechanisms.
Facts
Efferocytosis is a carefully orchestrated process by which phagocytes are recruited to sites of cell death, recognize and engulf dying cells, and clear them in an ‘immunologically silent' manner.
Dying cells have an active role in their own clearance; via the production of ‘find-me' signals to attract phagocytes and exposure of ‘eat-me' signals that engage phagocytic receptors to facilitate engulfment.
Defects in the efferocytosis machinery are associated with inflammation and autoimmune disorders, such as systemic lupus erythematosus (SLE).
Microtubule-associated protein 1A/1B-light chain 3 (LC3)-associated phagocytosis (LAP) is required for the effective clearance of dying cells.
Open Questions
Given the variety of ‘find-me' and ‘eat-me' signals, as well as their cognate receptors, how do these signals coordinate for effective efferocytosis?
How does LAP promote the anti-inflammatory response to dying cells, and what role does macrophage metabolism have?
Do defects in LAP contribute to inflammatory or autoimmune pathogenesis?
What role does LAP have in oncogenesis? What role does LAP have in tumor-associated macrophages?
An Introduction: Can I Interest You in Any Appetizers?
Even from our earliest developmental stages, the process of generating and maintaining a multicellular, functional organism is characterized by the creation and unceremonious destruction of billions of cells.1 Programmed cell death, such as apoptosis, necroptosis, or pyroptosis, are active mechanisms designed to sculpt, control, and aid the body in its development and survival. Much of our knowledge on the role of apoptosis in development comes from the study of Caenorhabditis elegans, wherein the first wave of cell death occurs ~4 h after fertilization, and of the 1090 cells that are generated, 131 of them are destined for death.2 In mammalian embryos, apoptosis is seen during cavitation and has a dynamic role in shaping the embryo.3 It is now well understood that proper apoptosis is fundamental for the proper development of the organism, as deficiencies in mediators of apoptosis result in embryonic lethality or animals with severe malformations.4 Conversely, other forms of cell death, such as necroptosis and pyroptosis, are not required during development, as evidenced by the normal development of mice deficient for receptor interacting protein kinase3 or caspase-1/11, respectively.5, 6 Once formed, the organism's relationship with cell death is far from over. Cellular turnover is a constant, genetically programmed process in the adult animal, and removal of these unwanted and unneeded cellular corpses is vital to prevent unwanted inflammation and immune activation.7 Although damage can certainly cause unwanted cellular death, most cellular death is an active process, and perturbations in the cell death programs can promote cell accumulation, autoimmunity, oncogenesis, attrition, and/or degeneration.
Within tissues, professional, non-professional, and specialized phagocytes are tasked with the clearance of dying cells. The best-characterized population of professional phagocytes, macrophages, is composed of tissue-specific, differentiated subsets of resident macrophages that clear dying cells and debris.8 For example, Kupffer cells in the liver clear aged red blood cells;9 aveolar macrophages of the lung clear apoptotic airway epithelial cells,8 and microglia in the central nervous system clear dying neurons.10 Other types of resident cells, such as epithelial cells and fibroblasts, have been termed non-professional phagocytes; though this designation may be a misnomer as these cells have a major role when professional phagocytes are rare, such as in the mammary gland or intestinal epithelium. In addition, airway epithelial cells are critical for the clearance of apoptotic airway epithelial cells, and epithelial cell-specific deletion of Rac1 results in increased allergen-induced airway inflammation.11 Still other types of tissue-specific, multifunctional cells exist as specialized phagocytes. In the testes, Sertoli cells are responsible for clearing apoptotic germ cells that arise during spermatogenesis.12 In the eye, retinal pigment epithelial (RPE) cells are critical for the homeostatic, daily removal of the photoreceptor outer segments (POSs), and the generation of 11-cis-retinal for the visual cycle. Defects in RPE cell-mediated removal of outer segments (or processing of outer segments via LAP, discuss below) can lead to a predisposition to conditions, such as age-related macular degeneration or retinitis pigmentosa.13
Like the death process itself, the innate immune system has tolerance systems in place to manage these morbid, yet necessary events. Although the generation and subsequent destruction of these cells is necessary for normal cellular homeostasis, wound healing, and immune responses in the adult organism, the ruin left in its wake would be catastrophic if not for the efficient work of the phagocytic system.14 Despite the constant, homeostatic turnover of cells, as well as cell death induced by stress, damage, or infection, it is rare to observe apoptotic cells under normal physiological conditions. Considering the average one million adult human cells that undergo apoptosis every second, one can appreciate the magnitude of the job facing phagocytes15 Moreover, as this is a reoccurring and normal event in the lifespan of an organism, this process of dead cell clearance must occur in a quiescent manner, so as to not inappropriately alert the immune system.16
We now appreciate the critical role that efferocytosis has on modulating immunity, as well as the impact that different types of cell death have on the immune response. In this review, we discuss the process of efferocytosis, chemoattraction of phagocytes, recognition of dying cells, engulfment of cellular corpses, and the processing of engulfed cellular cargo, specifically the role of LAP in clearance of dying cells and control of inflammation. Finally, we explore the effect of defective efferocytosis on pathology and disease states.
The Mechanisms of Efferocytosis: Would You Like to Hear the Specials?
As the focus of this review is the aftermath of cell death, we have summarized the four most well-defined modes of cell death (apoptosis, necrosis, necroptosis, and pyroptosis) in Table 1, as the roles and mechanisms of cell death have been studied and reviewed extensively.1
Table 1. Summary of the four major modes of cell death: apoptosis, necrosis, necroptosis and pyroptosis.
| Description | Characteristics | References |
|---|---|---|
| Apoptosis | ||
| Active cellular death, largely controlled by a family of cysteine proteases called caspases Apoptotic caspases are broadly grouped into initiator caspases (caspase-8 and -9) and executioner caspases (caspases-3, -6, and -7) Intrinsic or mitochondrial pathway Activated by stress-inducing stimuli (i.e., DNA damage, accumulation of unfolded proteins, and hypoxia) and developmental signals Signals converge on the mitochondria, where pro-apoptotic and anti-apoptotic members of the BCL2 family mediate the release of cytochrome c, formation of the apoptosome with caspase-9 and APAF-1, which leads to the activation of the downstream executioner caspases, such as caspase-3 and caspase-7 Extrinsic pathway Triggered by signals that engage extracellular death receptors (DR) Tumor necrosis factor (TNF) and TNF receptor-1 (TNFR1) CD95-ligand (CD95-L or Fas-L) and CD95 (or Fas) TNF-related apoptosis-inducing ligand (TRAIL) and TRAIL-R1/2 (DR4/5) Recruitment of pro-caspase-8 to the death-inducing signaling complex (DISC) at the DR (with the adapter proteins FADD or TRADD), resulting in dimerization and activation of caspase-8, leading to caspase-3 and caspase-7 activity Caspase-8 activity can also feed into the intrinsic pathway by cleaving and activating BCL2 family proteins | Membrane ‘blebbing,' often with separation of apoptotic bodies DNA fragmentation Chromatin condensation Considered immunologically silent due to the packaging of possible danger-associated molecular patterns (DAMPs) into discreet, tolerogenic pieces Active phosphatidylserine (PtdSer) exposure (Annexin V positive) Propidium iodide or 7-AAD negative at early stages | 1, 99, 100, 101, 102, 103 |
| | ||
| Necrosis | ||
| Characterized as an passive type of cell death that occurs independently of caspase activation Triggered in response to catastrophic damage or pathology, including infarction, mechanical trauma, ischemia, frostbite, and animal venom Apoptotic cells that are not efficiently cleared by phagocytes can undergo secondary necrosis independently of any apoptotic machinery | Cellular swelling (oncosis) Organelle swelling Nuclear distention Cellular rupture Releases inflammatory cellular contents (DAMPs) or alarmins that can activate neighboring immune cells via Toll-like receptor (TLR) signaling and other mechanisms Annexin V positive due to membrane rupture Propidium iodide or 7-AAD positive | 1, 104, 105, 106 |
| | ||
| Necroptosis | ||
| Genetically programed cell death with the morphological features of necrosis Triggered by diverse forms of neurodegeneration, ischemia, or infection. Engagement of the extrinsic pathway (i.e., TNF–TNFR pathway) in the absence of caspase-8 can result in a necrotic cell death that requires the kinase activity of receptor interacting protein kinase1 (RIPK1) and RIPK3 RIPK3 phosphorylates and activates the pseudokinase, mixed lineage-kinase like (MLKL) Induces a conformational change that allows for its oligomerization and interaction with the plasma membrane through binding to phosphatidylinositol lipids to directly disrupt membrane integrity RIPK1 is required for a variety of innate immune signaling pathways, such as TLRs, interferons, and the RIG-I-MAVS pathway | Loss of plasma membrane integrity Swollen cellular organelles Releases inflammatory cellular contents (DAMPs) or alarmins that can activate neighboring immune cells via TLR signaling and other mechanisms Active PtdSer exposure (Annexin V positive) Propidium iodide or 7-AAD positive | 5, 107, 108, 109, 110, 111, 112 |
| | ||
| Pyroptosis | ||
| Non-apoptotic, genetically programmed cellular death mediated by excessive inflammatory caspase activity (human caspase-1, -4, and -5; rodent caspase-1 and -11) Required for cell death in a variety of experimental settings, including in the immune system, the cardiovascular system, and the central nervous system Caspase-1 is activated by dimerization at complexes termed inflammasomes that form in the cytosol and detect a diverse repertoire of pathogenic molecules, including bacterial toxins and viral RNA Activated caspase-1 in turn cleaves pro-IL-1β and pro-IL-18, which facilitates the secretion of these pro-inflammatory cytokines Characterized by caspase-1-dependent formation of plasma membrane pores, which leads to pathological ion fluxes that ultimately result in cellular lysis and release of inflammatory intracellular contents Caspase-1 can also activate caspase-7 | Cellular rupture DNA fragmentation Releases inflammatory cellular contents (DAMPs) or alarmins that can activate neighboring immune cells via TLR signaling and other mechanisms Often Annexin V positive due to membrane rupture Propidium iodide or 7-AAD positive | 1, 4, 6, 51, 113, 114 |
Efferocytosis is not merely a passive event, but a carefully orchestrated process designed to efficiently eliminate cellular corpses and limit exposure to their potentially damaging components, with the goal being immunological tolerance.17 Efferocytosis can be generally categorized into 4 steps: 1) the release of ‘find-me' signals by dying cells to recruit phagocytes, 2) phagocyte recognition and engagement of ‘eat-me' signals on dying cells, 3) the engulfment of the cellular corpse, and 4) the processing, degradation, and immune response to the engulfed corpse. We now recognize that defects in any of these four steps can contribute to unwanted inflammation and autoimmune disorders, such as systemic lupus erythematosus18 (Table 2).
Table 2. Components of the efferocytosis machinery and their association with inflammatory and autoimmune diseases.
| Molecule | Role in efferocytosis | Disease(s) | References |
|---|---|---|---|
| Molecules associated with increased incidence or risk of disease | |||
| Nucleotides (ATP/UTP) | ‘Find-me' | MS/EAE | 115 |
| Pannexin-1 | ‘Find-me' | MS/EAE, seizure disorders | 116, 117 |
| S1P | ‘Find-me' | MS/EAE | 118 |
| LPC | ‘Find-me' | Atherosclerosis, SLE/systemic autoimmunity | 119 |
| S1PR1-5 | ‘Find-me' | MS/EAE | 118 |
| G2A | ‘Find-me' | SLE/systemic autoimmunity, atherosclerosis | 120, 121 |
| CX3CR | ‘Find-me' | Autoimmune uveitis, MS/EAE | 122, 123 |
| ICAM3 | ‘Eat-me' | RA, SLE/systemic autoimmunity, GBS, MS/EAE | 124 |
| CRT | ‘Eat-me' | RA, Sjogren's syndrome, Celiac disease, SLE/systemic autoimmunity | 125 |
| C1q | ‘Eat-me' | SLE/systemic autoimmunity, RA, atherosclerosis | 126 |
| TIM1 | ‘Eat-me' | SLE/systemic autoimmunity, airway inflammation | 127, 128 |
| TIM3 | ‘Eat-me' | Airway inflammation, MS/EAE | 128 |
| TIM4 | ‘Eat-me' | SLE/systemic autoimmunity | 37 |
| BAI1 | ‘Eat-me' | Glioblastoma, neurological disorders | 129 |
| Integrins (avβ3) | ‘Eat-me' | Scleroderma, ulcerative colitis | 52, 130 |
| MerTK | ‘Eat-me' | SLE/systemic autoimmunity, retinisis pigmentosa, atherosclerosis | 53, 131, 132 |
| MFG-E8 | ‘Eat-me' | SLE/systemic autoimmunity, atherosclerosis | 31, 133 |
| ProteinS | ‘Eat-me' | Thrombosis, SLE/systemic autoimmunity | 134, 135 |
| CD300f | ‘Eat-me' | SLE/glomerulonephritis | 136 |
| ELMO1 | Engulfment | Testicular pathology, impaired neurogenesis | 12, 137 |
| DOCK180 | Engulfment | Cardiovascular abnormalities, myogenesis abnormalities | 138, 139 |
| GRK6 | Engulfment | SLE/systemic autoimmunity | 140 |
| RAC1 | Engulfment | RA, airway inflammation | 11, 141 |
| DNAse II | Processing | Polyarthritis | 72 |
| LXRα/β | Processing | MS/EAE, SLE/systemic autoimmunity, autoimmune uveitis, type I diabetes, atherosclerosis | 142, 143, 144, 145, 146, 147, 148 |
| PPARδ/γ | Processing | MS/EAE, SLE/glomerulonephritis, atherosclerosis, osteoarthritis | 95, 96, 149, 150, 151 |
| ABCA1 | Processing | SLE/glomerulonephritis | 152 |
| Molecules associated with decreased incidence or risk of disease | |||
| Fractalkine (CX3CL1) | ‘Find-me' | Sjogren's syndrome, airway inflammation, RA | 153, 154, 155 |
| Purigenic receptors (P2Y2) | ‘Find-me' | Sjogren's syndrome, autoimmune uveitis | 156, 157 |
| Integrins (avβ3) | ‘Eat-me' | MS/EAE | 158 |
| CD91 (LRP) | ‘Eat-me' | RA, SLE/systemic autoimmunity | 159 |
| RAGE | ‘Eat-me' | MS/EAE, DTH | 160, 161 |
| GAS6 | ‘Eat-me' | Thrombosis, nephrotoxic nephritis, SLE/systemic autoimmunity | 162, 163, 164 |
Abbreviations: BA1, brain-specific angiogenesis inhibitor 1; DTH, delayed-type hypersensitivity; EAE, experimental autoimmune encephalitis; C1q, complement 1q; CRT, calreticulin; GBS, Guillain-Barré syndrome; LPC, lysophosphatidylcholine; LXR, liver X receptor; MFG-E8, milk fat globule-EGF factor 8; MS, multiple sclerosis; PPARδ/γ; peroxisome proliferator-activated receptor γ/δ; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; S1P, sphingosine-1-phosphate; TIM, T-cell immunoglobulin mucin receptor
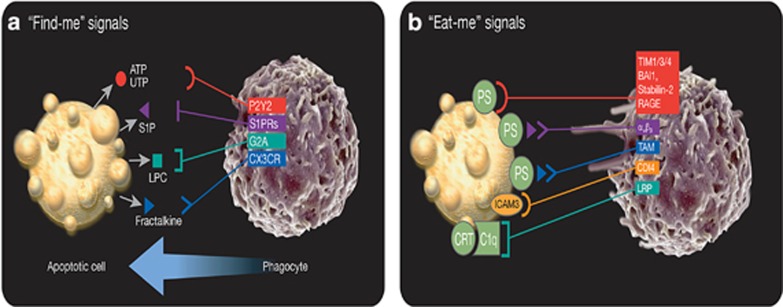
As phagocytes are often not proximal to sites of cell death or even reside in the tissues they must survey, dying cells must ‘advertise' their presence to phagocytes.19 Recruitment of phagocytes to sites of cell death in C. elegans occurs before the completion of apoptosis, indicating that one of the first acts of a dying cell is to prepare for its own elimination.20, 21 During this process, apoptotic cells release ‘find-me' signals, distinct molecules that establish a chemotactic gradient to attract phagocytic cells.22 Nucleotides, such as ATP, are released in a caspase-dependent manner via activation of pannexin-1 channels and are perhaps the most well-defined ‘find-me' signals.23 These nucleotides are detected by phagocytes via purinergic receptors, like P2Y2, and disruption of the nucleotide/P2Y2 interaction results in an accumulation of dying cells in vivo.19 Apoptotic cells also release the membrane-associated molecule fractalkine (or CX3CL1), which is sensed by CX3CR1 and mediates the migration of macrophages to the dying cells. Mice deficient for CX3CR1, however, do not display a defect in apoptotic cell clearance, suggesting that multiple factors are required to recruit effectively phagocytes.24 Molecules of lipid origin can also act as ‘find-me' signals. Lysophosphatidylcholine is generated and released via caspase-3-dependent activation of phospholipase A, and is sensed by the G-protein-coupled receptor G2A.25 Sphingosine-1-phosphate (S1P), also a lipid ‘find-me' signal, is released by dying cells and sensed by multiple G-protein-coupled receptors S1P-R1-5. These lipid signals have been demonstrated to mediate phagocyte chemotaxis26 (Figure 1a).
Figure 1.
The recruitment of phagocytes and recognition of dying cells by phagocytes. (a) Dying cells release ‘find-me' signals, such as ATP, UTP, S1P, lysophosphatidylcholine (LPC), or fractalkine, that recruit phagocytes to sites of cell death. Phagocytes sense these ‘find-me' signals via cognate receptors (P2Y2, S1PRs, G2A, and CXCR3, respectively). (b) Phagocytes express a variety of receptors and bridging molecules that recognize and engage dying cells via ‘eat-me' signals exposed on apoptotic cell surfaces. The most common ‘eat-me' signal, phosphatidylserine (PtdSer or PS), engages the PtdSer-specific receptors, TIM1, TIM3, TIM4, BAI1, stabilin-2, and RAGE, as well as the PS-specific bridging molecules MFG-E8, Gas6, and protein S. These bridging molecules engage other surface engulfment receptors (αvβ3 or TAM) to facilitate uptake. Other ‘eat-me' signals, such as calreticulin (CRT) and ICAM3, exist and mediate recognition and engulfment via the receptors LRP (via C1q) and CD14, respectively
There are significant caveats to the ability of ‘find-me' signals to efficiently act as chemoattractants in physiologically scenarios. The idea of an apoptotic cell's purposeful release of a ‘find-me' signal to actively recruit phagocytes is undermined by the relatively low level of signal released by apoptotic cells compared with necrotic cells. ‘Find-me' signals are often released in an active, caspase-dependent manner, yet these molecules are also released (and in greater quantities) during other forms of cell death, such as necrosis or necroptosis.27 Indeed, <2% of intracellular ATP is released during apoptosis.15 In addition, these released nucleotides must also survive degradation by extracellular nucleotidases, indicating that they most likely act in a short-range capacity.19 Similarly, ‘find-me' signals of lipid origin are present ubiquitously in the circulation at a concentration higher than that released by apoptotic cells.7 The mechanisms by which ‘find-me' signals, which can be recognized by a wide variety of cells, specifically recruit phagocytes, the majority of which are macrophages, are unknown. The counteractive effect of ‘keep out' signals has been proposed to aid in the coordinated recruitment of phagocytes. For example, lactoferrin, a glycoprotein released by apoptotic cells, has been shown to actively exclude neutrophils and eosinophils from sites of cell death.28, 29 Further complicating the matter is the dual role that ‘find-me' signals can have, as danger-associated molecular patterns (DAMPs)30 or as activating factors to prime phagocytes.31
Cell death does not occur in a vacuum; sites of cell death are a conglomeration of dying cells, healthy cells, and immune cells. The phagocyte must distinguish living cells from dying cells in order to maintain homeostasis, promote proper development, and prevent unwanted inflammation. Just as dying cells must recruit phagocytes, they must also transform themselves into targets for engulfment, displaying distinct signals that differentiate them from viable cells.32, 33 The extracellularly exposed lipid, phosphatidylserine (PtdSer), is the most well-characterized ‘eat-me' signal and an essential factor in effective efferocytosis.34 Normally confined to the inner leaflet of the plasma membrane lipid bilayer of living cells (and in a relatively minor amount), PtdSer is actively and rapidly externalized in a caspase-dependent manner during apoptosis.34 Caspase 3-mediated cleavage of the scramblase Xkr8 facilitates exposure of PtdSer during apoptosis,35 an event normally reversed by the activity of the flippase ATP11C, which is inactivated by caspase-3 cleavage.36 Extracellularly exposed PtdSer is recognized by multiple, bona fide membrane receptors, such as T-cell immunoglobulin mucin receptor 4 (TIM4), brain-specific angiogenesis inhibitor 1 (BAI1), and stabilin-2,37, 38, 39 and bridging molecules, such as milk fat globule-EGF factor 8 (MFG-E8) and Gas6, that recognize PtdSer and then engage phagocytic cell surface receptors such as integrin αvβ3, αvβ5, or Tryo3-Axl-Mer (TAM) receptors40, 41, 42 for engulfment.
Other ‘eat-me' signals have also been identified, which are likely to have a ‘tethering' function, facilitating the above events. ICAM3 can bind to membrane-associated CD14,43 and externalized calreticulin bound to complement C1q can engage CD91 (or LRP1).44 Oxidized LDL-like moieties and glycosylated surface proteins can serve as ‘eat-me' signals, binding to scavenger receptors45 and lectins,46 respectively (Figure 1b).
Similar to the ‘find-me'/'keep out' signal paradigm, there is evidence of a negative signal to discourage phagocytosis. Although PtdSer is considered a hallmark of cell death, forced PtdSer exposure47 or physiologically normal exposure on activated, living cells does not mediate recognition or engulfment.48 Thus, the simultaneous presence of ‘don't eat-me' signals, such as CD31, CD47, and CD61, on viable cells, may negatively regulate phagocytosis, indicating to the phagocyte that despite the extracellular PtdSer, this cell is not intended for clearance.18, 49, 50 Therefore, a coordinated effort between the dying cell and the phagocyte must exist to facilitate efferocytosis.
Although the majority of work on ‘find-me' and ‘eat-me' signals stems from apoptotic cells, these signals also function during other types of cell death, such as necroptosis and pyroptosis.7 As previously mentioned, ‘find-me' signals, such as ATP, are released (and in greater quantities) during necrosis, necroptosis, and pyropotosis.6, 14, 51 Similarly, necrotic and pyroptotic cells also stain positive for Annexin V, although in these cases, externalized PtdSer can be attributed to rupture of the plasma membrane rather than an active exposure process.52, 53, 54 These dead cells can still be recognized and engaged by PtdSer receptors;55 however, owing to the lytic nature of their demise, DAMPs have already been released into the milieu and can activate inflammatory programs. Therefore, although apoptotic cells actively coordinate their own clearance, necrotic and pyroptotic cells passively utilize these systems as well.
The tissue specificity of PtdSer receptors may help to explain why multiple receptors are required for efficient efferocytosis.17, 18, 56 Stabilin-2 is highly expressed in endothelial cells within atherosclerotic plaques,57 although defects in BAI1, highly expressed in glial and neuronal cells, are associated with neurodegenerative disorders.58 Despite a common ligand and a common goal of engulfment, the mechanisms by which PtdSer receptors mediate phagocytosis are often varied. Once engaged by PtdSer-bound integrins, bridging molecules, such as αvβ3 or TAM, associates with the adapter proteins ELMO1 and DOCK180 (via CrkII) at the site of phagoytosis.59, 60 The PtdSer receptor BAI1 also requires the activity of the DOCK180/ELMO1 complex for engulfment, but BAI1 is able to recruit the complex independently.38 Stabilin-2 and CD91/LRP, however, require the activity of the adapter protein, GULP, to facilitate phagocytosis.61, 62, 63 One of the most well-known PtdSer receptors, TIM4, contains a very short cytoplasmic region and currently its signaling components are unknown.64
Dying cell engulfment involves active membrane ruffling by a process similar to macropinocytosis.65, 66 Engagement of PtdSer receptors results in cytoskeletal reorganization to facilitate phagocytosis, which is mediated by the Rho family of small GTPases, including members RhoA, ROCK, Rac, Rab5, and Cdc42.67 These GTPases cycle between the resting, inactive GDP-bound state and the active GTP-bound state, mediated by specific guanine-nucleotide-exchange factors (GEFs), such as the bipartite GEF formed by DOCK180 and ELMO1.68 Ultimately, signaling during efferocytosis converges on the activation of evolutionarily conserved Rac1, acting at the phagocytic cup to promote actin polymerization and cytoskeletal rearrangement via the Scar/WAVE complex.67, 69, 70 Similarly, CDC42 has been linked to the engulfment of apoptotic cells, although its precise role is unclear.71 Once encased within the phagocyte, however, the dying cell is now capable of exerting its effect on critical downstream immunological and metabolic pathways.
Degradation After Phagocytosis: Did You Save Room for Dessert?
The mechanisms by which phagocytes handle the burden of processing an engulfed cellular corpse are currently of great interest. Not only must a phagocyte interpret its ingested cargo in an immunologically tolerant manner, it must also contend with the excess lipid, cholesterol, and protein that an entire engulfed cell brings. Acidic proteases and nucleases in mature phagolysosomal compartments degrade dying cells into their basic cellular components, including fats, sterols, peptides, and nucleotides. For example, DNAse II, a lysosomal enzyme, is required for the degradation of DNA, and DNAse II deficiency results in an accumulation of undigested DNA fragments within phagocytic cells, capable of activating intracellular nucleic acid sensors.72
The recent discovery of LAP has shed some light on this issue. The two ancient systems of phagocytosis and autophagy represent two modes of nutrient acquisition, during abundance and scarcity, respectively. These two evolutionarily conserved pathways converge, however, during the engulfment of pathogens or dead cells.73 LAP is a process that marries the processes of phagocytosis and autophagy into a fundamentally new concept, allowing us to reinterpret the impact of the autophagy machinery on innate host defense mechanisms (Table 3).
Table 3. Components of the autophagic machinery and their association with inflammatory and autoimmune diseases.
| Molecule | Confirmed pathway(s) | Associated Disease(s) | References |
|---|---|---|---|
| NOX2 | LAP | CGD, Alzheimer's disease, Creuzfeldt–Jakob disease | 165, 166, 167 |
| Rubicon | LAP | Ataxia | 168 |
| Beclin1 | Autophagy LAP | Ovarian cancer, breast cancer, lung cancer, cystic fibrosis, Alzheimer's disease, RA | 169, 170, 171, 172, 173, 174 |
| VPS34 | Autophagy LAP | Schizophrenia | 175 |
| UVRAG | Autophagy LAP | Stomach cancer, non-segmental vitiligo, colorectal cancer, cardiomyopthay | 85, 176, 177, 178 |
| ATG5 | Autophagy LAP | Airway inflammation, SLE/systemic autoimmunity, MS/EAE, RA, Alzheimer's disease, atherosclerosis | 179, 180, 181, 182, 183, 184 |
| ATG16L | Autophagy LAP | Crohn's disease, atheroclerosis | 185, 186 |
| ATG7 | Autophagy LAP | SLE/systemic autoimmunity, MS/EAE, type I diabetes, RA, Alzheimer's disease, cardiomyopathy | 187, 188, 189, 190 |
| ATG4 | Autophagy LAP | Otoconia | 191 |
| LC3 | Autophagy LAP | Nasu-Hakola disease | 192 |
| LAMP2 | Autophagy LAP | Danon disease, type II diabetes | 193 |
| ULK1 | Autophagy | Crohn's disease | 194 |
| FIP200 | Autophagy | Inflammatory skin disorders | 195 |
| p62 | Autophagy | Huntingtin's disease | 196 |
| EPG5 | Autophagy | Vici syndrome | 197 |
| IRGM | Autophagy | Crohn's disease, MS/EAE | 198 |
| SMURF1 | Autophagy | Ulcerative colitis | 199 |
| WDR45 | Autophagy | Encephalopathy | 200 |
| Parkin | Mitophagy | Parkinson's disease | 201 |
| PINK1 | Mitophagy | Parkinson's disease | 202 |
Abbreviations: ATG, autophagy-related gene; CGD, chronic granulomatous disease; EAE, experimental autoimmune encephalitis; LAP, LC3-associated phagocytosis; MS, Multiple sclerosis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus
Confirmed activity in autophagy, LAP, and/or mitophagy is indicated
LAP is triggered when an extracellular particle, such as a pathogen, immune complex, or dead cell, is sensed by an extracellular receptor, including Toll-like receptor1/2 (TLR1/2), TLR2/6, TLR4, FcR, and TIM4, and phagocytosed.55, 74, 75, 76 This engulfment recruits some, but not all, members of the autophagy machinery to the cargo-containing vesicle.55, 77 It is the activity of these autophagic players that facilitates the rapid processing of the cargo via fusion with the lysosomal pathway, which can have a critical role in the degradation of engulfed cargo,77, 78 as well as modulate the resulting immune response.55, 75, 78
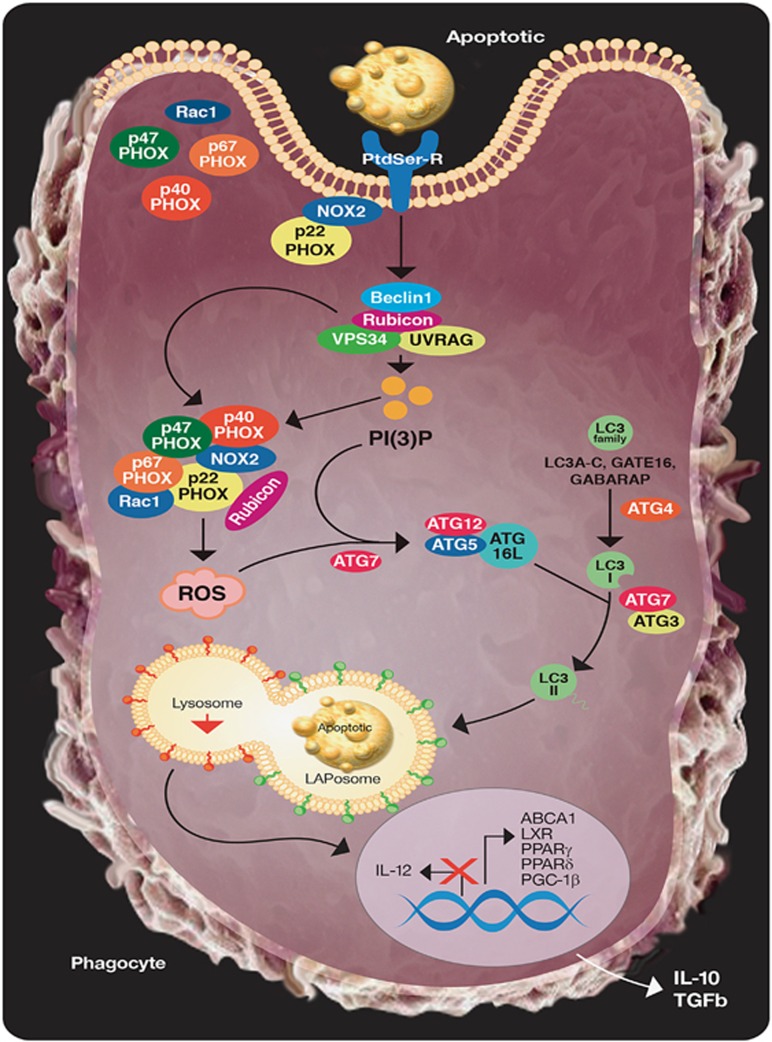
Despite sharing common molecular machinery, there currently exist several distinctions that differentiate LAP from canonical autophagy (Figure 2). Originally, LAP and autophagy were distinguished by the structure of the LC3-decorated phagosome (or LAPosome) and the rapidity with which LAP occurs. EM analysis revealed that LAP results in single-membrane structures,77 as opposed to the double-membrane autophagosomes surrounding autophagic cargo.79 Whereas LC3-decorated autophagosomes can take hours to form, LC3-II can be detected on LAPosomes in as few as 10 min after phagocytosis, and phosphatidylinositol 3-phosphate (PI(3)P) activity can be seen at the LAPosome within minutes after phagocytosis.55, 75, 77
Figure 2.
The processing of engulfed dying cells requires LC3-associated phagocytosis (LAP) and promotes an anti-inflammatory response. Upon engulfment of dying cells, components of the LAP pathway are recruited to dead cell-containing phagosome (or LAPosome). The class III PI3 K complex, comprised of Beclin 1, VPS34, UVRAG, and Rubicon, is critical to the sustained and localized production of PI(3)P at the LAPosome. PI(3)P serves two roles – the recruitment of the downstream LAP machinery (such as ATG5, ATG12, ATG16L, and ATG7) and stabilization of the NOX2 complex for the production of ROS. Rubicon is also required for the stabilization of the NOX2 complex. Both ROS and PI(3)P are required for successful LC3-II decoration of the LAPosome, and LC3-II is required for fusion to the lysosome and maturation of LAPosome. The anti-inflammatory effects of efferocytosis are mediated by the activity of lipid and cholesterol sensors, such as ABCA1, LXR, PPARγ/δ, and PGC-1β, leading to the production of IL-10 and TGFβ, whereas pro-inflammatory mediators, such as IL-12, are actively repressed
Recent studies have elucidated the molecular mechanisms governing LAP.78 Although a majority of the core autophagy components are required for LAP, there exist some critical differences that can distinguish the two processes. Under basal conditions, mTOR inhibits the pre-initiation complex, comprised of FIP200, autophagy-related gene13 (ATG13), and ULK1/2, and hence autophagy. However, the pre-initiation complex is dispensable for LAP.55, 75, 78 Furthermore, canonical autophagy requires the ULK1-dependent release of a Beclin1-activating cofactor, Ambra1, from the dynein motor complex,80 and the function of WIPI2,81 whereas LAP does not.78
Both LAP and canonical autophagy require the class III PI3K complex, which contains the core components Beclin1, VPS34, and VPS15.82 It can, however, differ in its additional composition. ATG14 and UVRAG are mutually exclusive in their association with the class III PI3K complex during autophagy,83 and silencing of either ATG1483, 84 or UVRAG85 inhibits canonical autophagy. LAP, on the other hand, only requires the activity of the UVRAG-containing class III PI3K complex, whereas ATG14 is dispensible.78
Rubicon (RUN domain protein as Beclin 1 interacting and cysteine-rich containing) is a protein that associates constitutively with the UVRAG-containing class III PI3K complex.86 Rubicon is a negative regulator of autophagy (via its inhibition of VPS3484, 86 or by blocking GTPase Rab7 activation87), and silencing of Rubicon results in an increase in the number of autophagosomes.78 During LAP, Rubicon is uniquely associated with LAPosomes (but not conventional phagosomes), and Rubicon-deficient cells are completely defective in LAP.78 Thus, Rubicon is a molecule that is uniquely required for LAP, but dispensable for canonical autophagy.
Studies suggest that the role for Rubicon in LAP is twofold. First, Rubicon promotes the association of the active class III PI3K complex with the LAPosome, thereby aiding in the localization of VPS34-mediated PI(3)P at the LAPosome. In both canonical autophagy and LAP, PI(3)P is required for the recruitment of the downstream ubiquitin-like conjugation systems, the ATG5-12 and LC3-PE conjugation systems.78 In LAP, Rubicon and PI(3)P have an additional role. Rubicon stabilizes NOX2, the predominant NADPH oxidase in phagocytes, by interacting with its p22phox subunit via its serine-rich domain (aa 567–625), a domain separate from the CCD domain (aa 515–550) responsible for its interaction with Beclin188 and the RUN domain (aa 49–180) responsible for its interaction with VPS34.89 Moreover, PI(3)P binds and stabilizes the p40phox subunit of NOX2.90 Collectively, Rubicon promotes the association of the active class III PI3K complex with the LAPosome and the production of PI(3)P. Rubicon and PI(3)P stabilize the active NOX2 complex to promote optimal reactive oxygen species (ROS) production, which is also required for successful LAP.78 Indeed, NOX2-deficeint cells fail to undergo LAP78, 91 and scavenging of ROS by antioxidants, such as resveratrol, Tiron, or alpha-tocopherol is also an effective way to inhibit LAP.78, 88, 91 Thus, LAP and canonical autophagy are molecularly distinct processes.13, 55, 75, 76
In addition, LAP and canonical autophagy are functionally distinct as well. There is mounting evidence that LAP is a critical regulator of inflammation in vivo and under physiologically relevant conditions. Not only is LAP critical for the degradation of engulfed organisms, such as intraphagosomal yeast77 or Aspergillus fumigatus,74 but LAP can have a profound effect on the immune response to the engulfed material. Upon intranasal challenge with A. fumigatus, a TLR2 ligand, LAP-deficient animals fail to efficiently clear the pathogen and display increased levels of pro-inflammatory cytokines both locally (lung) and systemically (serum).74 Thus, many of the autophagic defects associated with control of pathogens could actually be defects in LAP.
LAP can also be triggered in specialized phagocytes, such as the RPE. On a daily basis and regulated by circadian rhythm, RPE cells phagocytose and digest shed POSs, a process crucial for supplying nutrients and O2 to the retina and the metabolism of vitamin A for the visual cycle. However, the receptor(s) that recognize shed POS and trigger LAP remains unknown. What is known is the requirement for LAP in the proper processing of POS and promotion of the visual cycle, a series of biochemical reactions within the RPE and retina that ultimately results in the production of the chromophore 11-cis-retinal (RAL) necessary for the phototransduction signaling cascade. RPE cells deficient for LAP (ATG5, Beclin1), but not canonical autophagy (ULK1, FIP200, ATG13) displayed defective POS degradation, diminished production of 11-cis-retinal, and decreased visual function with age. Thus, LAP functions to support chromophore regeneration through the efficient processing of POS by the RPE.13
LAP is also required for establishing specific signaling compartment and is a critical regulator of the type I interferon response in some cases. In plasmacytoid dendritic cells, LAP is induced by engagement of the FcγR by immune complexes (IC), complexes of self-antigen (such as DNA) and autoantibodies commonly found in patients with SLE. In cells deficient for LAP, failure to lipidate LC3 on the DNA-IC-containing LAPosome results in a failure to acquire a late-endolysosomal phenotype. Subsequently, these LAP-deficient cells fail to form the specialized interferon regulatory factor 7 (IRF7)-signaling compartment required for TLR9-mediated activation of IRF7, and therefore fail to produce IFN-α. This suggests that LAP could affect the functional immune response elicited by autoantigens and have an important role in autoimmunity.75
Unwanted inflammation and autoimmunity is held in check by the efficient clearance of dying cells every day.55, 92 It is the responsibility of the phagocytes to first clear the dying cell from circulation and then instigate an anti-inflammatory response. Phagocytes that have engulfed apoptotic cells have been shown to secrete anti-inflammatory cytokines, such as TGFβ and interleukin-10 (IL-10),54 whereas actively suppressing pro-inflammatory cytokines, such as tumor necrosis factor, IL-1, and IL-12.93 How the phagocyte achieves this feat is of great interest. LAP is triggered during efferocytosis, and apoptotic, necrotic, and necroptotic cells can engage the PS receptor, TIM4, resulting in a recruitment of the LAP machinery to the dead-cell-containing, single-membrane LAPosome. LAP-deficient macrophages fail to recruit LC3 to the LAPosome, leading to a failure in phagosomal acidification and subsequent corpse degradation. Whereas the paradigm of efferocytosis is ‘immunologically silent', LAP-deficient macrophages produce markedly increased levels of IL-1β and IL-6 when fed dying cells, yet produce significantly less anti-inflammatory cytokines, such as IL-10, upon such engulfment.55 LAP is engaged by a variety of receptors and is critical for directing a variety of different immune response, including preventing an unwanted inflammatory response and promoting the formation of the interferon signaling compartment.55, 75 Although these functions may appear contradictory, it suggests that the fundamental role of LAP is to shape the appropriate response, and absence of this pathway seems to result in aberrant inflammation and pathogen control.
How the LAP pathway modulates the immune response to apoptotic cells remains to be elucidated, though clues may lie in the mechanisms by which the phagocyte handles the metabolic stress of doubling its content of cellular components. The sensing of one such component, cholesterol, can have a significant effect on the phagocyte's response to engulfed dead cells and their increase in basal cholesterol efflux activity.94 Members of the peroxisome proliferator-activated receptor γ/δ (PPARγ/δ) and liver X receptor (LXR) families, both important regulators of cellular lipid homeostasis, are activated during efferocytosis, and results in a positive feedback signal wherein the phagocytic receptors, such as members of the TAM family, are upregulated.95, 96 Furthermore, cholesterol efflux machinery, such as 12-transmembrane protein ABCA1 (ATP-binding cassette sub-family A, member 1), is upregulated to accommodate the increase in cholesterol load.92
The non-immunogenic nature of efferocytosis of apoptotic cells is one of its key characteristics, and cholesterol homeostasis has a critical role in establishing this tolerance.18, 22 PPARγ/δ are central players in the polarization of anti-inflammatory (‘M2') macrophages, and agonizts for both PPARγ and LXR have been shown to inhibit inflammatory responses.18, 96 Conversely, PPARγ−/− and PPARδ−/− macrophages are defective in efferocytosis. The dual functions of PPARs and LXRs in both lipid apoptotic cell clearance and lipid homeostasis suggest the interconnectedness between efferocytosis and metabolism.
Despite all types of dying cells providing excess cholesterol for the engulfing cells, uptake of necrotic cells does not induce enhanced cholesterol efflux in the phagocytes, suggesting that engagement of ligands on apoptotic cells, not extra cholesterol, induces a 'prophylactic' cholesterol efflux from phagocytes.97 Studies have shown that mere co-culture of macrophages with PtdSer liposomes can induce the cholesterol efflux, anti-inflammatory cytokine production, and suppression of pro-inflammatory genes.93, 98 These data suggest that metabolic sensors, in conjunction with engagement of ‘eat-me' signals, such as PtdSer, contribute to the immunological tolerance associated with efferocytosis.
Conclusions: Check Please!
Defects at multiple points in the efferocytosis pathway have been reported to result in unchecked inflammation or autoimmunity, and understanding the mechanisms by which dying cells are effectively cleared can pave the way for therapies that target these processes. Although many studies have examined inflammatory disorders in the context of defective attraction, recognition, and physical engulfment of dead cells, we now recognize that aberrant processing of dead cells, potentially via deviations in LAP, can also result in inflammation. Although systemic disorders, such as SLE, have been long linked to defects in dying cell clearance and the autophagy machinery, more definitive roles for these pathways in ‘localized' inflammatory diseases, such as ulcerative colitis, atherosclerosis, neurodegeneration, and rheumatoid arthritis should be described. Moreover, the intricate link between inflammation and cancer raises the question of what the role of efferocytosis is during tumor development, metastasis, and chemotherapy-mediated tumor clearance. Although clearance of dying cells is a common occurrence in healthy and diseased cells, recent studies describe the process of entosis, wherein living cells are engulfed by phagocytes. Although some entotic cells can escape from their engulfment unscathed, most are targeted for destruction by LAP.76 Entosis events are common in human cancers, but their role remains unclear.76 The mechanisms by which entosis occurs, and its similarity to efferocytosis, implies that the burden that lays before the phagocytic system is a daunting one.
Glossary
- ATG
autophagy-related gene
- DAMP
danger associated molecular pattern
- IC
immune complex
- IRF
interferon regulatory factor
- LAP
LC3-associated phagocytosis
- LC3
microtubule-associated protein 1A/1B-light chain 3
- LXR
liver X receptor
- PI(3)P
phosphatidylinositol 3-phosphate
- POS
photoreceptor outer segment
- PPAR
peroxisome proliferator-activated receptor
- PtdSer or PS
phosphatidylserine
- ROS
reactive oxygen species
- RPE
retinal pigment epithelium
- SLE
systemic lupus erythematosus
- TAM
Tyro-3, Axl, and Mer
- TLR
Toll-like receptor
The authors declare no conflict of interest.
Footnotes
Edited by G Kroemer
References
- Green DR. Means To An End: Apoptosis And Other Cell Death Mechanisms. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2011, p 220. [Google Scholar]
- Kinchen JM. A model to die for: signaling to apoptotic cell removal in worm, fly and mouse. Apoptosis 2010; 15: 998–1006. [DOI] [PubMed] [Google Scholar]
- Penaloza C, Lin L, Lockshin RA, Zakeri Z. Cell death in development: shaping the embryo. Histochem Cell Biol 2006; 126: 149–158. [DOI] [PubMed] [Google Scholar]
- McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 2013; 5: a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev 2011; 243: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. Prix fixe: efferocytosis as a four-course meal. Curr Top Microbiol Immunol 2015: 1–36. [DOI] [PMC free article] [PubMed]
- Arandjelovic S, Ravichandran KS. Phagocytosis of apoptotic cells in homeostasis. Nat Immunol 2015; 16: 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini L, Pagliara P, Carla EC. Phagocytosis of apoptotic cells by liver: a morphological study. Microsc Res Tech 2002; 57: 530–540. [DOI] [PubMed] [Google Scholar]
- Bilimoria PM, Stevens B. Microglia function during brain development: new insights from animal models. Brain Res 2015; 1617: 7–17. [DOI] [PubMed] [Google Scholar]
- Juncadella IJ, Kadl A, Sharma AK, Shim YM, Hochreiter-Hufford A, Borish L et al. Apoptotic cell clearance by bronchial epithelial cells critically influences airway inflammation. Nature 2013; 493: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Zheng S, Park D, Woodson RI, Reardon MA, Juncadella IJ et al. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature 2010; 467: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH et al. Noncanonical autophagy promotes the visual cycle. Cell 2013; 154: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C, Wesselborg S, Herrmann M, Lauber K. Dangerous attraction: phagocyte recruitment and danger signals of apoptotic and necrotic cells. Apoptosis 2010; 15: 1007–1028. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J Exp Med 2010; 207: 1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SP, Dransfield I, Rossi AG. Phagocytosis of apoptotic cells. Methods 2008; 44: 280–285. [DOI] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell 2010; 140: 619–630. [DOI] [PubMed] [Google Scholar]
- Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol 2014; 14: 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 2009; 461: 282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol 2000; 2: 131–136. [DOI] [PubMed] [Google Scholar]
- Hoeppner DJ, Hengartner MO, Schnabel R. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 2001; 412: 202–206. [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb Perspect Biol 2013; 5: a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER et al. Pannexin 1 channels mediate 'find-me' signal release and membrane permeability during apoptosis. Nature 2010; 467: 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman LA, Ford CA, Pasikowska M, Pound JD, Wilkinson SJ, Dumitriu IE et al. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood 2008; 112: 5026–5036. [DOI] [PubMed] [Google Scholar]
- Peter C, Waibel M, Radu CG, Yang LV, Witte ON, Schulze-Osthoff K et al. Migration to apoptotic ‘find-me' signals is mediated via the phagocyte receptor G2A. J Biol Chem 2008; 283: 5296–5305. [DOI] [PubMed] [Google Scholar]
- Gude DR, Alvarez SE, Paugh SW, Mitra P, Yu J, Griffiths R et al. Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a ‘come-and-get-me' signal. FASEB J 2008; 22: 2629–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, Pulskens WP, Sadler JJ, Butter LM, Teske GJ, Ulland TK et al. Necrotic cells trigger a sterile inflammatory response through the Nlrp3 inflammasome. Proc Natl Acad Sci USA 2009; 106: 20388–20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazou I, Mackenzie KJ, Duffin R, Rossi AG, Gregory CD. Inhibition of eosinophil migration by lactoferrin. Immunol Cell Biol 2010; 88: 220–223. [DOI] [PubMed] [Google Scholar]
- Bournazou I, Pound JD, Duffin R, Bournazos S, Melville LA, Brown SB et al. Apoptotic human cells inhibit migration of granulocytes via release of lactoferrin. J Clin Invest 2009; 119: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H, Chen CJ, Ontiveros F, Rock KL. Uric acid promotes an acute inflammatory response to sterile cell death in mice. J Clin Invest 2010; 120: 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miyasaka K, Aozasa K, Koike M, Uchiyama Y et al. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 2004; 304: 1147–1150. [DOI] [PubMed] [Google Scholar]
- Toda S, Hanayama R, Nagata S. Two-step engulfment of apoptotic cells. Mol Cell Biol 2012; 32: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysko DV, Vandenabeele P. Clearance of dead cells: mechanisms, immune responses and implication in the development of diseases. Apoptosis 2010; 15: 995–997. [DOI] [PubMed] [Google Scholar]
- Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: a matter of life and death. Ann Rev Physiol 2003; 65: 701–734. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 2013; 341: 403–406. [DOI] [PubMed] [Google Scholar]
- Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 2014; 344: 1164–1168. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzanet R, Sanjuan MA, Wu HY, Quintana FJ, Xiao S, Anderson AC et al. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proc Natl Acad Sci USA 2010; 107: 8706–8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007; 450: 430–434. [DOI] [PubMed] [Google Scholar]
- Park SY, Jung MY, Kim HJ, Lee SJ, Kim SY, Lee BH et al. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ 2008; 15: 192–201. [DOI] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002; 417: 182–187. [DOI] [PubMed] [Google Scholar]
- Ishimoto Y, Ohashi K, Mizuno K, Nakano T. Promotion of the uptake of PS liposomes and apoptotic cells by a product of growth arrest-specific gene, gas6. J Biochem 2000; 127: 411–417. [DOI] [PubMed] [Google Scholar]
- Zizzo G, Hilliard BA, Monestier M, Cohen PL. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol 2012; 189: 3508–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CD, Devitt A, Moffatt O. Roles of ICAM-3 and CD14 in the recognition and phagocytosis of apoptotic cells by macrophages. Biochem Soc Trans 1998; 26: 644–649. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE et al. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 2005; 123: 321–334. [DOI] [PubMed] [Google Scholar]
- Gordon S. Macrophage-restricted molecules: role in differentiation and activation. Immunol Lett 1999; 65: 5–8. [DOI] [PubMed] [Google Scholar]
- Ezekowitz RA, Sastry K, Bailly P, Warner A. Molecular characterization of the human macrophage mannose receptor: demonstration of multiple carbohydrate recognition-like domains and phagocytosis of yeasts in Cos-1 cells. J Exp Med 1990; 172: 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, Suzuki J, Nagata S. Constitutive exposure of phosphatidylserine on viable cells. Proc Natl Acad Sci USA 2011; 108: 19246–19251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Eijnde SM, van den Hoff MJ, Reutelingsperger CP, van Heerde WL, Henfling ME, Vermeij-Keers C et al. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J Cell Sci 2001; 114(Pt 20): 3631–3642. [DOI] [PubMed] [Google Scholar]
- Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science 2000; 288: 2051–2054. [DOI] [PubMed] [Google Scholar]
- Elward K, Griffiths M, Mizuno M, Harris CL, Neal JW, Morgan BP et al. CD46 plays a key role in tailoring innate immune recognition of apoptotic and necrotic cells. J Biol Chem 2005; 280: 36342–36354. [DOI] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 2009; 7: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy-Hulbert A, Smith AM, Tissire H, Barry M, Crowley D, Bronson RT et al. Ulcerative colitis and autoimmunity induced by loss of myeloid alphav integrins. Proc Natl Acad Sci USA 2007; 104: 15823–15828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PL, Caricchio R, Abraham V, Camenisch TD, Jennette JC, Roubey RA et al. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J Exp Med 2002; 196: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Guthrie L, Henson PM. Differential effects of apoptotic versus lysed cells on macrophage production of cytokines: role of proteases. J Immunol 2001; 166: 6847–6854. [DOI] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA 2011; 108: 17396–17401. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Camins A, Pallas M, Silvestre JS. Apoptotic mechanisms involved in neurodegenerative diseases: experimental and therapeutic approaches. Methods Find Exp Clin Pharmacol 2008; 30: 43–65. [DOI] [PubMed] [Google Scholar]
- Lee GY, Kim JH, Oh GT, Lee BH, Kwon IC, Kim IS. Molecular targeting of atherosclerotic plaques by a stabilin-2-specific peptide ligand. J Control Release 2011; 155: 211–217. [DOI] [PubMed] [Google Scholar]
- Sokolowski JD, Mandell JW. Phagocytic clearance in neurodegeneration. Am J Pathol 2011; 178: 1416–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert ML, Kim JI, Birge RB. alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol 2000; 2: 899–905. [DOI] [PubMed] [Google Scholar]
- Wu Y, Singh S, Georgescu MM, Birge RB. A role for Mer tyrosine kinase in alphavbeta5 integrin-mediated phagocytosis of apoptotic cells. J Cell Sci 2005; 118(Pt 3): 539–553. [DOI] [PubMed] [Google Scholar]
- Park SY, Kang KB, Thapa N, Kim SY, Lee SJ, Kim IS. Requirement of adaptor protein GULP during stabilin-2-mediated cell corpse engulfment. J Biol Chem 2008; 283: 10593–10600. [DOI] [PubMed] [Google Scholar]
- Park SY, Kim SY, Kang KB, Kim IS. Adaptor protein GULP is involved in stabilin-1-mediated phagocytosis. Biochem Bophys Res Communi 2010; 398: 467–472. [DOI] [PubMed] [Google Scholar]
- Su HP, Nakada-Tsukui K, Tosello-Trampont AC, Li Y, Bu G, Henson PM et al. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J Biol Chem 2002; 277: 11772–11779. [DOI] [PubMed] [Google Scholar]
- Park D, Hochreiter-Hufford A, Ravichandran KS. The phosphatidylserine receptor TIM-4 does not mediate direct signaling. Curr Biol 2009; 19: 346–351. [DOI] [PubMed] [Google Scholar]
- Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Revi Mol Cell Biol 2003; 4: 446–456. [DOI] [PubMed] [Google Scholar]
- Olazabal IM, Caron E, May RC, Schilling K, Knecht DA, Machesky LM. Rho-kinase and myosin-II control phagocytic cup formation during CR, but not FcgammaR, phagocytosis. Curr Biol 2002; 12: 1413–1418. [DOI] [PubMed] [Google Scholar]
- Nakaya M, Tanaka M, Okabe Y, Hanayama R, Nagata S. Opposite effects of rho family GTPases on engulfment of apoptotic cells by macrophages. J Biol Chem 2006; 281: 8836–8842. [DOI] [PubMed] [Google Scholar]
- Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol 2002; 4: 574–582. [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J 1998; 17: 6932–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano F, Montcourrier P, Chavrier P. Membrane recruitment of Rac1 triggers phagocytosis. J Cell Sci 2000; 113(Pt 17): 2955–2961. [DOI] [PubMed] [Google Scholar]
- Leverrier Y, Lorenzi R, Blundell MP, Brickell P, Kinnon C, Ridley AJ et al. Cutting edge: the Wiskott-Aldrich syndrome protein is required for efficient phagocytosis of apoptotic cells. J Immunol 2001; 166: 4831–4834. [DOI] [PubMed] [Google Scholar]
- Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 2006; 443: 998–1002. [DOI] [PubMed] [Google Scholar]
- Martinez J, Verbist K, Wang R, Green DR. The relationship between metabolism and the autophagy machinery during the innate immune response. Cell Metab 2013; 17: 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S et al. Molecular characterization of LC3-associated phagocytosis (LAP) reveals distinct roles for Rubicon, NOX2, and autophagy proteins. Nat Cell Biol 2015; 17: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 2012; 37: 986–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 2011; 13: 1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007; 450: 1253–1257. [DOI] [PubMed] [Google Scholar]
- Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 2015; 17: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct 2002; 27: 421–429. [DOI] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447: 1121–1125. [DOI] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010; 6: 506–522. [DOI] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 2007; 7: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19: 5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11: 468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, An L, Ye Y, Wu J, Zou Y, He L et al. Essential role for UVRAG in autophagy and maintenance of cardiac function. Cardiovasc Res 2014; 101: 48–56. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11: 385–396. [DOI] [PubMed] [Google Scholar]
- Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci USA 2010; 107: 19338–19343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Lee JS, Rodgers M, Min CK, Lee JY, Kim HJ et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe 2012; 11: 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem 2011; 286: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama T, Nakakita J, Nakamura T, Kobayashi T, Kobayashi T, Son J et al. Cooperation of p40(phox) with p47(phox) for Nox2-based NADPH oxidase activation during Fcgamma receptor (FcgammaR)-mediated phagocytosis: mechanism for acquisition of p40(phox) phosphatidylinositol 3-phosphate (PI(3)P) binding. J Biol Chem 2011; 286: 40693–40705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci USA 2009; 106: 6226–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CZ, Ravichandran KS. Metabolic connections during apoptotic cell engulfment. Cell 2011; 147: 1442–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity 2004; 21: 643–653. [DOI] [PubMed] [Google Scholar]
- A-Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 2009; 31: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszer T, Menendez-Gutierrez MP, Lefterova MI, Alameda D, Nunez V, Lazar MA et al. Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator-activated receptor gamma or retinoid X receptor alpha deficiency. J Immunol 2011; 186: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan L, Odegaard JI, Morel CR, Heredia JE, Mwangi JW, Ricardo-Gonzalez RR et al. PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat Med 2009; 15: 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss RS, Elliott MR, Ma Z, Marcel YL, Ravichandran KS. Apoptotic cells induce a phosphatidylserine-dependent homeostatic response from phagocytes. Curr Biol 2006; 16: 2252–2258. [DOI] [PubMed] [Google Scholar]
- Huynh ML, Fadok VA, Henson PM. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-beta1 secretion and the resolution of inflammation. J Clin Invest 2002; 109: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol 2009; 21: 871–877. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 2010; 11: 621–632. [DOI] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell 2011; 44: 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM et al. A unified model for apical caspase activation. Mol Cell 2003; 11: 529–541. [DOI] [PubMed] [Google Scholar]
- Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol 2009; 9: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys acta 2013; 1833: 3448–3459. [DOI] [PubMed] [Google Scholar]
- Leist M, Jaattela M. Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol 2001; 2: 589–598. [DOI] [PubMed] [Google Scholar]
- Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol 2008; 8: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinlich R, Green DR. The two faces of receptor interacting protein kinase-1. Mol Cell 2014; 56: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidere N, Su HC, Lenardo MJ. Genetic disorders of programmed cell death in the immune system. Ann Revi Immunol 2006; 24: 321–352. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K. RIPK1 and RIPK3: critical regulators of inflammation and cell death. Trends Cell Biol 2015; 25: 347–353. [DOI] [PubMed] [Google Scholar]
- Rodriguez DA, Weinlich R, Brown S, Guy C, Fitzgerald P, Dillon CP et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ 2015; 23: 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock CN, Cookson BT. Burning down the house: cellular actions during pyroptosis. PLoS Pathog 2013; 9: e1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev 2015; 265: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute C, Torre I, Perez-Cerda F, Perez-Samartin A, Alberdi E, Etxebarria E et al. P2X(7) receptor blockade prevents ATP excitotoxicity in oligodendrocytes and ameliorates experimental autoimmune encephalomyelitis. J Neurosci 2007; 27: 9525–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz SE, Gonzalez-Fernandez E, Ventura JC, Perez-Samartin A, Tarassishin L, Negoro H et al. Contribution of pannexin1 to experimental autoimmune encephalomyelitis. PloS ONE 2013; 8: e66657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Kang TC. The P2X7 receptor-pannexin-1 complex decreases muscarinic acetylcholine receptor-mediated seizure susceptibility in mice. J Clin Invest 2011; 121: 2037–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris CS, Wu L, Acharya S, Arac A, Blaho VA, Huang Y et al. Defective sphingosine 1-phosphate receptor 1 (S1P1) phosphorylation exacerbates TH17-mediated autoimmune neuroinflammation. Nat Immunol 2013; 14: 1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Kobayashi T, Kamata K. Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr Med Chem 2007; 14: 3209–3220. [DOI] [PubMed] [Google Scholar]
- Le LQ, Kabarowski JH, Weng Z, Satterthwaite AB, Harvill ET, Jensen ER et al. Mice lacking the orphan G protein-coupled receptor G2A develop a late-onset autoimmune syndrome. Immunity 2001; 14: 561–571. [DOI] [PubMed] [Google Scholar]
- Bolick DT, Skaflen MD, Johnson LE, Kwon SC, Howatt D, Daugherty A et al. G2A deficiency in mice promotes macrophage activation and atherosclerosis. Circ Res 2009; 104: 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagkalis A, Wallace C, Hing B, Liversidge J, Crane IJ. CX3CR1-deficiency is associated with increased severity of disease in experimental autoimmune uveitis. Immunology 2009; 128: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme SA, Lio FM, Maciejewski-Lenoir D, Bacon KB, Conlon PJ. The chemokine fractalkine inhibits Fas-mediated cell death of brain microglia. J Immunol 2000; 165: 397–403. [DOI] [PubMed] [Google Scholar]
- Martin S, Rieckmann P, Melchers I, Wagner R, Bertrams J, Voskuyl AE et al. Circulating forms of ICAM-3 (cICAM-3). Elevated levels in autoimmune diseases and lack of association with cICAM-1. J Immunol 1995; 154: 1951–1955. [PubMed] [Google Scholar]
- Sontheimer R, Racila D, Racila E, Eggleton P, Donnelly S. Calreticulin's role(s) in autoimmune disorders. In: Eggleton P, Michalak M (eds). Calreticulin Molecular Biology Intelligence Unit. Chap 17. Springer: New York City, NY, USA, 2003, pp 180–192. [Google Scholar]
- Walport MJ. Complement. Second of two parts. N Engl J Med 2001; 344: 1140–1144. [DOI] [PubMed] [Google Scholar]
- Xiao S, Brooks CR, Zhu C, Wu C, Sweere JM, Petecka S et al. Defect in regulatory B-cell function and development of systemic autoimmunity in T-cell Ig mucin 1 (Tim-1) mucin domain-mutant mice. Proc Natl Acad Sci USA 2012; 109: 12105–12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire JJ, Umetsu SE, Akbari O, Potter M, Kuchroo VK, Barsh GS et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol 2001; 2: 1109–1116. [DOI] [PubMed] [Google Scholar]
- Zhu D, Li C, Swanson AM, Villalba RM, Guo J, Zhang Z et al. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J Clin Invest 2015; 125: 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber EE, Gallo EM, Fontana SC, Davis EC, Wigley FM, Huso DL et al. Integrin-modulating therapy prevents fibrosis and autoimmunity in mouse models of scleroderma. Nature 2013; 503: 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet 2000; 26: 270–271. [DOI] [PubMed] [Google Scholar]
- Thorp E, Cui D, Schrijvers DM, Kuriakose G, Tabas I. Mertk receptor mutation reduces efferocytosis efficiency and promotes apoptotic cell accumulation and plaque necrosis in atherosclerotic lesions of apoe-/- mice. Arterioscler Thromb Vasc Biol 2008; 28: 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Oufella H, Kinugawa K, Zoll J, Simon T, Boddaert J, Heeneman S et al. Lactadherin deficiency leads to apoptotic cell accumulation and accelerated atherosclerosis in mice. Circulation 2007; 115: 2168–2177. [DOI] [PubMed] [Google Scholar]
- Engesser L, Broekmans AW, Briet E, Brommer EJ, Bertina RM. Hereditary protein S deficiency: clinical manifestations. Ann Intern Med 1987; 106: 677–682. [DOI] [PubMed] [Google Scholar]
- Suh CH, Hilliard B, Li S, Merrill JT, Cohen PL. TAM receptor ligands in lupus: protein S but not Gas6 levels reflect disease activity in systemic lupus erythematosus. Arthritis Res Ther 2010; 12: R146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Choi SC, Murakami Y, Allen J, Morse HC 3rd, Qi CF et al. p85alpha recruitment by the CD300f phosphatidylserine receptor mediates apoptotic cell clearance required for autoimmunity suppression. Nat Commun 2014; 5: 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Elliott MR, Chen Y, Walsh JT, Klibanov AL, Ravichandran KS et al. Phagocytic activity of neuronal progenitors regulates adult neurogenesis. Nat Cell Biol 2011; 13: 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanematsu F, Hirashima M, Laurin M, Takii R, Nishikimi A, Kitajima K et al. DOCK180 is a Rac activator that regulates cardiovascular development by acting downstream of CXCR4. Circ Res 2010; 107: 1102–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M, Fradet N, Blangy A, Hall A, Vuori K, Cote JF. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci USA 2008; 105: 15446–15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Tajima M, Kosako H, Nakaya T, Hashimoto A, Watari K et al. GRK6 deficiency in mice causes autoimmune disease due to impaired apoptotic cell clearance. Nat Commun 2013; 4: 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu JR, Dontje W, Krausz S, de Launay D, van Hennik PB, van Stalborch AM et al. A Rac1 inhibitory peptide suppresses antibody production and paw swelling in the murine collagen-induced arthritis model of rheumatoid arthritis. Arthritis Res Ther 2010; 12: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wagoner G, Douglas JC, Drew PD. Liver X receptor agonist regulation of Th17 lymphocyte function in autoimmunity. J Leuk Biol 2009; 86: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Qin X, Wu L, Zhang Y, Sheng X, Yu Q et al. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J Clin Invest 2011; 121: 658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Kidani Y, A-Gonzalez N, Phung T, Ito A, Rong X et al. Coordinate regulation of neutrophil homeostasis by liver X receptors in mice. J Clin Invest 2012; 122: 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JY, Nam JY, Kim HA, Park YB, Bae SC, Suh CH. Liver X receptors alpha gene (NR1H3) promoter polymorphisms are associated with systemic lupus erythematosus in Koreans. Arthritis Res Ther 2014; 16: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zheng S, Qiu Y, Yang Y, Wang C, Yang P et al. Activation of liver X receptor alleviates ocular inflammation in experimental autoimmune uveitis. Invest Ophthalmol Vis Sci 2014; 55: 2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Liang Y, Broderick CL, Oldham BA, Beyer TP, Schmidt RJ et al. Antidiabetic action of a liver x receptor agonist mediated by inhibition of hepatic gluconeogenesis. J Biol Chem 2003; 278: 1131–1136. [DOI] [PubMed] [Google Scholar]
- Joseph SB, McKilligin E, Pei L, Watson MA, Collins AR, Laffitte BA et al. Synthetic LXR ligand inhibits the development of atherosclerosis in mice. Proc Natl Acad Sci USA 2002; 99: 7604–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med 2001; 7: 48–52. [DOI] [PubMed] [Google Scholar]
- Babaev VR, Yancey PG, Ryzhov SV, Kon V, Breyer MD, Magnuson MA et al. Conditional knockout of macrophage PPARgamma increases atherosclerosis in C57BL/6 and low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol 2005; 25: 1647–1653. [DOI] [PubMed] [Google Scholar]
- Vasheghani F, Monemdjou R, Fahmi H, Zhang Y, Perez G, Blati M et al. Adult cartilage-specific peroxisome proliferator-activated receptor gamma knockout mice exhibit the spontaneous osteoarthritis phenotype. Am J Pathol 2013; 182: 1099–1106. [DOI] [PubMed] [Google Scholar]
- Christiansen-Weber TA, Voland JR, Wu Y, Ngo K, Roland BL, Nguyen S et al. Functional loss of ABCA1 in mice causes severe placental malformation, aberrant lipid distribution, and kidney glomerulonephritis as well as high-density lipoprotein cholesterol deficiency. Am J Pathol 2000; 157: 1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota K, Nishiyama T, Mishima K, Inoue H, Doi T, Hattori Y et al. The role of fractalkine as an accelerating factor on the autoimmune exocrinopathy in mice. Invest Ophthalmol Vis Sci 2009; 50: 4753–4760. [DOI] [PubMed] [Google Scholar]
- Rimaniol AC, Till SJ, Garcia G, Capel F, Godot V, Balabanian K et al. The CX3C chemokine fractalkine in allergic asthma and rhinitis. J Allergy Clin Immunol 2003; 112: 1139–1146. [DOI] [PubMed] [Google Scholar]
- Nanki T, Imai T, Nagasaka K, Urasaki Y, Nonomura Y, Taniguchi K et al. Migration of CX3CR1-positive T cells producing type 1 cytokines and cytotoxic molecules into the synovium of patients with rheumatoid arthritis. Arthritis Rheum 2002; 46: 2878–2883. [DOI] [PubMed] [Google Scholar]
- Baker OJ, Camden JM, Rome DE, Seye CI, Weisman GA. P2Y2 nucleotide receptor activation up-regulates vascular cell adhesion molecule-1 [corrected] expression and enhances lymphocyte adherence to a human submandibular gland cell line. Mol Immunol 2008; 45: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relvas LJ, Makhoul M, Dewispelaere R, Caspers L, Communi D, Boeynaems JM et al. P2Y2R deficiency attenuates experimental autoimmune uveitis development. PLoS ONE 2015; 10: e0116518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya M, Mukhopadhyay S, Paidassi H, Jamil T, Chow C, Kissler S et al. alphav Integrin expression by DCs is required for Th17 cell differentiation and development of experimental autoimmune encephalomyelitis in mice. J Clin Invest 2010; 120: 4445–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaiah R, Moudgil KD. Heat-shock proteins can promote as well as regulate autoimmunity. Autoimmun Rev 2009; 8: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SS, Wu ZY, Zhang HP, Furtado G, Chen X, Yan SF et al. Suppression of experimental autoimmune encephalomyelitis by selective blockade of encephalitogenic T-cell infiltration of the central nervous system. Nat Med 2003; 9: 287–293. [DOI] [PubMed] [Google Scholar]
- Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell 1999; 97: 889–901. [DOI] [PubMed] [Google Scholar]
- Angelillo-Scherrer A, de Frutos P, Aparicio C, Melis E, Savi P, Lupu F et al. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nat Med 2001; 7: 215–221. [DOI] [PubMed] [Google Scholar]
- Yanagita M, Ishimoto Y, Arai H, Nagai K, Ito T, Nakano T et al. Essential role of Gas6 for glomerular injury in nephrotoxic nephritis. J Clin Invest 2002; 110: 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Nam JY, Jeon JY, An JM, Jung JY, Bae CB et al. Serum growth arrest-specific protein 6 levels are a reliable biomarker of disease activity in systemic lupus erythematosus. J Clin Immunol 2013; 33: 143–150. [DOI] [PubMed] [Google Scholar]
- Holland SM. Chronic granulomatous disease. Hematol Oncol Clin North Am 2013; 27: 89–99, viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimohama S, Tanino H, Kawakami N, Okamura N, Kodama H, Yamaguchi T et al. Activation of NADPH oxidase in Alzheimer's disease brains. Biochem Biophys Res Commun 2000; 273: 5–9. [DOI] [PubMed] [Google Scholar]
- Sorce S, Nuvolone M, Keller A, Falsig J, Varol A, Schwarz P et al. The role of the NADPH oxidase NOX2 in prion pathogenesis. PLoS Pathog 2014; 10: e1004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assoum M, Salih MA, Drouot N, Hnia K, Martelli A, Koenig M. The Salih ataxia mutation impairs Rubicon endosomal localization. Cerebellum 2013; 12: 835–840. [DOI] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999; 59: 59–65. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402: 672–676. [DOI] [PubMed] [Google Scholar]
- Jiang ZF, Shao LJ, Wang WM, Yan XB, Liu RY. Decreased expression of Beclin-1 and LC3 in human lung cancer. Mol Biol Rep 2012; 39: 259–267. [DOI] [PubMed] [Google Scholar]
- Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol 2010; 12: 863–875. [DOI] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 2008; 118: 2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Goronzy JJ, Weyand CM. Autophagy in autoimmune disease. J Mol Med (Berl) 2015; 93: 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Zhao X, Fang C, Tang W, Huang K, Wang L et al. Investigation of variants in the promoter region of PIK3C3 in schizophrenia. Neurosci Lett 2008; 437: 42–44. [DOI] [PubMed] [Google Scholar]
- Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol 2008; 39: 1059–1063. [DOI] [PubMed] [Google Scholar]
- Jeong TJ, Shin MK, Uhm YK, Kim HJ, Chung JH, Lee MH. Association of UVRAG polymorphisms with susceptibility to non-segmental vitiligo in a Korean sample. Exp Dermatol 2010; 19: e323–e325. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006; 8: 688–699. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Gupta J, Jyothula SS, Butsch Kovacic M, Biagini Myers JM, Patterson TL et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS ONE 2012; 7: e33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XJ, Lu XL, Lv JC, Yang HZ, Qin LX, Zhao MH et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 2011; 70: 1330–1337. [DOI] [PubMed] [Google Scholar]
- Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med 2011; 208: 2625–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Bustos V, Flajolet M, Greengard P. A small-molecule enhancer of autophagy decreases levels of Abeta and APP-CTF via Atg5-dependent autophagy pathway. FASEB J 2011; 25: 1934–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009; 41: 1234–1237. [DOI] [PubMed] [Google Scholar]
- Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab 2012; 15: 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008; 456: 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne J, Gustafsson P, Jin H, Maegdefessel L, Hultenby K, Wernerson A et al. ATG16L1 expression in carotid atherosclerotic plaques is associated with plaque vulnerability. Arterioscler Thromb Vasc Biol 2015; 35: 1226–1235. [DOI] [PubMed] [Google Scholar]
- Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann Rheum Dis 2015; 74: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Parillon X, Zeng S, Han S, Eissa NT. Deficiency of autophagy in dendritic cells protects against experimental autoimmune encephalomyelitis. J Biol Chem 2014; 289: 26525–26532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet 2009; 41: 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM et al. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest 2013; 123: 5284–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till A, Subramani S. A balancing act for autophagin. J Clin Invest 2010; 120: 2273–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J, Motohashi N, Kino Y, Ishida T, Yagishita S, Jinnai K et al. LC3, an autophagosome marker, is expressed on oligodendrocytes in Nasu-Hakola disease brains. Orphanet J Rare Dis 2014; 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masini M, Bugliani M, Lupi R, del Guerra S, Boggi U, Filipponi F et al. Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia 2009; 52: 1083–1086. [DOI] [PubMed] [Google Scholar]
- Henckaerts L, Cleynen I, Brinar M, John JM, Van Steen K, Rutgeerts P et al. Genetic variation in the autophagy gene ULK1 and risk of Crohn's disease. Inflamm Bowel Dis 2011; 17: 1392–1397. [DOI] [PubMed] [Google Scholar]
- Wei H, Gan B, Wu X, Guan JL. Inactivation of FIP200 leads to inflammatory skin disorder, but not tumorigenesis, in conditional knock-out mouse models. J Biol Chem 2009; 284: 6004–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci 2010; 13: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet 2013; 45: 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet 2007; 39: 830–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Balschun T, Sina C, Ellinghaus D, Hasler R, Mayr G et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL). Nat Genet 2010; 42: 292–294. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 2013; 45: 445–449, 449e1. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 2004; 304: 1158–1160. [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010; 12: 119–131. [DOI] [PubMed] [Google Scholar]