Abstract
The turnover and clearance of cells is an essential process that is part of many physiological and pathological processes. Improper or deficient clearance of apoptotic cells can lead to excessive inflammation and autoimmune disease. The steps involved in cell clearance include: migration of the phagocyte toward the proximity of the dying cells, specific recognition and internalization of the dying cell, and degradation of the corpse. The ability of phagocytes to recognize and react to dying cells to perform efficient and immunologically silent engulfment has been well-characterized in vitro and in vivo. However, how apoptotic cells themselves initiate the corpse removal and also influence the cells within the neighboring environment during clearance was less understood. Recent exciting observations suggest that apoptotic cells can attract phagocytes through the regulated release of ‘find-me' signals. More recent studies also suggest that these find-me signals can have additional roles outside of phagocyte attraction to help orchestrate engulfment. This review will discuss our current understanding of the different find-me signals released by apoptotic cells, how they may be relevant in vivo, and their additional roles in facilitating engulfment.
Facts:
Apoptotic cells can induce phagocyte migration through the release of several ‘find-me' signals (soluble and membrane bound).
Find-me signals are released in an apoptosis-dependent manner.
These mediators are released in a regulated manner, as the membrane remains intact (in contrast to necrotic cells, which spill their intracellular contents).
Specific receptors on phagocytes mediate their migration to the find-me signals.
Open Questions:
What is the cell-type specificity of the different find-me signals?
What is the in vivo relevance of these signals during cell clearance in different physiological or pathological contexts?
At what concentrations and distances may these find-me signals operate in vivo?
Can metabolites from the breakdown of find-me signals continue to influence cell clearance?
How important are the additional actions of these released factors to engulfment and the microenvironment around dying cells?
Billions of cells are known to turnover in our body on a daily basis as part of development, homeostasis, and pathogenic conditions.1 For example, in the thymus and bone marrow, excess hematopoetic cells that are not considered to move further in development constantly undergo cell death. Whether dying cells come from physiological or pathological consequences, efficient clearance of these cells by professional (macrophages and dendritic cells) or non-professional (epithelial cells and fibroblasts) phagocytes must occur. In addition, the clearance of dying cells is actively immunosuppressive,2, 3 protecting the body from a break in tolerance to self-antigens. Failure to sustain efficient clearance results in secondary necrosis which can lead to exacerbated inflammation and autoimmune diseases.4 For these reasons both the dying cell and the phagocyte communicate and engage each other to ensure successful engulfment.
One way in which cellular turnover occurs is through apoptosis, a type of programmed cell death. Although different types of cell death exist,5 the majority of what is known about cell clearance involves the engulfment of caspase-driven apoptotic cells; therefore this review will focus on engulfment in the context of caspase-dependent apoptosis. Apoptosis can be initiated by intrinsic or extrinsic pathways, such as genotoxic stress or receptor-mediated death, respectively. The activation of caspases during apoptosis leads to many cellular changes such as DNA fragmentation, plasma membrane alterations, and the regulated release of cellular contents.6 It is now recognized that many of these processes, which an apoptotic cell undergoes, can help the overall process of cell clearance.
Much work has been done to understand how phagocytes mediate engulfment, as well as influence their immediate environment during clearance. However, before phagocytosis can occur, a key step is for the phagocyte to ‘find' the dying cells among the sea of living cells. How this is achieved is a fascinating question that is just beginning to be delineated. Recent studies suggest that apoptotic cells can release factors, termed ‘find-me' signals that advertise their presence to facilitate the migration of phagocytes toward the area of cell death.7 This review will focus on the find-me signals and their effects on phagocyte migration. Furthermore, we will examine how these mediators may influence cell engulfment and the environment around the dying cells to properly orchestrate corpse clearance.
Steps involved in clearance
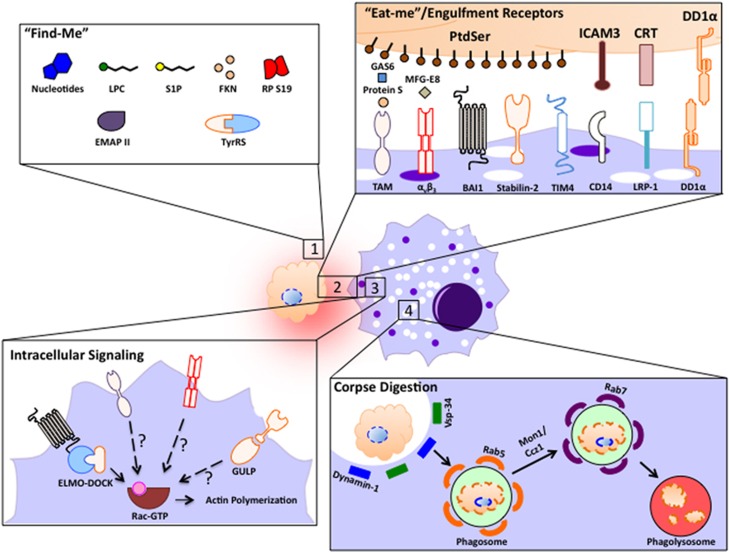
Proper apoptotic cell clearance occurs through several steps: migration toward the dying cell, recognition/binding to the apoptotic target, phagocytosis, and digestion of the corpse (Figure 1). Apoptotic cells first release find-me signals to promote phagocyte migration to the proximity of cells undergoing death. The phagocytes then specifically recognize the dying cells via ‘eat-me' signals, the best studied being phosphatidylserine (PtdSer). PtdSer is a phospholipid found on the inner leaflet of the plasma membrane, but flips to the outer membrane upon apoptosis induction, through the activation and deactivation of scramblases and flippases, respectively.8 Other less-characterized recognition signals as well as the respective receptors they engage on phagocytes are depicted in Figure 2.9, 10, 11 Factors such as the cell type undergoing death, the pathological or physiological stimulus, or the duration of this fatal process can affect the landscape of eat-me signals exposed.
Figure 1.
The following processes were involved in apoptotic cell engulfment: (i) apoptotic cells are capable of releasing several different ‘find-me' signals to attract phagocytes toward dying cells. (ii) Phagocytes express an array of different receptors that recognize ligands on apoptotic cells. This can occur through direct binding to the dying cell or through soluble intermediates called bridging molecules. (iii) Engulfment receptors that are bound to PtdSer can initiate intracellular signaling leading to Rac1 activation and cytoskeletal rearrangement. The specific mechanism by which signaling is mediated downstream of each receptor is not fully defined. (iv) Once internalized, phagosome maturation to the phagolysosome through recruitment of Vps34/Dynamin1, as well as Rab5, Mon1/Ccz1, and Rab7 proteins leads to eventual corpse degradation
Figure 2.
‘Find-me' signals. The different find-me signals released from apoptotic cells, their known or putative mechanism of release, and the possible receptors on phagocytes that can regulate chemotaxis
Different receptors and soluble molecules have been linked to PtdSer recognition and this recognition occurs either directly or indirectly. Direct recognition can occur via brain-specific angiogenesis inhibitor 1 (BAI1),12 Stabilin-2,13 and members of the T-cell immunoglobulin mucin domain;14, 15, 16 alternatively, bridging molecules such as GAS6/Protein S and milk-fat globule EGF factor 8 can bind to the PtdSer exposed on the apoptotic cells, and in turn be recognized by members of the Tyro-Axl-Mer family and αvβ3 integrin, respectively, on the phagocyte.17, 18 Although some of these receptors only serve to tether the apoptotic cells, others can initiate intracellular signaling to induce phagocytosis. Receptors such as BAI1 can signal intracellularly through the ELMO1-DOCK180-Rac signaling module to stimulate cytoskeletal rearrangement and engulfment.12 Stabilin-2 has been reported to require the adapter protein GULP and thymosinβ4 to engulf apoptotic cells.19, 20 Once internalized, phagolysosome maturation occurs through the action of several Rab-family GTPases eventually leading to the degradation of the ingested cell corpse.21
‘Find-me' signals
Both non-professional phagocytes and resident professional phagocytes can clear dying cells under homeostatic conditions. Although neighboring cells may not need to ‘migrate' toward an apoptotic cell to engulf, motile phagocytes such as a resident macrophage likely do. During situations where a large amount of apoptosis might be occurring, such as in an inflammatory setting, additional professional-phagocyte migration and their inherent increased-engulfment capability is necessary to cope with the death overload. Moreover, in many cases, healthy neighboring cells (e.g., thymocytes) are unable to clear their dying brethren, and therefore, migration of resident phagocytes within a tissue is required.
It is now well-established that apoptotic cells release mediators that induce phagocyte attraction to the proximity of the dying cells.22 Interestingly, many of these factors contain several extracellular signaling capabilities, indicating that they may have additional roles beyond simply inducing phagocyte migration. One possibility is that these recruitment/find-me signals also influence the cell clearance microenvironment, or the ability of phagocytes to engulf apoptotic cells. It is also possible that find-me signals not only recruit the right phagocytes but also ‘prepare' the environment for clearance.
Release and recognition
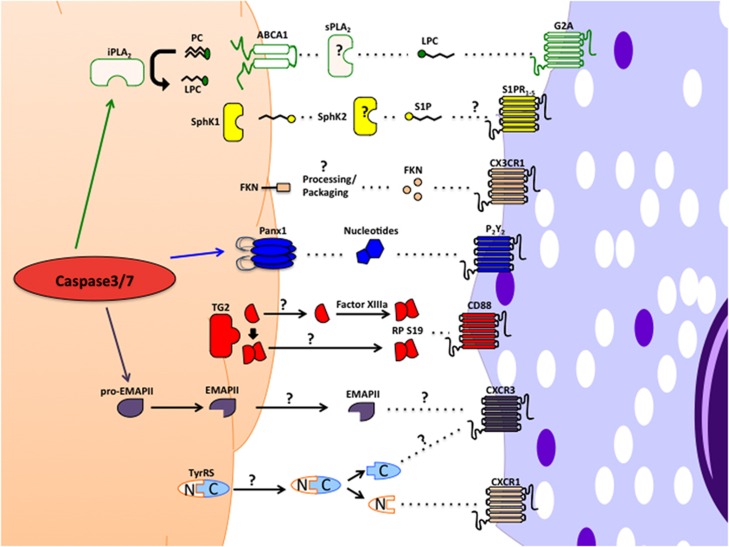
Apoptotic cells can release different mediators22 (Figure 2). Although some of these are better characterized, in-depth understanding of others is still required. The section below describes four find-me signals that are currently better detailed for their role in phagocyte attraction and beyond.
Lysophosphatidylcholine
One of the first identified recruitment signals for apoptotic cell engulfment was lysophosphatidylcholine (LPC).23 Lauber et. al.23 showed that the cell supernatants taken from apoptotic MCF7 cells could induce phagocyte migration. The authors linked soluble LPC as the factor responsible for recruitment. Unlike LPC, other lysophospholipids or metabolic derivatives of LPC were unable to induce phagocyte chemotaxis.24 This release of LPC was not simply leakage of cellular contents as the membrane integrity was intact. LPC release was dependent on caspase-3-mediated activation of calcium-independent phospholipase A2,23 which hydrolizes membrane-lipid phosphatidylcholine. However, it is not known whether LPC generation during apoptosis occurs intracellularly, extracellularly, or both, as the perturbed membrane structure is susceptible to secretory phospholipase A2 activity.25 Later, it was shown that ABCA1 might be important for the release of lysophospholipids during apoptosis, as knockdown of ABCA1 resulted in the reduced migration of macrophages toward apoptotic cell supernatants. However, the authors did not show that this was caused by the specific reduction of LPC in the supernatants.26 This linkage of LPC release to ABCA1 is of interest as among the many evolutionarily conserved genes linked to apoptotic cell clearance, the ABCA1 homolog CED-7 in Caenorhabditis elegans is the one player known to be required in both the apoptotic and engulfing cells to mediate clearance.27
Subsequently, the G-protein-coupled receptor G2A was suggested as the target for LPC. Knockdown of G2A could decrease the migratory capacity of phagocytes to apoptotic cell supernatants.24 However, other phospholipids were also able to neutralize migration to pure LPC in a G2A-dependent manner. It is possible that a balance of different lysophospholipids released from apoptotic cells can affect migration. There is also controversy as to whether LPC is a ligand for the G2A receptor,28 as LPC has been shown to inhibit G2A-mediated signaling, including actin polymerization.29 Furthermore, G2A receptor signaling may also depend on other oxidized fatty acids30 for specific interactions with intracellular G-proteins and GPCRs.31 Thus, the effects of LPC or other lysophospholipids on migration may depend on the specific phagocyte, owing to differential expression of GPCRs and G-proteins in different cell types.
Sphingosine-1-phosphate
Another lysophospholipid that has been shown to function as a find-me signal during apoptosis is sphingosine-1-phosphate (S1P). It had already been shown that apoptotic cells can release S1P,32 but its function as a lipid-attraction signal during cell death was not reported until 2008. Gude et al.33 suggested that sphingosine kinase 1 (SphK1) was upregulated during apoptosis and, in turn, led to the increased secretion of S1P. Concomitantly, purified S1P was able to induce phagocyte chemotaxis. However, it is important to note that it was never directly shown that S1P from the apoptotic supernatants was the chemotactic factor. In addition, the authors showed that there was no increase in extracellular SphK1,33 suggesting that the S1P generation occurred within a cell. However, they did not rule out any actions of SphK2 as it has since been shown to be secreted during apoptosis in a caspase-dependent manner, possibly leading to extracellular generation of S1P.34 Therefore, a complete understanding of S1P release during apoptosis and phagocyte migration remains to be characterized.
Although there are known receptors for S1P (S1PR1–5), the specific GPCR involved in phagocyte migration is unclear. Phagocytes can express several S1P receptors making it difficult to determine the receptor(s) involved. Cell type-specific expression, as well as concentration of S1P may be key factors in determining which receptor mediates phagocyte migration.
Fractalkine/CX3CL1
It has been observed that apoptotic cells can undergo membrane blebbing and release small vesicles during the death process. Truman et al.36 showed that these blebs can participate in monocyte chemotaxis toward apoptotic germinal center B cells.35 Later, it was shown that Fractalkine/CX3CL1 (FKN) associated with apoptotic microparticles was a chemotactic factor.36 This 90-kDa membrane-associated chemokine gets processed to a 60-kDa form during the early stages of apoptosis (presumed to occur via extracellular proteases) and released in PtdSer exposing microparticles. Although the precise mechanism of release for the microparticle-associated FKN is not known, phagocyte migration was dependent on the chemokine receptor CX3CR1, as CX3CR1-deficient macrophages failed to migrate toward apoptotic B cells.
Interestingly, it has been shown that FKN shedding or cleavage by ADAM17/ADAM10 can generate a soluble 90-kDa form regardless of apoptosis.36, 37 Why the 90-kDa fragment does not confer similar migratory effects during cell death remains to be determined. It is possible that PtdSer found on microparticles could further enhance chemotaxis. A 55 kDa soluble form of FKN is generated by cathepsin S processing, however, this is not microparticle bound.38 Therefore, the precise mechanism through which apoptosis is able to differentially regulate the 60-kDa FKN processing and packaging into microparticles currently remains unclear. Intriguingly, compared with the mature glycosylated FKN (90 kDa), the unglycosylated intracellular form is ~50–60 kDa.39 It is enticing to speculate that apoptosis may inhibit glycosylation of FKN, resulting in increased unglycosylated protein. In turn, intracellular vesicles liberated during membrane blebbing could entail a possible mechanism of FKN release. Alternatively, caspase-3 is also present in microparticles,40 and the cleavage could occur after the particles have been released.
Nucleotides
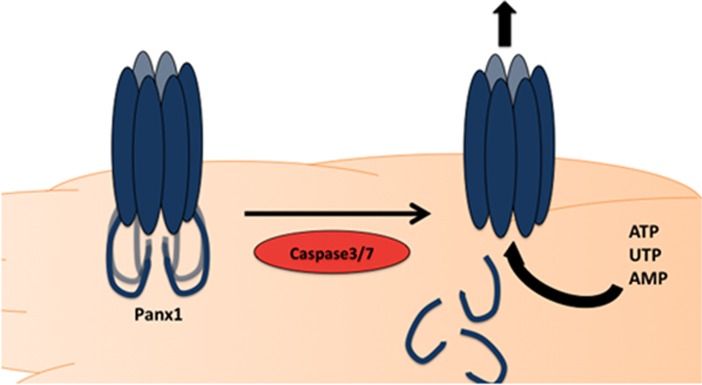
Nucleotides can also serve as find-me signals.41 Both adenosine triphosphate (ATP) and uridine triphosphate (UTP) can be released from apoptotic cells in a time- and caspase-dependent manner. Breakdown of these nucleotides by apyrase treatment, resulted in impaired migration (in vitro and in vivo) and defective cell clearance of apoptotic thymocytes.41 It was determined that the release of nucleotides from apoptotic cells occurred while the membrane was still intact, through plasma membrane Pannexin-1 (Panx1) channels.42 It was shown that during apoptosis, effector caspases 3 and 7 cleave the C-terminal tail of Panx1.42 Cleavage of the C-terminal tail resulted in a ‘open' channel that was able to release nucleotides into the extracellular space. Mutation of the caspase site on Panx1 rendered the pore incapable of nucleotide release, indicating the specific mechanism of action by which Panx1 released ATP/UTP.42 It was later shown that the C-terminal tail of Panx1 serves as a pore-associated auto-inhibitory region for the channel.43 Although nucleotide release through the pannexin channels is perhaps the best-detailed to date, among the find-me signals (Figure 3), there are still several questions that remain. For example, similar levels of UTP and ATP are released from apoptotic cells, even though the intracellular concentration of ATP is much higher.44 Whether Panx1 is more selective for UTP, if UTP is somehow more accessible for release, or if ATP is metabolized/degraded intracellularly45 during apoptosis remains to be sorted out. Although ATP remains the most studied metabolite released from Panx1, the channel forms a rather non-selective pore, allowing the passage of molecules up to 1 kDa in size.46 Therefore, it will be interesting to determine whether Panx1 is involved in the release of any other find-me signals that may either cooperate with nucleotides for phagocyte attraction, or alternatively, serve additional functions during cell clearance. In this context, intracellular AMP is also released through the pannexin channels during apoptosis.45
Figure 3.
Panx1 mediator release. During apoptosis, effector caspase 3/7 are capable of cleaving the C- terminal tail of Panx1, resulting in an ‘open' channel, that can release ATP, UTP, and AMP
Although there are many purinergic receptors that recognize extracellular nucleotides, including ionotropic P2X receptors (ATP-gated ion channels) and metabotropic P2Y receptors (G-protein-coupled receptor), they can have a wide variety of functions.47 Elliot et al.41 was able to demonstrate that the purinergic receptor P2Y2 was involved in mediating phagocyte chemotaxis, as P2Y2 receptor knock-out mice showed decreased migration and clearance toward apoptotic cell supernatants and thymocytes, respectively. It will be important to determine what other effects the release of ATP/UTP can have on the phagocytes during engulfment as cells are known to express several purinergic receptors.
Other find-me signals
Apoptotic cells also release other soluble mediators to induce phagocyte migration (Table 1), such as the dimer of ribosomal protein S19 (RP S19).48 Only the cross-linked dimer (formed by transglutaminase 2 intracellularly49 or by factor XIIIa extracellularly50), but not monomers,49 induced monocyte migration. CD88, the C5a receptor, was linked to sensing of S19 and the migration of monocytes.51 Although neutrophils also express CD88, the dimer RP S19 did not induce neutrophil chemotaxis. This is thought to be a consequence of a C-terminal moiety, which can antagonize the polymorphonuclear leukocyte receptor.52 Although the chemotactic properties of RP S19 have been established, how the factor is released during apoptosis remains unknown. In addition, whether this can be recapitulated under physiological apoptotic stimuli, rather then hyperthermia (43 °C for 60 min), remains to be shown.
Table 1. ‘Find-me' signals and possible supplementary actions they can have during apoptotic cell clearance.
| ‘Find-me' signal | Discovery context (cell types) | In vivo | Supplementary actions | Comments |
|---|---|---|---|---|
| LPC | MCF7caspase 3 | — | DC maturation, ↑MIP-2, TNFα, IFNγ release, ↑H2O2, superoxide release from neutrophils, ↓HMGB1 release, T-cell and NK cell chemotaxis, ↑ B-cell Ab release | LPC species and receptors may be important for specific functions during cell clearance |
| S1P | Jurkat Cells | — | Anti-apoptotic, lymphocyte migration, ↓TNFα, IL-6 and IL-12 production, polarization to M2 macrophage, ↑IL-10 and PGE2, suppress T-cell proliferation and activation responses | Unknown receptor, (SIP1/SIP3 have been implicated in migration) ABCA1 regulates lysophospholipid release during apoptosis (may be S1P) |
| FKN | Burkitt lymphoma cells Activated CD19-lymphocytes | ✓ | Chemotractant for NK cells, T cells, B cells, protective effects in CNS, ↑phagocytosis | Unknown protease for 60-kDa fragment generation Many auxiliary effects are a consequence of 90-kDa form |
| ATP/UTP | Thymocytes Jurkat Cells MCF7caspase3 | ✓ | Inflammatory at high concentrations and anti-inflammatory at low concentrations, ↑phagocytosis of microglia and macrophages through P2Y6 and P2X1/3, respectively | Effects of ATP metabolites on phagocytes are unclear Other factors released by Panx1 is also not known |
| RP S19 | AsPC-1 HL-60 NIH3T3 RA synovial tissue | ✓ | Inhibit neutrophil chemotaxis, responsible for adaptive immune response toward apoptotic cells, pro-apoptotic effects on non-macrophages | Chemotactic factor was not released until 24 h after apoptosis induction Release during physiological apoptotic death not examined |
| EMAPII | 32D Meth A cells MEFs | — | ↑Myeloperoxidase activity in neutrophils, neutrophil chemotaxis, upregulation of TNF-R1, pro-apoptotic effects on endothelial cells and lymphocytes, activation of monocytes | Released 10–12 h after apoptosis Unknown receptor on monocytes Pro-EMAPII (p43) can also be secreted (non-apoptotic) and has pro-inflammatory properties |
| TryRS | U-937 | — | Pro-angiogenic, C-terminal product-stimulated pro-inflammatory and chemotactic effects similar to EMAPII, N-terminal fragment only induced migration in neutrophils | C-terminal fragment shares homology with EMAPII Released 12 h after apoptosis |
The contexts in which these signals were discovered and the different cell types that are known to release them are indicated. The table also describes a list of some of the additional signaling capabilities that these mediators are capable of eliciting, as well as additional information that is not known or debated in the field.
Other attraction molecules include endothelial monocyte activating polypeptide II (EMAPII) and human tyrosyl tRNA synthetase (TyrRS). Both require proteolytic processing to gain their chemotactic properties.53 While EMAPII requires processing from the pro-EMAPII form, most likely through caspase 7 cleavage,54 TyrRS cleavage is thought to occur by neutrophil-released elastase.55 Of note, the C-terminal cleavage product of TyrRS shares homology (49%) with EMAPII.56 The N-terminal fragment of TyrRS could stimulate phagocyte migration via the membrane protein CXCR1, but the receptor for the EMAPII like C-terminal fragment of TryRS is unknown. EMAPII can facilitate the migration of endothelial progenitor cells, which are derived from monocytes, through CXCR3; however, this has not been shown in the context of apoptotic cell clearance.57 EMAPII and TyrRS release likely occur relatively late in the apoptotic process, suggesting that they may be released during secondary necrosis of these cells.53, 54 Last, although authors were able to show that apoptotic cells secreted EMAPII and TyrRS and that these could in turn mediate chemotaxis, these studies lacked direct evidence proving that they are the chemotactic component in apoptotic cell supernatants.
In vivo relevance
To fully understand the importance of these signals and their regulation of cell clearance, many of these must be better characterized in vivo. Unlike other components of the engulfment machinery (i.e., PtdSer receptors), where generation of transgenic and knock-out mice can help determine their involvement in vivo, many of the signals (LPC, ATP, S1P) released from apoptotic cells cannot be genetically knocked out. Instead, knockout of enzymes required for their production, release, or recognition (which can be indirect and less specific) has to be used in an attempt to address these issues.
To gain insight as to whether these find-me signals affects the overall process of cell clearance in vivo, and in turn physiological or pathological conditions, the examination of specific knock-out mice is required. For example, defective apoptotic cell clearance is thought to be involved in atherogenesis,58 and intriguingly FKN deficiency has been associated with this disease.59, 60 However, to what extent FKN released during apoptosis is involved in this pathogenesis has not been determined. The deficiency of CX3CR1 has also been associated with decreased numbers of macrophages in B-cell germinal centers.36 Although macrophage recruitment was affected, there was no increase in apoptotic B cells, indicating that the reduced number of macrophages was still capable of efficient clearance. This suggests that several find-me signals may be released by one cell and loss of one may be compensated sufficiently enough by others.
The channel involved in nucleotide find-me signals is Panx1. Under homeostatic conditions, Panx1-deficient mice have no overt phenotype,61 however, there may be compensation from other pannexin family members (Panx2/Panx3),62 or other nucleotide release channels. Alternatively, Panx1-deficient mice may need to be stressed to increase apoptotic load, as these mediators may be more readily compensated at basal levels of apoptosis. Studies suggest that Panx1 is involved in several pathological scenarios (Table 2), but whether this is dependent on nucleotide release from apoptotic cells awaits further clarification. In addition to their role in nucleotide release from apoptotic cells, the pannexin channels are also involved in regulating the integrity of the dying cells,63 regulation of vascular constriction via nucleotides released by non-apoptotic mechanisms,64 and cancer cell migration.65 Owing to the ubiquitous expression66 and vast regulatory mechanisms of Panx1 activation,61, 67 novel cell-type-specific knock-out and transgenic mice affecting apoptosis-specific activation may be necessary to understand its involvement during cell clearance in vivo.
Table 2. Efforts to define function of ‘find-me' signals in vivo.
| ‘Find-me' signal | KO mice | Phenotype | Mechanism | References |
|---|---|---|---|---|
| LPC | iPLA2 ABCA1 G2A | Neuronal dystrophy/neurodegeneration Male reduced fertility Increased neonatal death Decreased circulating HDL/congestive heart failure/kidney glomerulonephritis Increased atherosclerosis Autoimmune syndrome | Insufficient membrane remodeling Impaired spermatozoa motility Placenta malformation Improper cholesterol efflux/lipid-laden macrophages in areas of high turnover/immune complex deposition Macrophage-specific ABCA1−/− in ApoE−/− LDLR−/− mice T-cell hyperresponsiveness | 105, 106, 107 108, 109, 110 111 |
| S1P | SphK1 SphK2 SphK1/2 | Attenuated colon cancer Attenuate DSS-induced colitis Enhanced colitis-associated cancer Exacerbated collagen-induced arthritis Embryonic lethal | Has a role in tumor proliferation Decrease pro-inflammatory factors Increase SphK1/S1P which leads to inflammation Increased pro-inflammatory mediators Defects in neural and vascular development Increased numbers of apoptotic cells in fetal embryos | 112, 113 114, 115 116 |
| FKN | CX3CL1 CX3CR1 | Attenuated atherosclerotic lesions Decreased macrophages in B-cell germinal centers Attenuated atherosclerotic lesions | Fewer macrophages in lesions Impaired phagocyte migration Impaired monocyte recruitment to atherosclerotic lesions (ApoE−/−) | 36, 117 60 |
| ATP/UTP | Panx1 P2Y2 | Increased inflammation during peritonitis Delayed onset of disease in EAE Blood pressure regulation dysfunction Decreased allergic airway inflammation | Decrease release of anti- inflammatory mediator AMP Increased P2X7 expression Defective smooth muscle cell vasoconstriction Reduced eosinophil and dendritic cell migration | 45, 61, 64, 118, 119 120 |
A list of knock-out mice of proteins linked to specific find-me signal release or recognition. Some of phenotypes associated with these knock-out mice should be cautiously interpreted as the linkage between these phenotypes, the specific find-me signals, or apoptotic cell clearance is not detailed. This demonstrates the necessity to develop novel methodologies to truly understand the impact of find-me signals via combination of genetic and pharmacological methods in vivo.
Numerous knock-out studies have been performed on other proteins involved in different aspects of these attraction signals. Table 2 lists phenotypes of knock-out mice relevant for find-me signal release or recognition, however, whether find-me signals are having a role in these models requires clarification.
Another issue with several find-me signals is their in vivo concentration. As shown through investigations on LPC, apoptotic cell supernatants contained a concentration of 200 nM LPC, but the authors used 20–30 μM of purified LPC to detect cell migration.23, 24 Although plasma levels of LPC are known to be considerably high (100 μM–150 μM),68 extracellular actions of LPC have been reported;69 an observation likely in part because of the availability of free LPC, as the lipid is thought to be bound to albumin and other carrier proteins.70 Without detailed knowledge of the receptor for LPC during phagocyte migration, it is difficult to determine if physiological LPC is pertinent. Similarly, there is a discrepancy in the physiological S1P concentrations (>200 nM)71 compared with the S1P concentrations needed to stimulate phagocyte migration (1–1000 nM) and the much lower S1P levels measured in the supernatants of apoptotic cells (12–16 pmol).33 S1P is also bound in blood to albumin and lipoproteins.72 Therefore, how these signals are able to mediate migration in vivo requires elucidation, as phagocyte recruitment was not studied in vivo. Release of these components may be restricted to a local area, where higher concentrations could be achieved and thereby have an effect on nearby phagocytes. Alternatively, whether LPC or S1P bound to serum proteins or other extracellular components can affect their migratory capabilities is another possible factor in determining their ability to attract phagocytes.
Unlike lipid find-me signals, work on nucleotides and phagocyte migration could show that these mediators were indeed able to induce monocyte chemotaxis in vivo. Supernatants of apoptotic cells released concentrations of 100–200 nM ATP and UTP during cell death.41, 42 Studies have shown that resting ATP plasma levels are ~28 nM,73 indicating that the release of ATP from apoptotic cells is significant compared with physiological concentrations. Not surprisingly, the EC50 for many purinergic P2Y receptors are below 1 μM, including P2Y2, which recognizes ATP and UTP at concentration of 100 nM and 200 nM, respectively.74 Albiet, extracellular ATP can be rapidly degraded by nucleotidases,75 therefore limiting their availability to a short range. These observations indicate that nucleotides have the capacity to serve as find-me signals in vivo, but certain features of this nucleotide-mediated phagocyte attraction requires further characterization.
Additional functions of find-me signals
Find-me signals were first discovered based on their ability to promote phagocyte migration. However, continued studies on these factors has brought about interesting observations, suggesting other roles during engulfment. As cell clearance is an overall anti-inflammatory process, find-me signals can also contribute to the suppression of inflammation during cell clearance. In addition, the factors may stimulate the ‘appetite' of a phagocyte or increase their engulfment capacity. This section will cover some of the auxiliary functions of these apoptotic-released signals and how it may regulate engulfment (Table 1).
Nucleotides are well-known to have extracellular signaling capabilities. Although they were first considered pro-inflammatory, it is now appreciated that the situation is not that simple.47 Low levels of ATP can actually have anti-inflammatory properties such as decreasing secretion of pro-inflammatory cytokines from macrophages and dendritic cells.76, 77 Many of the inflammatory properties of ATP are linked to their stimulation of the P2X7R; however, activation of P2X7R requires >100 μM ATP,74 indicating that 100–200 nM released during cell death is unlikely to activate the P2X7R. In fact, this has been supported by studies that showed pannexin-mediated ATP release does not activate P2X7R.78 P2X receptors only engage ATP, while the P2Y receptors, which are implicated in migration, recognize ATP, UTP, and their metabolites.79 This recognition repertoire can have an important function in cell clearance, as apoptotic cells also release UTP. Furthermore, ATP and UTP can also elicit monocyte migration indirectly by affecting adhesion molecule expression on vascular endothelial cells.80 Outside of migration, nucleotides can increase microglial phagocytosis through UDP acting on P2Y6, although the mechanism is not known.81 In addition, stimulation of P2 purinergic receptors can increase the phagocytic capacity of macrophages, which is thought to be dependent on increased apoptotic cell-receptor expression, such as αvβ3. Nucleotide effects on phagocytosis could be detected as early as 30 min, indicating the swift communication that occurs between find-me signals and the phagocyte.82 Although ATP and UTP are released from apoptotic cells, their degradation products may be able to further substantiate their actions during clearance. Adenosine is a potent anti-inflammatory molecule and can suppress both innate and adaptive components of the immune system. It was recently suggested that apoptotic cells release substantial amounts of AMP (through Panx1), which can contribute to the anti-inflammatory effects associated with cell clearance.45 Therefore, nucleotides provide an exciting new avenue for clearance, both in migration and subsequent consequences.
FKN has been shown to enhance phagocytosis by macrophages via its ability to enhance MFG-E8 induction, however, this upregulation was seen 24 h after FKN treatment.83 In addition to increasing phagocytosis, FKN has also been associated with anti-inflammatory actions such as providing neuroprotection during glutamate toxicity,84, 85 modulating TNF-α secretion by microglia,86 and promoting proliferative effects,87, 88 which may all have important roles in different settings of cell clearance. However, clarification is needed as to which processed form is responsible for these FKN effects.
LPC may also have an indirect role in migration by upregulating vascular endothelial cell expression of different chemokines responsible for immune cell recruitment.89, 90 LPC has been shown to both increase production of pro-inflammatory cytokines,69 and have anti-inflammatory effects such as inhibiting high-mobility group protein B1 (HMGB1) secretion(16 h post-treatment with LPC).91 These effects are dependent on different receptors as well as cell types, but not yet fully understood. LPC can also act as an indirect eat-me signal by promoting the LPC-dependent binding of IgM to apoptotic,92 late apoptotic93, 94 and necrotic cells,95 ultimately leading to clearance. LPC-IgM-dependent binding to different apoptotic and necrotic states may serve as a backup to normal recognition in situations where large numbers of cells are undergoing apoptosis.
Although the receptor mediating S1P recognition during cell death is not known, S1P can have both anti-inflammatory and anti-apoptotic effects on macrophages.96 S1P can cause M2 polarization, leading to lower pro-inflammatory cytokines. Interestingly these actions are thought to be a consequence of S1P generation by SphK2,97 and not SphK1(suggested to be responsible for the S1P find-me signal). Further analysis of the different sphingosine kinase members and their activity during cell death will be important for the understanding of S1P. Lastly, although the death of circulating cells does not generally disrupt any tissues, epithelial death can affect the integrity of barrier surfaces. A recent study has shown that S1P released from apoptotic cells can act on neighboring cells through S1P2R to facilitate apical extrusion of the dying cell.98, 99
Apart from these well-studied mediators, other components released during apoptosis have been implicated in stimulating functions outside of chemotaxis. Apoptotic blebs can regulate migration indirectly by enhancing monocyte-endothelial interactions in the vasculature.100 Although apoptotic bleb-induced migration may be important for efficient clearance, unlike apoptotic bodies, phagocytosis of such blebs stimulates dendritic cell maturation. This can lead to dendritic cell-mediated T-cell activation, which can predispose to autoimmune disease.101, 102 RP S19 has been reported to antagonize neutrophil chemotaxis and induce their apoptosis, indicating its anti-inflammatory properties.103 Finally, EMAPII and TyrRS have been shown to elicit pro-inflammatory actions such as increased peroxidase activity in neutrophils and production of TNFα, respectively.55, 104 These observations do not coincide with the generally anti-inflammatory nature of apoptotic cell clearance; therefore, a deeper understanding of these mediators is needed.
Conclusion
Apoptotic cells are capable of the controlled release of different soluble and/or membrane-bound mediators. These factors can have several effects during physiology and pathology, including their function as find-me signals to attract phagocytes toward apoptotic cells.7 It has become evident that not only are the find-me signals involved in chemotaxis, but they may also have additional roles that promote efficient and anti-inflammatory engulfment of dying cells. Although our understanding of find-me signals has moved rapidly, a number of questions remain. First, defining how the many different find-me signals are released by a given apoptotic cell in vivo is not known; perhaps this depends on the type of apoptotic cells and the type of apoptosis. Second, what is the attraction radius for a released find-me signal before its original properties are altered, such as metabolic conversion? This is a rather challenging question; as sensitive probes are needed to determine the low levels of find-me signals that might be released from a few dying cells in vivo (rather than in tissue culture contexts where a large number of cells can be induced to undergo death synchronously). Third, what is the effect of metabolites that are derived from the original find-me signals; for example, is there a positive or negative feedback from neighboring cells? Fourth, another substantial layer of complexity arises when one considers different settings such as inflammation, tumors, or wound healing. In these states, different cell types may simultaneously and continually undergo death, and also different phagocytes may be simultaneously responding to the find-me signal landscape. This heterogeneity of phagocytes can differentially react to the find-me signals; also, the kinetics of their response can range from minutes to hours, a phenomenon that may depend on the find-me signal or reaction elicited. This type of complexity is both beautiful and daunting, but further understanding of these signals in vivo will help determine their roles in apoptotic cell clearance and in turn physiological and pathological conditions.
Glossary
- (LPC)
Lysophosphatidylcholine
- (S1PR)
sphingosine-1-phosphate
- (FKN)
fractalkine/CX3CL1
- (RP S19)
adenosine ribosomal protein S19
- (EMAPII)
endothelial monocyte activating polypeptide II
- (TyrRS)
tyrosyl tRNA synthetase
- (CRT)
calreticulin
- (ICAM3)
the intracellular cell adhesion molecule 3
- (DD1α)
death Domain 1α
- (LRP1)
LDL receptorrelated protein 1
- (BAI1)
brain-specific angiogenesis inhibitor 1
- (TIM)
T cell immunoglobulin mucin domain
- (GAS6)
growth arrestin-specific
- (MFGE8)
milk fat globule EGF factor 8
- (TAM)
Tyro-Axl-Mer
- (ELMO-DOCK180)
engulfment and cell motility–downstream of Crk
- (Vsp)
vacuolar sorting protein
- (iPLA2)
calcium-independent phospholipase A2
- (sPLA2)
secretory PLA2
- (ABCA1)
ATP-binding cassette transporter
- (SphK1/2)
sphingosine kinase 1/2
- (G2A)
S1P receptor (S1PR) G-protein coupled receptor 2A
- (CX3CR1)
CX3C chemokine receptor 1
- (Panx)
pannexin
- (P2X/P2Y)
purinergic receptors
- (UTP)
Uridine triphosphate
- (AMP)
Adenosin monophosphate
- (ADAM17/ADAM10)
A Disintegrin and metalloproteinase domain-containing protein
- (HMGB1)
high mobility group protein B1
- (EAE)
Experimental autoimmune encephalomyelitis
- (ApoE)
Apolipoprotein
- (LDL)
low-density lipoprotein
- (HDL)
high density lipoprotein
Footnotes
Edited by G Kroemer
References
- Henson PM. Dampening inflammation. Nat Immunol 2005; 6: 1179–1181. [DOI] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest 1998; 101: 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh M-LN, Fadok VA, Henson PM. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-beta1 secretion and the resolution of inflammation. J Clin Invest 2002; 109: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell 2010; 140: 619–630. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 2009; 16: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007; 35: 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran KS. “Recruitment Signals” from apoptotic cells. Cell 2003; 113: 817–820. [DOI] [PubMed] [Google Scholar]
- Segawa K, Nagata S. An apoptotic “Eat Me” signal: phosphatidylserine exposure. Trends Cell Biol 2015; 25: 639–650. [DOI] [PubMed] [Google Scholar]
- Chang MK, Bergmark C, Laurila A, Hörkkö S, Han KH, Friedman P et al. Monoclonal antibodies against oxidized low-density lipoprotein bind to apoptotic cells and inhibit their phagocytosis by elicited macrophages: evidence that oxidation-specific epitopes mediate macrophage recognition. Proc Natl Acad Sci USA 1999: 6353–8. [DOI] [PMC free article] [PubMed]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE et al. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 2005; 123: 321–334. [DOI] [PubMed] [Google Scholar]
- Yoon KW, Byun S, Kwon E, Hwang S-Y, Chu K, Hiraki M et al. Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science 2015; 349: 1261669–1261669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont A-C, Elliott MR, Lu M, Haney LB, Ma Z et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007; 450: 430–434. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Jung M-Y, Kim H-J, Lee S-J, Kim S-Y, Lee B-H et al. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ 2008; 15: 192–201. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Karisola P, Peña-Cruz V, Dorfman DM, Jinushi M, Umetsu SE et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 2007; 27: 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Akiba H, Takeda K, Kojima Y, Hashiguchi M, Azuma M et al. Tim-3 mediates phagocytosis of apoptotic cells and cross-presentation. Blood 2009; 113: 3821–3830. [DOI] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S Identification of Tim4 as a phosphatidylserine receptor Nature 2007; 450: 435–439. [DOI] [PubMed]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002; 417: 182–187. [DOI] [PubMed] [Google Scholar]
- Ishimoto Y, Ohashi K, Mizuno K, Nakano T. Promotion of the uptake of PS liposomes and apoptotic cells by a product of growth arrest-specific gene, gas6. J Biochem 2000; 127: 411–417. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Kang K-B, Thapa N, Kim S-Y, Lee S-J, Kim I-S. Requirement of adaptor protein GULP during stabilin-2-mediated cell corpse engulfment. J Biol Chem 2008; 283: 10593–10600. [DOI] [PubMed] [Google Scholar]
- Lee S-J, So I-S, Park S-Y, Kim I-S. Thymosin beta4 is involved in stabilin-2-mediated apoptotic cell engulfment. FEBS Lett 2008; 582: 2161–2166. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol 2008; 10: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C, Wesselborg S, Herrmann M, Lauber K. Dangerous attraction: phagocyte recruitment and danger signals of apoptotic and necrotic cells. Apoptosis 2010; 15: 1007–1028. [DOI] [PubMed] [Google Scholar]
- Lauber K, Bohn E, Kröber SM, Xiao Y-J, Blumenthal SG, Lindemann RK et al. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell 2003; 113: 717–730. [DOI] [PubMed] [Google Scholar]
- Peter C, Waibel M, Radu CG, Yang LV, Witte ON, Schulze-Osthoff K et al. Migration to apoptotic “find-me” signals is mediated via the phagocyte receptor G2A. J Biol Chem 2008; 283: 5296–5305. [DOI] [PubMed] [Google Scholar]
- Atsumi G, Murakami M, Tajima M, Shimbara S, Hara N, Kudo I. The perturbed membrane of cells undergoing apoptosis is susceptible to type II secretory phospholipase A2 to liberate arachidonic acid. Biochim Biophys Acta 1997; 1349: 43–54. [DOI] [PubMed] [Google Scholar]
- Peter C, Waibel M, Keppeler H, Lehmann R, Xu G, Halama A et al. Release of lysophospholipid “find-me” signals during apoptosis requires the ATP-binding cassette transporter A1. Autoimmunity 2012; 45: 568–573. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 1998; 93: 951–960. [DOI] [PubMed] [Google Scholar]
- Witte ON, Kabarowski JH, Xu Y, Le LQ, Zhu K. Retraction. Science 2005; 307: 206–206b. [DOI] [PubMed] [Google Scholar]
- Murakami N, Yokomizo T, Okuno T, Shimizu T. G2A is a proton-sensing G-protein-coupled receptor antagonized by lysophosphatidylcholine. J Biol Chem 2004; 279: 42484–42491. [DOI] [PubMed] [Google Scholar]
- Obinata H, Hattori T, Nakane S, Tatei K, Izumi T. Identification of 9-hydroxyoctadecadienoic acid and other oxidized free fatty acids as ligands of the G protein-coupled receptor G2A. J Biol Chem 2005; 280: 40676–40683. [DOI] [PubMed] [Google Scholar]
- Lin P, Ye RD. The lysophospholipid receptor G2A activates a specific combination of G proteins and promotes apoptosis. J Biol Chem 2003; 278: 14379–14386. [DOI] [PubMed] [Google Scholar]
- Hait NC, Oskeritzian CA, Paugh SW, Milstien S, Spiegel S. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochim Biophys Acta 2006; 1758: 2016–2026. [DOI] [PubMed] [Google Scholar]
- Gude DR, Alvarez SE, Paugh SW, Mitra P, Yu J, Griffiths R et al. Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a “come-and-get-me” signal. FASEB J 2008; 22: 2629–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert A, Cremer S, Schmidt MV, Knethen von A, Angioni C, Geisslinger G et al. Cleavage of sphingosine kinase 2 by caspase-1 provokes its release from apoptotic cells. Blood 2010; 115: 3531–3540. [DOI] [PubMed] [Google Scholar]
- Segundo C, Medina F, Rodríguez C, Martínez-Palencia R, Leyva-Cobián F, Brieva JA. Surface molecule loss and bleb formation by human germinal center B cells undergoing apoptosis: role of apoptotic blebs in monocyte chemotaxis. Blood 1999; 94: 1012–1020. [PubMed] [Google Scholar]
- Truman LA, Ford CA, Pasikowska M, Pound JD, Wilkinson SJ, Dumitriu IE et al. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood 2008; 112: 5026–5036. [DOI] [PubMed] [Google Scholar]
- Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 2003; 102: 1186–1195. [DOI] [PubMed] [Google Scholar]
- Fonović UP, Jevnikar Z, Kos J. Cathepsin S generates soluble CX3CL1 (fractalkine) in vascular smooth muscle cells. Biol Chem 2013; 394: 1349–1352. [DOI] [PubMed] [Google Scholar]
- Garton KJ, Gough PJ, Blobel CP, Murphy G, Greaves DR, Dempsey PJ et al. Tumor necrosis factor-alpha-converting enzyme (ADAM17) mediates the cleavage and shedding of fractalkine (CX3CL1). J Biol Chem 2001; 276: 37993–38001. [DOI] [PubMed] [Google Scholar]
- Böing AN, Hau CM, Sturk A, Nieuwland R. Platelet microparticles contain active caspase 3. Platelets 2008; 19: 96–103. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 2009; 461: 282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER et al. Pannexin 1 channels mediate “find-me” signal release and membrane permeability during apoptosis. Nature 2010; 467: 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilos JK, Chiu Y-H, Chekeni FB, Armstrong AJ, Walk SF, Ravichandran KS et al. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J Biol Chem 2012; 287: 11303–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C, Wesselborg S, Lauber K. Apoptosis: opening PANdora's BoX. Curr Biol 2010; 20: R940–R942. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Maruyama T, Urade Y, Nagata S. Immunosuppression via adenosine receptor activation by adenosine monophosphate released from apoptotic cells. Elife 2014; 3: e02172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma M, Locovei S, Keane RW, Dahl G. Modulation of membrane channel currents by gap junction protein mimetic peptides: size matters. American Journal of Physiology - Cell Physiology 2007; 293: C1112–C1119. [DOI] [PubMed] [Google Scholar]
- Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature 2014; 509: 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horino K, Nishiura H, Ohsako T, Shibuya Y, Hiraoka T, Kitamura N et al. A monocyte chemotactic factor, S19 ribosomal protein dimer, in phagocytic clearance of apoptotic cells. Lab Invest 1998; 78: 603–617. [PubMed] [Google Scholar]
- Nishimura T, Horino K, Nishiura H, Shibuya Y, Hiraoka T, Tanase S et al. Apoptotic cells of an epithelial cell line, AsPC-1, release monocyte chemotactic S19 ribosomal protein dimer. J Biochem 2001; 129: 445–454. [DOI] [PubMed] [Google Scholar]
- Nishiura H, Tanase S, Sibuya Y, Nishimura T, Yamamoto T. Determination of the cross-linked residues in homo-dimerization of S19 ribosomal protein concomitant with exhibition of monocyte chemotactic activity. Lab Invest 1999; 79: 915–923. [PubMed] [Google Scholar]
- Nishiura H, Shibuya Y, Yamamoto T. S19 ribosomal protein cross-linked dimer causes monocyte-predominant infiltration by means of molecular mimicry to complement C5a. Lab Invest 1998; 78: 1615–1623. [PubMed] [Google Scholar]
- Shrestha A, Shiokawa M, Nishimura T, Nishiura H, Tanaka Y, Nishino N et al. Switch moiety in agonist/antagonist dual effect of S19 ribosomal protein dimer on leukocyte chemotactic C5a receptor. Am J Pathol 2003; 162: 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies UE, Behrensdorf HA, Mitchell CA, Deutsch U, Risau W, Drexler HC et al. Regulation of endothelial monocyte-activating polypeptide II release by apoptosis. Proc Natl Acad Sci USA 1998; 95: 12322–12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrensdorf HA, van de Craen M, Knies UE, Vandenabeele P, Clauss M. The endothelial monocyte-activating polypeptide II (EMAP II) is a substrate for caspase-7. FEBS Lett 2000; 466: 143–147. [DOI] [PubMed] [Google Scholar]
- Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 1999; 284: 147–151. [DOI] [PubMed] [Google Scholar]
- Kleeman TA, Wei D, Simpson KL, First EA. Human tyrosyl-tRNA synthetase shares amino acid sequence homology with a putative cytokine. J Biol Chem 1997; 272: 14420–14425. [DOI] [PubMed] [Google Scholar]
- Hou Y, Plett PA, Ingram DA, Rajashekhar G, Orschell CM, Yoder MC et al. Endothelial-monocyte-activating polypeptide II induces migration of endothelial progenitor cells via the chemokine receptor CXCR3. Exp Hematol 2006; 34: 1125–1132. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol 2010; 189: 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Chan L, Lira SA, Charo IF. Fractalkine deficiency markedly reduces macrophage accumulation and atherosclerotic lesion formation in CCR2-/- mice: evidence for independent chemokine functions in atherogenesis. Circulation 2008; 117: 1642–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combadiere C, Potteaux S, Gao J-L, Esposito B, Casanova S, Lee EJ et al. Decreased atherosclerotic lesion formation in CX3CR1/apolipoprotein E double knockout mice. Circulation 2003; 107: 1009–1016. [DOI] [PubMed] [Google Scholar]
- Bond SR, Naus CC. The pannexins: past and present. Front Physiol 2014; 5: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiotas P, Krenz A, Hormuzdi SG, Ridder DA, Herb A, Barakat W et al. Pannexins in ischemia-induced neurodegeneration. Proc Natl Acad Sci USA 2011; 108: 20772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IKH, Chiu Y-H, Armstrong AJ, Kinchen JM, Juncadella IJ, Bayliss DA et al. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature 2014; 507: 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billaud M, Chiu Y-H, Lohman AW, Parpaite T, Butcher JT, Mutchler SM et al. A molecular signature in the pannexin1 intracellular loop confers channel activation by the α1 adrenoreceptor in smooth muscle cells. Sci Signal 2015; 8: ra17–ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlow PW, Zhang S, Soong TD, Halberg N, Goodarzi H, Mangrum C et al. Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival. Nat Cell Biol 2015; 17: 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova A, Ivanov D, Petrash N, Pestova A, Skoblov M, Kelmanson I et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 2004; 83: 706–716. [DOI] [PubMed] [Google Scholar]
- Chiu Y-H, Ravichandran KS, Bayliss DA. Intrinsic properties and regulation of Pannexin 1 channel. Channels (Austin) 2014; 8: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M, Ojala PJ, Hrzenjak A, Graier WF, Malli R, Tritscher M et al. Acyl chain-dependent effect of lysophosphatidylcholine on endothelial prostacyclin production. J Lipid Res 2010; 51: 2957–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Schäfer-Elinder L, Wu R, Claesson HE, Frostegård J. Lysophosphatidylcholine (LPC) induces proinflammatory cytokines by a platelet-activating factor (PAF) receptor-dependent mechanism. Clin Exp Immunol 1999; 116: 326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala PJ, Hermansson M, Tolvanen M, Polvinen K, Hirvonen T, Impola U et al. Identification of alpha-1 acid glycoprotein as a lysophospholipid binding protein: a complementary role to albumin in the scavenging of lysophosphatidylcholine. Biochemistry 2006; 45: 14021–14031. [DOI] [PubMed] [Google Scholar]
- Okajima F. Plasma lipoproteins behave as carriers of extracellular sphingosine 1-phosphate: is this an atherogenic mediator or an anti-atherogenic mediator? Biochim Biophys Acta 2002; 1582: 132–137. [DOI] [PubMed] [Google Scholar]
- Murata N, Sato K, Kon J, Tomura H, Yanagita M, Kuwabara A et al. Interaction of sphingosine 1-phosphate with plasma components, including lipoproteins, regulates the lipid receptor-mediated actions. Biochem J 2000; 352(Pt 3): 809–815. [PMC free article] [PubMed] [Google Scholar]
- Gorman MW, Feigl EO, Buffington CW. Human plasma ATP concentration. Clin Chem 2007; 53: 318–325. [DOI] [PubMed] [Google Scholar]
- Junger WG Immune cell regulation by autocrine purinergic signalling Nat Rev Immunol 2011; 11: 201–212. [DOI] [PMC free article] [PubMed]
- Zimmermann H, Zebisch M, Sträter N. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal 2012; 8: 437–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Sala A, Ferrari D, Corinti S, Cavani A, Di Virgilio F, Girolomoni G. Extracellular ATP induces a distorted maturation of dendritic cells and inhibits their capacity to initiate Th1 responses. J Immunol 2001; 166: 1611–1617. [DOI] [PubMed] [Google Scholar]
- Haskó G, Kuhel DG, Salzman AL, Szabó C. ATP suppression of interleukin-12 and tumour necrosis factor-alpha release from macrophages. Br J Pharmacol 2000; 129: 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Misaghi S, Newton K, Gilmour LL, Louie S, Cupp JE et al. Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J Immunol 2011; 186: 6553–6561. [DOI] [PubMed] [Google Scholar]
- Kronlage M, Song J, Sorokin L, Isfort K, Schwerdtle T, Leipziger J et al. Autocrine purinergic receptor signaling is essential for macrophage chemotaxis. Sci Signal 2010; 3: ra55–ra55. [DOI] [PubMed] [Google Scholar]
- Seye CI, Yu N, Jain R, Kong Q, Minor T, Newton J et al. The P2Y2 nucleotide receptor mediates UTP-induced vascular cell adhesion molecule-1 expression in coronary artery endothelial cells. J Biol Chem 2003; 278: 24960–24965. [DOI] [PubMed] [Google Scholar]
- Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, Shinozaki Y, Ohsawa K, Tsuda M et al. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature 2007; 446: 1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-da-Silva C, Burnstock G, Ojcius DM, Coutinho-Silva R. Purinergic receptor agonists modulate phagocytosis and clearance of apoptotic cells in macrophages. Immunobiology 2011; 216: 1–11. [DOI] [PubMed] [Google Scholar]
- Miksa M, Amin D, Wu R, Ravikumar TS, Wang P. Fractalkine-induced MFG-E8 leads to enhanced apoptotic cell clearance by macrophages. Mol Med 2007; 13: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M, Doi Y, Liang J, Kawanokuchi J, Sonobe Y, Takeuchi H et al. Fractalkine attenuates excito-neurotoxicity via microglial clearance of damaged neurons and antioxidant enzyme heme oxygenase-1 expression. J Biol Chem 2011; 286: 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Kawanokuchi J, Numata K, Suzumura A. Production and neuroprotective functions of fractalkine in the central nervous system. Brain Res 2003; 979: 65–70. [DOI] [PubMed] [Google Scholar]
- Zujovic V, Benavides J, Vigé X, Carter C, Taupin V. Fractalkine modulates TNF-alpha secretion and neurotoxicity induced by microglial activation. Glia 2000; 29: 305–315. [PubMed] [Google Scholar]
- White GE, Tan TCC, John AE, Whatling C, McPheat WL, Greaves DR. Fractalkine has anti-apoptotic and proliferative effects on human vascular smooth muscle cells via epidermal growth factor receptor signalling. Cardiovasc Res 2010; 85: 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White GE, Greaves DR. Fractalkine: a survivor's guide: chemokines as antiapoptotic mediators. Arterioscler Thromb Vasc Biol 2012; 32: 589–594. [DOI] [PubMed] [Google Scholar]
- Quinn MT, Parthasarathy S, Steinberg D. Lysophosphatidylcholine: a chemotactic factor for human monocytes and its potential role in atherogenesis. Proc Natl Acad Sci USA 1988; 85: 2805–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes U, Hinder M, Scheuer W, Friebe WG, Schramm S, Kaiser B. Phospholipase A2 is involved in chemotaxis of human leukocytes. Adv Exp Med Biol 1999; 469: 189–197. [DOI] [PubMed] [Google Scholar]
- Chen G, Li J, Qiang X, Czura CJ, Ochani M, Ochani K et al. Suppression of HMGB1 release by stearoyl lysophosphatidylcholine:an additional mechanism for its therapeutic effects in experimental sepsis. J Lipid Res 2005; 46: 623–627. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Gershov D, Ma X, Brot N, Elkon KB. I-PLA (2) activation during apoptosis promotes the exposure of membrane lysophosphatidylcholine leading to binding by natural immunoglobulin M antibodies and complement activation. J Exp Med 2002; 196: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart B, Ciurana C, Rensink I, Manoe R, Hack CE, Aarden LA. Complement activation by apoptotic cells occurs predominantly via IgM and is limited to late apoptotic (secondary necrotic) cells. Autoimmunity 2004; 37: 95–102. [DOI] [PubMed] [Google Scholar]
- Fu M, Fan P-S, Li W, Li C-X, Xing Y, An J-G et al. Identification of poly-reactive natural IgM antibody that recognizes late apoptotic cells and promotes phagocytosis of the cells. Apoptosis 2007; 12: 355–362. [DOI] [PubMed] [Google Scholar]
- Ciurana CLF, Zwart B, van Mierlo G, Hack CE. Complement activation by necrotic cells in normal plasma environment compares to that by late apoptotic cells and involves predominantly IgM. Eur J Immunol 2004; 34: 2609–2619. [DOI] [PubMed] [Google Scholar]
- Weigert A, Johann AM, Knethen von A, Schmidt H, Geisslinger G, Brüne B. Apoptotic cells promote macrophage survival by releasing the antiapoptotic mediator sphingosine-1-phosphate. Blood 2006; 108: 1635–1642. [DOI] [PubMed] [Google Scholar]
- Weigert A, Tzieply N, Knethen von A, Johann AM, Schmidt H, Geisslinger G et al. Tumor cell apoptosis polarizes macrophages role of sphingosine-1-phosphate. Mol Biol Cell 2007; 18: 3810–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Shea J, Slattum G, Firpo MA, Alexander M, Mulvihill SJ et al. Defective apical extrusion signaling contributes to aggressive tumor hallmarks. Elife 2015; 4: e04069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Forostyan T, Sabbadini R, Rosenblatt J. Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. J Cell Biol 2011; 193: 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J, Vales A, Mitulovic G, Blumer M, Schmid R, Witztum JL et al. Oxidized membrane vesicles and blebs from apoptotic cells contain biologically active oxidized phospholipids that induce monocyte-endothelial interactions. Arterioscler Thromb Vasc Biol 2002; 22: 101–107. [DOI] [PubMed] [Google Scholar]
- Fransen JH, Hilbrands LB, Jacobs CW, Adema GJ, Berden JH, Van der Vlag J. Both early and late apoptotic blebs are taken up by DC and induce IL-6 production. Autoimmunity 2009; 42: 325–327. [DOI] [PubMed] [Google Scholar]
- Fransen JH, Hilbrands LB, Ruben J, Stoffels M, Adema GJ, van der Vlag J et al. Mouse dendritic cells matured by ingestion of apoptotic blebs induce T cells to produce interleukin-17. Arthritis Rheum 2009; 60: 2304–2313. [DOI] [PubMed] [Google Scholar]
- Yamamoto T. Roles of the ribosomal protein S19 dimer and the C5a receptor in pathophysiological functions of phagocytic leukocytes. Pathol Int 2007; 57: 1–11. [DOI] [PubMed] [Google Scholar]
- Kao J, Fan YG, Haehnel I, Brett J, Greenberg S, Clauss M et al. A peptide derived from the amino terminus of endothelial-monocyte-activating polypeptide II modulates mononuclear and polymorphonuclear leukocyte functions, defines an apparently novel cellular interaction site, and induces an acute inflammatory response. J Biol Chem 1994; 269: 9774–9782. [PubMed] [Google Scholar]
- Shinzawa K, Sumi H, Ikawa M, Matsuoka Y, Okabe M, Sakoda S et al. Neuroaxonal dystrophy caused by group VIA phospholipase A2 deficiency in mice: a model of human neurodegenerative disease. J Neurosci 2008; 28: 2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck G, Sugiura Y, Shinzawa K, Kato S, Setou M, Tsujimoto Y et al. Neuroaxonal dystrophy in calcium-independent phospholipase A2β deficiency results from insufficient remodeling and degeneration of mitochondrial and presynaptic membranes. J Neurosci 2011; 31: 11411–11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Miller DJ, Ma Z, Wohltmann M, Eng G, Ramanadham S et al. Male mice that do not express group VIA phospholipase A2 produce spermatozoa with impaired motility and have greatly reduced fertility. J Biol Chem 2004; 279: 38194–38200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen-Weber TA, Voland JR, Wu Y, Ngo K, Roland BL, Nguyen S et al. Functional loss of ABCA1 in mice causes severe placental malformation, aberrant lipid distribution, and kidney glomerulonephritis as well as high-density lipoprotein cholesterol deficiency. Am J Pathol 2000; 157: 1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins JM, Lee J-Y, Boudyguina E, Kluckman KD, Brunham LR, Mulya A et al. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J Clin Invest 2005; 115: 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello RJ, Brees D, Bourassa P-A, Royer L, Lindsey S, Coskran T et al. Increased atherosclerosis in hyperlipidemic mice with inactivation of ABCA1 in macrophages. Arterioscler Thromb Vasc Biol 2002; 22: 630–637. [DOI] [PubMed] [Google Scholar]
- Le LQ, Kabarowski JH, Weng Z, Satterthwaite AB, Harvill ET, Jensen ER et al. Mice lacking the orphan G protein-coupled receptor G2A develop a late-onset autoimmune syndrome. Immunity 2001; 14: 561–571. [DOI] [PubMed] [Google Scholar]
- Kohno M, Momoi M, Oo ML, Paik J-H, Lee Y-M, Venkataraman K et al. Intracellular role for sphingosine kinase 1 in intestinal adenoma cell proliferation. Mol Cell Biol 2006; 26: 7211–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider AJ, Kawamori T, Bradshaw SG, Orr KA, Gilkeson GS, Hannun YA et al. A role for sphingosine kinase 1 in dextran sulfate sodium-induced colitis. FASEB J 2009; 23: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Nagahashi M, Kim EY, Harikumar KB, Yamada A, Huang W-C et al. Sphingosine-1-phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis-associated cancer. Cancer Cell 2013; 23: 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W-Q, Irwan AW, Goh HH, Melendez AJ, McInnes IB, Leung BP. Distinct roles of sphingosine kinase 1 and 2 in murine collagen-induced arthritis. J Immunol 2009; 183: 2097–2103. [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Yamashita T, Olivera A, Miller GF, Spiegel S, Proia RL. Essential role for sphingosine kinases in neural and vascular development. Mol Cell Biol 2005; 25: 11113–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teupser D, Pavlides S, Tan M, Gutierrez-Ramos J-C, Kolbeck R, Breslow JL. Major reduction of atherosclerosis in fractalkine (CX3CL1)-deficient mice is at the brachiocephalic artery, not the aortic root. Proc Natl Acad Sci USA 2004; 101: 17795–17800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuela S, Harland L, Simek J, Laird DW. Pannexin channels and their links to human disease. Biochem J 2014; 461: 371–381. [DOI] [PubMed] [Google Scholar]
- Lutz SE, González-Fernández E, Ventura JCC, Pérez-Samartín A, Tarassishin L, Negoro H et al. Contribution of pannexin1 to experimental autoimmune encephalomyelitis. PLoS ONE 2013; 8: e66657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T, Robaye B, Vieira RP, Ferrari D, Grimm M, Jakob T et al. The purinergic receptor P2Y2 receptor mediates chemotaxis of dendritic cells and eosinophils in allergic lung inflammation. Allergy 2010; 65: 1545–1553. [DOI] [PubMed] [Google Scholar]