Significance
The activity of many proteins is dependent on molecular chaperones and their accessory proteins, cochaperones. The ability of a cohort of kinases, many of which are oncogenic, to transduce signals is promoted by the heat shock protein 90 (Hsp90) chaperone and Cdc37 cochaperone, and requires the removal of a phosphate from Cdc37 by protein phosphatase 5 (PP5). We present the crystal structure of PP5 with Cdc37 trapped in the active site. The structure reveals how PP5 can associate with different substrates and previously unknown determinants of specificity. Our findings show kinase release from the chaperone complex is critically dependent on this dephosphorylation and that combined inhibition of both Hsp90 and PP5 could provide an effective therapeutic strategy for treating cancers addicted to these kinases.
Keywords: Hsp90, PP5, Cdc37, chaperone, phosphatase
Abstract
The serine/threonine phosphatase protein phosphatase 5 (PP5) regulates hormone- and stress-induced cellular signaling by association with the molecular chaperone heat shock protein 90 (Hsp90). PP5-mediated dephosphorylation of the cochaperone Cdc37 is essential for activation of Hsp90-dependent kinases. However, the details of this mechanism remain unknown. We determined the crystal structure of a Cdc37 phosphomimetic peptide bound to the catalytic domain of PP5. The structure reveals PP5 utilization of conserved elements of phosphoprotein phosphatase (PPP) structure to bind substrate and provides a template for many PPP–substrate interactions. Our data show that, despite a highly conserved structure, elements of substrate specificity are determined within the phosphatase catalytic domain itself. Structure-based mutations in vivo reveal that PP5-mediated dephosphorylation is required for kinase and steroid hormone receptor release from the chaperone complex. Finally, our data show that hyper- or hypoactivity of PP5 mutants increases Hsp90 binding to its inhibitor, suggesting a mechanism to enhance the efficacy of Hsp90 inhibitors by regulation of PP5 activity in tumors.
Protein phosphatase 5 (PP5) has pleiotropic roles in cellular signaling, including DNA damage repair, proliferation of breast cancer cells, circadian cycling, response to cytotoxic stresses, Rac-dependent potassium ion channel activity, and activation of steroid hormone receptors [e.g., glucocorticoid receptor (GR) and estrogen receptor] (1, 2). It is a member of the phosphoprotein phosphatase (PPP) family of serine/threonine phosphatases, which has members that share a highly conserved catalytic core and catalytic mechanism dependent on two metal ions, commonly Mn2+. Most PPP family members exhibit high, nonspecific phosphatase activity. Specificity is provided by a large cohort of regulatory and other interacting proteins that function to inhibit basal activity and recruit substrates, thereby finely tuning the enzymes (3). This combinatorial approach enables a small number of catalytic subunits to have the breadth of specificity equivalent to that seen in kinases, which are greater in number by an order of magnitude. Structures of complexes between regulatory and catalytic domains have illuminated the importance of regulatory subunits in facilitating substrate recruitment (3). However, to date, there is no structural information describing how a substrate binds at the active site of a PPP; therefore, a central question remains of how local interactions between the substrate and the catalytic domain contribute to the molecular basis of dephosphorylation.
PP5 is unique among the PPP family because it has a low basal activity caused by an autoinhibitory N-terminal tetratricopeptide (TPR) domain (4). Its activity is promoted by a number of cellular factors, including fatty acids and the molecular chaperone heat shock protein 90 (Hsp90) (5), both of which release autoinhibition by interacting with the TPR domain (6, 7). Many established PP5 substrates are dependent on Hsp90 for their activation (known as Hsp90 clients). In addition to a requirement for Hsp90’s chaperone activity, it is likely that these PP5 substrates require Hsp90 to act as a molecular bridge to bring the catalytic domain of PP5 in close proximity to enable dephosphorylation, which has been shown for the Hsp90 cochaperone Cdc37 (8). In such cases, Hsp90 performs a role similar to that observed by the regulatory subunits of the PP1 and PP2A family (3).
The cochaperone Cdc37 regulates the activation of Hsp90 client kinases by distinguishing between client and nonclient kinases (9) and recruiting the former to Hsp90 (10). Many of these kinases are oncogenes; therefore, the molecular details of their activation are of considerable interest in cancer therapy. Activation is dependent on a cycle of Cdc37-Ser13 phosphorylation by the constitutively active kinase CK2 (11, 12) and dephosphorylation by PP5 (8). The mechanisms by which Cdc37 phosphorylation and dephosphorylation regulate kinase activation are not understood.
To understand the molecular determinants of the phospho-Ser13 Cdc37-PP5 interaction, we determined the 2.3-Å crystal structure of a Cdc37 phosphomimetic peptide bound to the catalytic domain of PP5. The structure reveals how PP5 uses conserved elements of PPP structure to bind substrate, whereas in vitro and in vivo analyses indicate that, despite being highly conserved, elements of substrate specificity are determined within the phosphatase catalytic domain itself.
Results
Crystal Structure of the PP5-Cdc37 Complex.
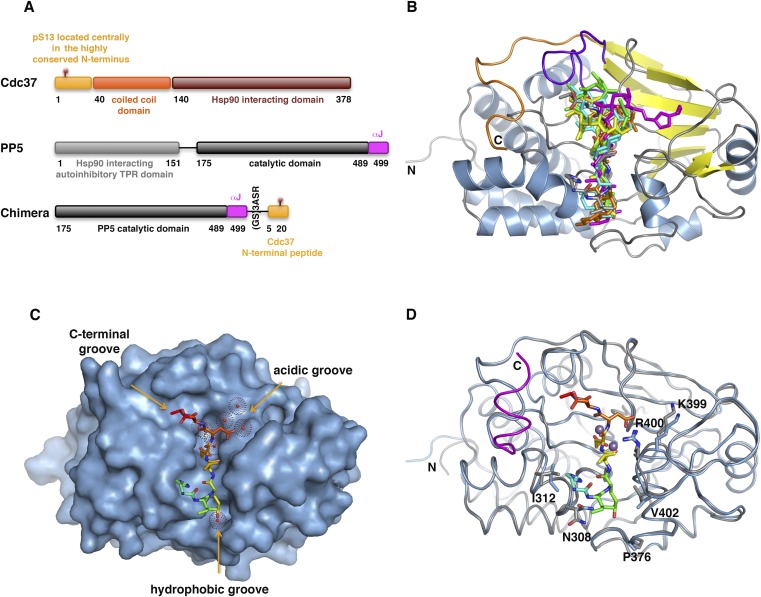
Cocrystallization of the PP5 catalytic domain with Cdc37 peptides, in which the phosphomimetic mutation Ser13→Glu was introduced to trap the substrate in the phosphatase active site, yielded crystals with either no or very low peptide occupancy. In vivo and in vitro PP5-mediated dephosphorylation of phospho–Cdc37-Ser13 requires Hsp90 to act as both a PP5 activator and a PP5 substrate recruiter. To obtain a construct suitable for crystallization, we circumvented the need for Hsp90 by using a chimera construct in which the TPR domain of PP5 was deleted and a peptide comprising residues 5–20 of Cdc37, including the Ser13→Glu mutation, was covalently linked to the catalytic domain of PP5 through a flexible linker of nine residues (Fig. S1A). This construct yielded crystals that diffracted to 2.3 Å (Table S1).
Fig. S1.
Construct design and structural analysis for the PP5-Cdc37 chimera (related to Fig. 1). (A) Schematic representation of human Cdc37, PP5, and the chimeric construct used to determine the crystal structure. (B) Structural alignment of the Cdc37-bound PP5 with toxin inhibitor-bound PP1 structures. The PP1 backbone is omitted for clarity, because the phosphatase catalytic domains are highly structurally homologous. The cartoon representation of PP5 is colored as in Fig. 1B. The Cdc37 substrate, okadaic acid (PDB ID code 1JK7), microcystin (PDB ID code 1FJM), Caliculin (PDB ID code 1IT6), Nodularin-R (PDB ID code 3E7A), and Tautomycin (PDB ID code 3E7B) are shown as gray, cyan, green, magenta, yellow, and pink sticks, respectively. (C) Surface representation of the PP5 catalytic domain. Cdc37 substrate (sticks) binds in the hydrophobic groove and extends toward the C-terminal groove. The hydrophobic, acidic, and C-terminal grooves are highlighted. (D) Least squares fit of the phosphate-bound catalytic domain (PDB ID code 1S95) in gray, with the chimera structure in blue. The αJ helix in 1S95 is highlighted in magenta. The residues that show the greatest change in position are highlighted in stick format. The N and C termini are labeled.
Table S1.
Data collection and refinement statistics for the PP5-Cdc37 chimera structure
| Crystal parameters | PP5-Cdc37 chimera |
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 42.31, 65.16, 132.12 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 46.39–2.27 (2.34–2.27)* |
| Rmerge | 0.164 (0.657) |
| I/σI | 9.1 (2.9) |
| Completeness (%) | 95.3 (89.5) |
| Redundancy | 5.3 (4.3) |
| Refinement | |
| Resolution (Å) | 2.3 |
| No. of reflections | 16,806 |
| Rwork/Rfree | 0.168/0.235 |
| No. of atoms | |
| Protein | 2,626 |
| Ligand/ion | 14/2 |
| Water | 105 |
| B factors | |
| Protein | 18.1 |
| Ligand/ion | 29.1/8.7 |
| Water | 19.0 |
| rmsds | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 1.63 |
The values for the highest resolution shell are shown in parentheses.
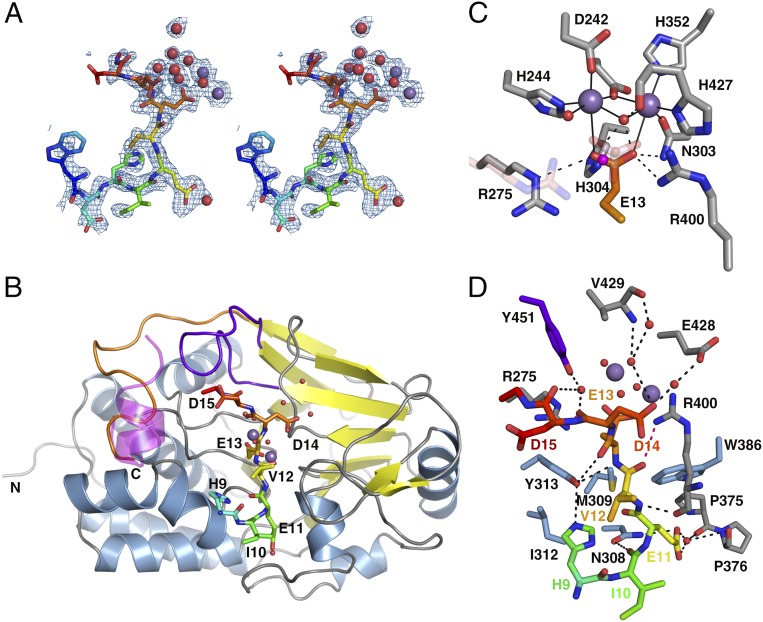
In the resulting structure, there is clear electron density for the Cdc37 peptide from Trp7 to Asp15 (Fig. 1A) bound in an extended conformation across the dimetallic active site of PP5 (Fig. 1B). The entire catalytic domain of PP5 is also clearly defined in the electron density, with the exception of residues 491–499 (see below) and several side chains on the surface of the protein remote from the catalytic center. The overall orientation of the substrate peptide is identical to that of the microcystin (13), okadaic acid (14), nodularin, and tautomycin (15) classes of PPP toxin inhibitors (Fig. S1B). Three putative substrate binding clefts radiate from the catalytic center of the PPP family member PP1 (16). In PP5, two of these are occupied by Cdc37. Substrate residues at the N-terminal end of S13E are in the “hydrophobic groove,” whereas the C-terminal end of the substrate occupies the “C-terminal groove” (Fig. S1C).
Fig. 1.
Structure of Cdc37 phosphomimetic substrate bound in the PP5 catalytic cleft. (A) Stereo image of the electron density map contoured at 1σ for the Cdc37 phosphomimetic peptide. The peptide is colored from blue (Trp7) to red (Asp15). Water molecules (red spheres) and Mn2+ ions (purple spheres) are shown. (B) The Cdc37-bound PP5 catalytic domain. Cdc37 is shown as sticks (colored as in A); PP5 is in cartoon representation. Cdc37-Trp7 and Asp8 do not make contact with the catalytic domain and are omitted for clarity. Sheets, helices, and loops are colored yellow, blue, and gray, respectively; the β12–13 loop is purple, and the PP5-specific C terminus is orange. The location of the αJ helix from the apo PP5 catalytic domain structure (PDB ID code 1S95) is shown in transparent magenta generated by an rms fit of 1S95 and the chimera structure. The rest of 1S95 is omitted for clarity (Fig. S1D shows the complete fit). (C) Interactions between residues of the catalytic site of PP5 (gray) and the phosphomimetic Glu13 of Cdc37 (orange), including water molecules (red) and Mn2+ ions (purple). The new water molecule found in the phosphomimetic structure is colored magenta. Differences in the PO43−-bound apo structure (PDB ID code 1S95) are shown in transparent pink. (D) Hydrogen bonds (black dashed lines) between the Cdc37 peptide and PP5 (colored as in B). The catalytic residues interacting with E13 are omitted for clarity. Those hydrogen bonds involving the catalytic residues R275 and R400 are shown in pink.
There are no significant global structural changes in the PP5 catalytic domain on binding the Cdc37 substrate peptide. The rmsd between the Cdc37- and PO43−-bound [Protein Data Bank (PDB) ID code 1S95] (17) structures is 0.391 Å (Fig. S1D). With the exception of the N and C termini of the domain and regions involved in crystal packing, the principal differences in main and side chain orientation are around the substrate binding cleft, including Asn308, Ile312, Pro376, Gly401, and Val402, all of which provide either van der Waals or hydrogen bond contacts with the peptide. The largest difference between the apo and holo crystal structures is the absence of density for the αJ helix of the PP5-specific C-terminal subdomain. This disorder of the helix may be a result of the design of the chimera, because both the αJ helix and the substrate are parallel to each other (Fig. 1B). Nonetheless, the location of this subdomain in the PO43−-bound enzyme is not compatible with the direction of the substrate peptide downstream of Asp15, suggesting that the αJ helix plays a role in regulation of PP5 activity, not only through stabilizing the autoinhibitory state of the TPR domain (6), but also by steric exclusion of the substrate.
The phosphomimetic residue S13E is bound deep in the active site of the PP5 catalytic domain, and the carboxylate group occupies the same location as the bound phosphate in the holo structure (Fig. 1C). A new water molecule fills the cavity left by one of the missing oxygen atoms of the phosphate. The conformations of almost all PP5 residues coordinating Cdc37-S13E, the active site water molecules, and the Mn2+ ions are unchanged compared with the phosphate-bound holo structure (17). The exception is Arg275, for which the guanadinium group has flipped. The conformation of the Cdc37 peptide is, therefore, that of a trapped substrate, because all residues and active site waters are in orientations that are suitable for the inline nucleophilic attack proposed for the catalytic mechanism (17).
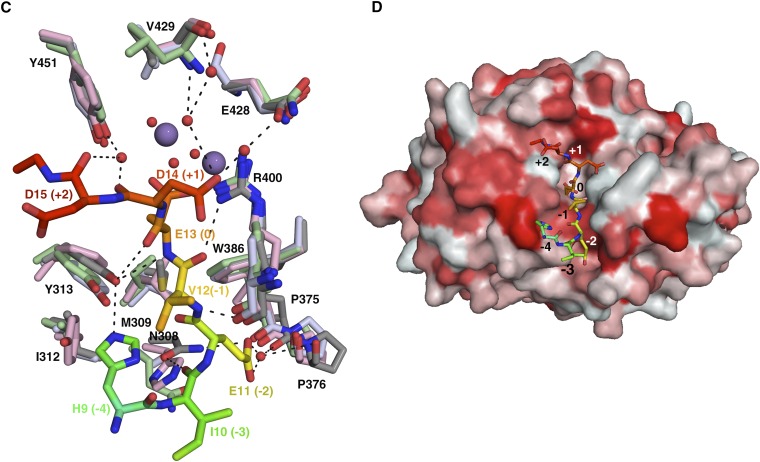
The majority of H bonds that determine the substrate conformation are from the substrate backbone amide groups, with all residues, except His9 and Glu11, making contributions (Fig. 1D). In the phosphatase domain, these interactions are contributed by PP5-Arg275, Asn308, Tyr313, Arg400, and Tyr451. Of these residues, Arg275 and Arg400 are also involved in the catalytic mechanism, and Tyr451 is part of the β12–13 loop, which has a conformation that is sensitive to the binding of some toxin inhibitors (16). The main chain of the phosphomimetic Cdc37-S13E is coordinated by both its amino and carbonyl groups to the hydroxyl of PP5-Tyr313.
The peptide adopts an “in–out–in–out” conformation, with the side chains of His9, Glu11, and Glu14 orienting toward and Ile10, Val12, and Asp15 orienting away from the surface of the PP5 catalytic domain. The side chains of His9, Glu11, and Asp14 make direct or water-mediated H bonds to PP5. Cdc37-His9 takes a highly unusual rotamer conformation to make a direct H bond with the hydroxyl group of Tyr313. Cdc37-Glu11 is buried in a deep groove and makes water-mediated hydrogen bonds with the backbone carbonyl groups of PP5-Pro375 and Pro376. The carboxylate group of Asp14 makes water-mediated interactions with the main chain of Val429 and the side chain of Glu428 within the PP5 acidic groove. Significant van der Waals interactions are contributed from PP5-Trp386, Met309, Ile312, and Asn308.
Conservation Analysis of the PP5-Cdc37 Interaction.
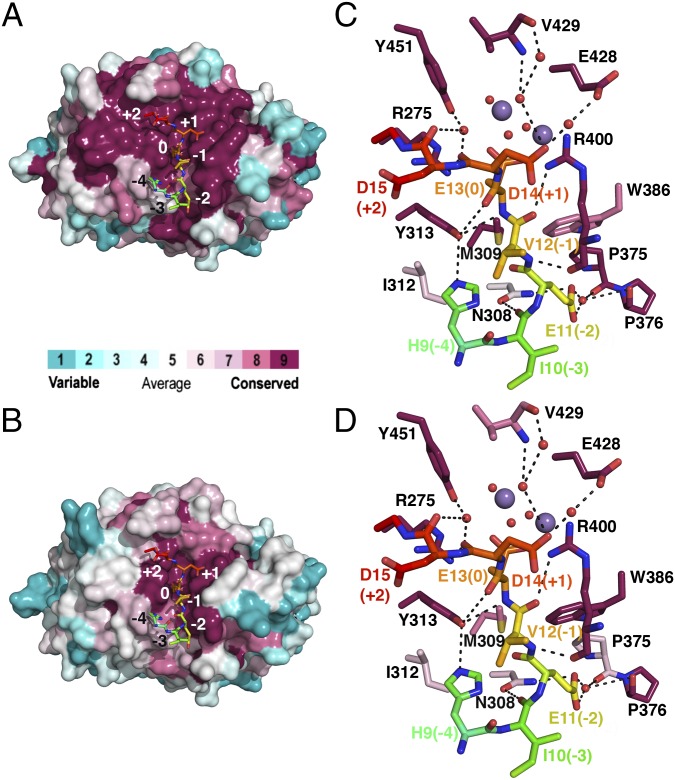
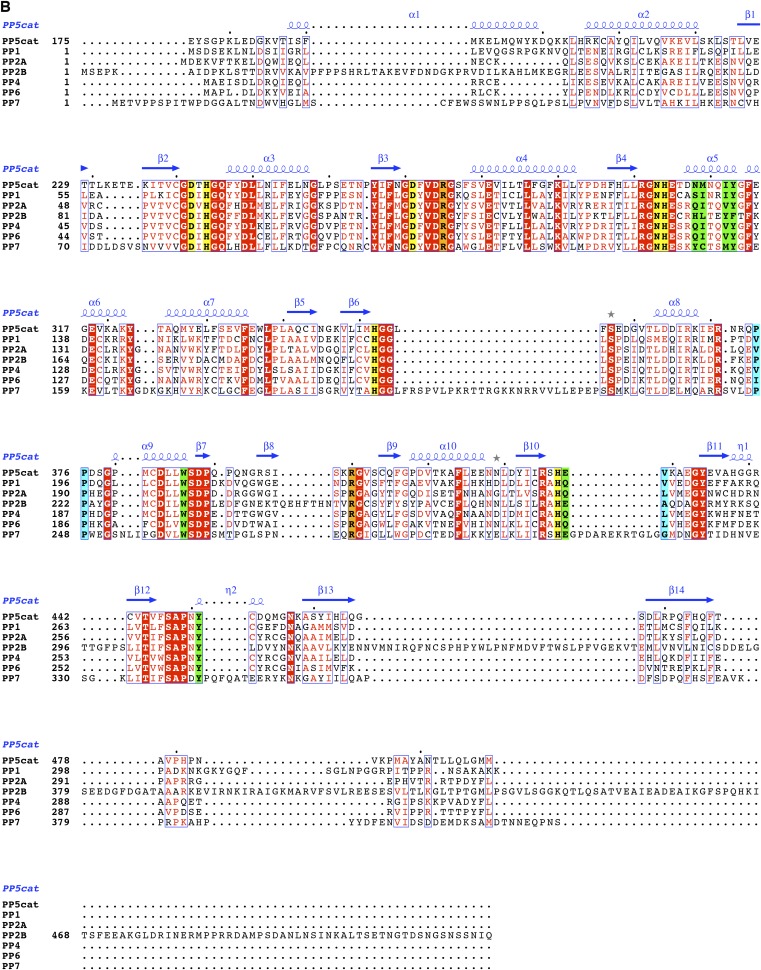
In addition to almost complete conservation of the catalytic residues within the PPP family of serine/threonine phosphatases (3), the substrate binding cleft in PP5 is strongly conserved both among PP5 homologs (Fig. 2A) and between PPP family members (Fig. 2B). A detailed analysis reveals that the interactions that dictate Cdc37 backbone conformation involve either PP5 main chain groups, with orientation that is likely to be invariant in PP5 homologs, or side chains, with greater than 98% sequence identity (of 69 homologs analyzed) (Fig. 2C, Fig. S2A, and Table S2). In addition, because sequence and structural conservation of PPP catalytic domains is extremely high (Fig. S2 B and C), interactions involving backbone groups are likely to be conserved not only among PP5 homologs but also, between PPP family members (Fig. 2D). This striking conclusion also holds for the side chains that interact with the substrate main chain: these residues have greater than 79% sequence similarity among 87 PPP family members analyzed (Fig. 2D, Fig. S2C, and Table S2). The only exception to this strong conservation is PP5-N308: although there is a predominant ability of the residue at this position to form hydrogen bonds (94% of PP5 homologs and 99% of PPP family members), Asn308 is not highly conserved within the PPP superfamily or even the PP5 family (Fig. S2 A and B). Therefore, there is a greater likelihood of differences in the backbone conformation of the substrate in the −3 position. We conclude from this analysis that it is possible that many PPP substrates that occupy the hydrophobic and C-terminal grooves will adopt the main chain conformation seen for Cdc37, at least from the −2 to +2 positions.
Fig. 2.
Substrate main chain conformation is likely to be conserved throughout PP5 and PPP family members. Surface representation of (A and B) PP5 and (C and D) Cdc37 substrate binding residues colored according to sequence conservation using ConSurf (Methods). A and C show conservation between PP5 homologs; B and D show conservation among PPP family phosphatases. Cdc37 substrate is shown as sticks, colored as in Fig. 1A, and labeled from −4 to +2 to identify location relative to phosphomimetic S13E (position 0).
Fig. S2.
Sequence and structural alignment of phosphatase families (related to Fig. 2). Sequence alignment of the catalytic domain of (A) PP5 homologs and (B) PPP family members. Residues that contribute H bonds or van der Waals contacts to Cdc37 through their side chain are colored green, and those that contribute H bonds or van der Waals contacts to Cdc37 through their main chain are colored cyan. Residues involved in interactions at the catalytic site are colored yellow. Arg400 and Arg275, colored orange, both coordinate the substrate and play a role in the catalytic center. The αJ helix is colored magenta. Alignments were generated using ClustalW2 (42) and illustrated with ESPRipt3.0 (45). Sequences used for the PPP family alignment in B are all human and α-isoform, with the exception of PP7 from Arabidopsis. The accession numbers are P62136, PP1A; P67775, PP2Aα; Q08209, PP2Bα; P60510, PP4; P53041, PP5; O00743, PP6; and Q9FN02, PP7. (C) Least squares fit of PP1 (light blue; PDB ID code 4MOV), PP2A (light green; PDB ID code 2IE4), and PP2B (pink; PDB ID code 4F0Z) structures to PP5 (gray). Only residues that line the substrate binding cleft are shown. Hydrogen bonds made between PP5 and Cdc37 (colored rainbow) are shown. Structures with the highest resolution from human PPP superfamily members were used in this analysis. Cdc37 substrate is shown as sticks, colored as in Fig. 1, and labeled from −4 through +2 to identify location relative to S13E (position 0). (D) Hydrophobicity of the PP5 surface from red (most hydrophobic) to white (least hydrophobic). The grooves in which the side chains of Glu11 and Asp14 are bound display intermediate properties. Cdc37 is colored and labeled as in C.
Table S2.
Residue variety (%) by amino acid position generated using ConSurf for residues lining the substrate binding cleft (Methods)
| PP5 amino acid | Ala | Cys | Asp | Glu | Phe | Gly | His | Ile | Leu | Met | Asn | Pro | Gln | Arg | Ser | Thr | Val | Trp | Tyr | Maximum, % identity/similarity |
| PP5 family* | ||||||||||||||||||||
| R275 | 100 | R 100.000 | ||||||||||||||||||
| N308 | 7.246 | 1.449 | 62.319 | 4.348 | 18.841 | 4.348 | 1.449 | N 62.319 | ||||||||||||
| M309 | 100 | M 100.000 | ||||||||||||||||||
| I312 | 49.275 | 1.449 | 46.377 | 2.899 | I/M/V 98.511 | |||||||||||||||
| Y313 | 100 | Y 100.000 | ||||||||||||||||||
| P375 | 1.449 | 98.551 | P 98.551 | |||||||||||||||||
| P376 | 1.449 | 98.551 | P 98.551 | |||||||||||||||||
| W386 | 1.449 | 98.551 | W 98.551 | |||||||||||||||||
| R400 | 100 | R 100.000 | ||||||||||||||||||
| E428 | 100 | E 100.000 | ||||||||||||||||||
| V429 | 1.471 | 1.471 | 97.059 | V 97.059 | ||||||||||||||||
| Y451 | 100 | Y 100.000 | ||||||||||||||||||
| PPP superfamily† | ||||||||||||||||||||
| R275 | 100 | R 100.000 | ||||||||||||||||||
| N308 | 20.69 | 5.747 | 39.08 | 31.034 | 1.149 | 1.149 | 1.149 | Q 39.080 | ||||||||||||
| M309 | 1.149 | 67.816 | 20.69 | 10.345 | I/L/M 98.851 | |||||||||||||||
| I312 | 34.483 | 5.747 | 39.08 | 20.69 | I/M/V 79.31 | |||||||||||||||
| Y313 | 20.69 | 79.31 | Y 79.310 | |||||||||||||||||
| P375 | 14.943 | 29.885 | 55.172 | I/V 70.115 | ||||||||||||||||
| P376 | 100 | P 100.000 | ||||||||||||||||||
| W386 | 100 | W 100.000 | ||||||||||||||||||
| R400 | 100 | R 100.000 | ||||||||||||||||||
| E428 | 32.184 | 67.816 | Q/E 100.00 | |||||||||||||||||
| V429 | 20.69 | 1.149 | 40.23 | 37.931 | L/V 78.161 | |||||||||||||||
| Y451 | 100 | Y 100.000 |
Numbering is the same as for PP5 catalytic domain.
In total, 69 homologs were identified using BLAST with identity greater than 56% throughout both the TPR and catalytic domains.
In total, 87 homologs were identified using UniProt (>10 homologs for each of the PPP family members). The exception is PP7, for which only Arabidopsis was included.
The Cdc37 side chains Glu11 and Asp14 at the −2 and +1 positions, respectively, both contribute water-mediated hydrogen bonds with PP5 and sit in grooves that are conserved and spacious relative to the size of the side chain itself. Analysis of the hydrophobic nature of these grooves (Fig. S2D) and crystal structures of the toxin inhibitors of the PPP family illustrate that both polar and nonpolar groups can be found in each of these sites (13–15). Therefore it is likely that PP5, and indeed members of the PPP superfamily in general, can accommodate substrates with side chains of different sizes and properties at the −2 and +1 positions and that water molecules will play a role in molding the substrate to the enzyme.
Mutational Analysis of PP5–Substrate Interactions in Vitro and in Vivo.
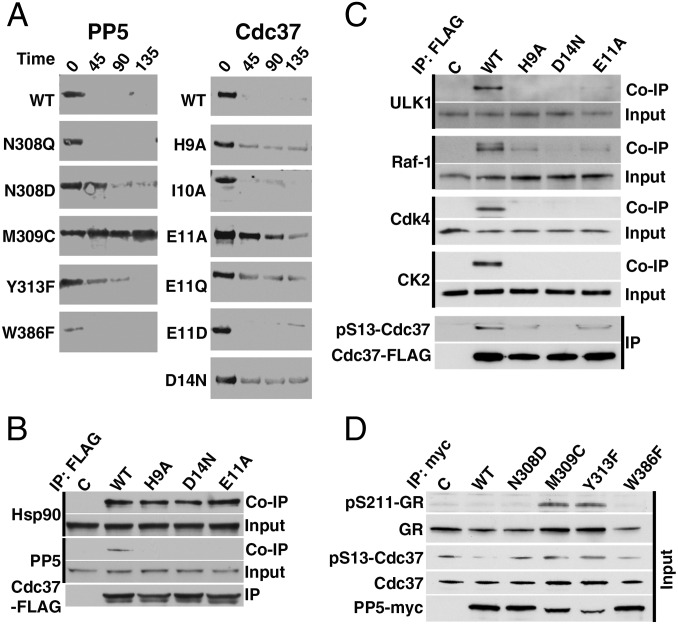
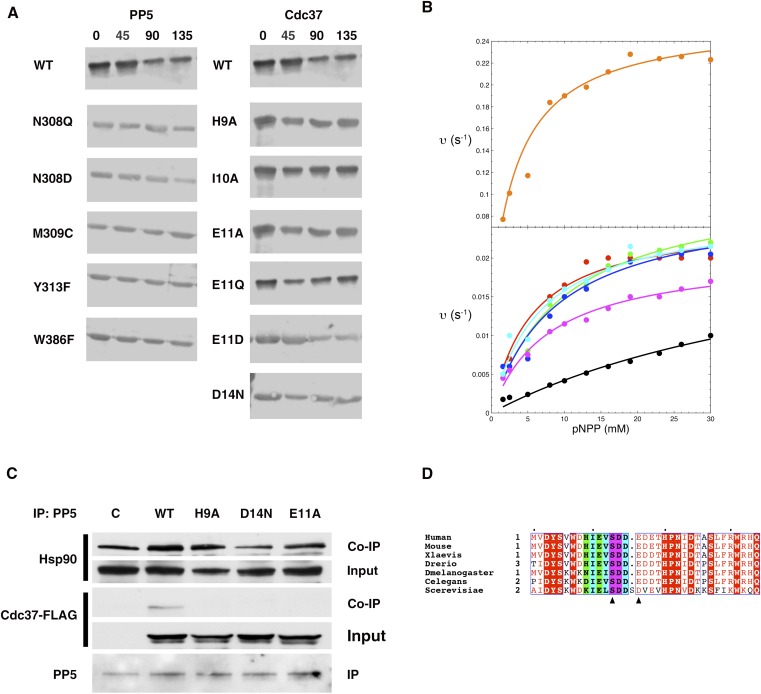
To test our structure- and conservation-based predictions of the relative importance of residues in both the enzyme and the substrate for determining specificity, we introduced a series of mutations in full-length PP5 and Cdc37 and assessed their activity in a dephosphorylation reaction in the presence or absence of Hsp90 (Fig. 3A and Fig. S3A). PP5 residues previously identified as involved in coordination of phosphate or Mn2+ ions or proposed to have a role in the mechanism of dephosphorylation were not modified. To confirm that the chosen mutations did not disrupt the catalytic mechanism of PP5, all PP5 mutants were initially tested for their ability to dephosphorylate para-nitrophenyl phosphate, a commonly used small molecule for assaying nonspecific phosphatase activity (Fig. S3B). With the exception of W386F (hyperactive) and Y313F (approximately one-half as active as the WT), all other mutants displayed similar kinetics of dephosphorylation to the WT, indicating that the mutations did not cause significant structural disruption to the active site.
Fig. 3.
Structure-based mutations reveal the functional importance of specific interactions between PP5 and substrate in vitro and in vivo. (A) Dephosphorylation of phospho–Cdc37-Ser13 in the context of purified, full-length WT and indicated mutants of Cdc37 and PP5 in the presence of Hsp90. Activity was assessed using a phosphospecific antibody over time (minutes). (B) HEK293 cells were transiently transfected with empty plasmid (C), WT Cdc37-FLAG, or indicated mutants. Cdc37-FLAG proteins were subject to immunoprecipitation (IP). Co-IP of Hsp90 and PP5 was examined by immunoblotting. (C) HEK293 cells were transiently transfected with C, WT Cdc37-FLAG, or indicated mutants. Cdc37-FLAG proteins were subject to IP, and the level of phospho-Ser13 was examined by immunoblotting. Co-IP of CK2 and Hsp90 client kinases CDK4, Raf-1, and ULK1 was also examined by immunoblotting. (D) Total protein lysates were prepared from HEK293 cells transiently transfected with empty plasmid (C), WT PP5–c-myc, or indicated mutants. Cdc37, phospho–Ser13-Cdc37, GR, and phospho GR-211 protein levels were examined by immunoblotting.
Fig. S3.
Related to Fig. 3. (A) Dephosphorylation of phospho–Cdc37-Ser13 by PP5 for purified WT and indicated mutants of Cdc37 and PP5 in the absence of Hsp90. Activity was assessed using a phosphospecific antibody over time (minutes). WT control panels (row 1) are the same experiment. (B) Rate of dephosphorylation of the model substrate pNPP by PP5 mutants: W386F, orange; WT, red; N308Q, green; N308D, cyan; M309C, blue; Y313F, magenta; without Hsp90, black. (C) HEK293 cells were transiently transfected with empty plasmid (C), WT Cdc37-FLAG, or indicated mutants. PP5 was subject to immunoprecipitation (IP) using beads conjugated with anti-PP5 antibody, and co-IP of Hsp90 and Cdc37-FLAG was examined by immunoblotting. (D) Sequence alignment of the extreme N terminus of Cdc37. Residues that contribute H bonds to PP5 through their side chain are colored green, residues that contribute H bonds to PP5 through their main chain are colored cyan, and residues that contribute H bonds to PP5 through both are colored magenta. For the remaining residues, those that are completely conserved are shown as white text in red, whereas residues that are strongly conserved are colored red text in white. Residues of the CK2 consensus motif SXXE/D are indicated with triangles. Alignments were generated using ClustalW2 (42) and illustrated with ESPRipt3.0 (45).
Mutations of PP5 residues that are involved in direct coordination (N308D and Y313F) or providing van der Waals interactions (M309C) to the Cdc37 substrate main chain result in loss of activity, indicated by phospho–Cdc37-Ser13 detection using purified proteins in vitro, whereas the conservative mutation N308Q has no effect on Cdc37-Ser13 dephosphorylation (Fig. 3A). For the substrate, all of the changes to residues that interact either directly or through water-mediated interactions with PP5 (H9A, E11A/Q, and D14N) result in loss of dephosphorylation. By contrast, mutation of the conserved residue Ile10 to alanine, which has a side chain that is oriented away from the surface of PP5, has no effect on dephosphorylation. A careful balance of polar and charged groups is essential in directing the PP5-Cdc37 interaction, because the mutations PP5-N308Q and Cdc37-E11D have minimal effect on dephosphorylation, whereas altering the electrostatic environment with the mutations PP5-N308D or Cdc37-E11Q causes loss of activity.
To investigate the consequences of these mutations for the substrate–phosphatase interaction in vivo, FLAG-Cdc37 WT and mutants (H9A, E11A, and D14N) (Fig. 3 B and C) were transiently expressed and immunoprecipitated from HEK293 cells.
Cdc37 mutants that were not dephosphorylated in vitro (H9A, E11A, and D14N) lose their interaction with PP5 in vivo while retaining WT-like interaction with Hsp90 (Fig. 3B). Reciprocal coimmunoprecipitation experiments show that this is a consequence of the direct interaction between PP5 and Cdc37, because the Cdc37 mutations do not alter the interaction between PP5 and Hsp90 (Fig. S3C).
However, loss of interaction with PP5 does not result in an increase of phospho–Cdc37-Ser13. Instead, phosphorylation of Cdc37 is diminished (Fig. 3C). A positive feedback loop between CK2 and Cdc37 regulates the activation of the former and the phosphorylation of the latter (18, 19). His9, Glu11, and Asp14 form part of the highly conserved N-terminal region of Cdc37 (Fig. S3D) that is required for client kinase binding to Cdc37 (9, 20–23). These mutations disrupt this feedback mechanism, diminishing or preventing phosphorylation of Cdc37 by CK2, because they prevent recruitment of CK2 to Cdc37 (Fig. 3C), despite not overlapping with the CK2 consensus sequence (SXXE/D) (Fig. S3D) or residues implicated in direct association with the client kinase B-Raf (9). Consequently, recruitment of ULK1, Raf-1, and Cdk4, known Hsp90 client kinases, is also affected, because their association is dependent on Cdc37 phosphorylation (Fig. 3C). These results emphasize the essential nature of the polypeptide sequence surrounding Cdc37-Ser13 in client kinase activation and expand the known residues in this N-terminal region that are essential for productive client kinase chaperoning (9, 20).
Mutations in the catalytic cleft of myc-tagged PP5 (N308D, M309C, Y313F, and W386F) strongly diminish dephosphorylation of phospho–Cdc37-Ser13 in cells compared with myc-tagged WT PP5 (Fig. 3D), as seen in vitro. These mutations also impair PP5’s ability to dephosphorylate phospho–GR-Ser211, another known target of PP5 discussed in detail later (2). These data are in agreement with our previous work, in which overexpression of PP5 in HCT116 colon cancer cells enhanced Cdc37 dephosphorylation (8).
Implications for Hsp90-Dependent Activation of Client Proteins.
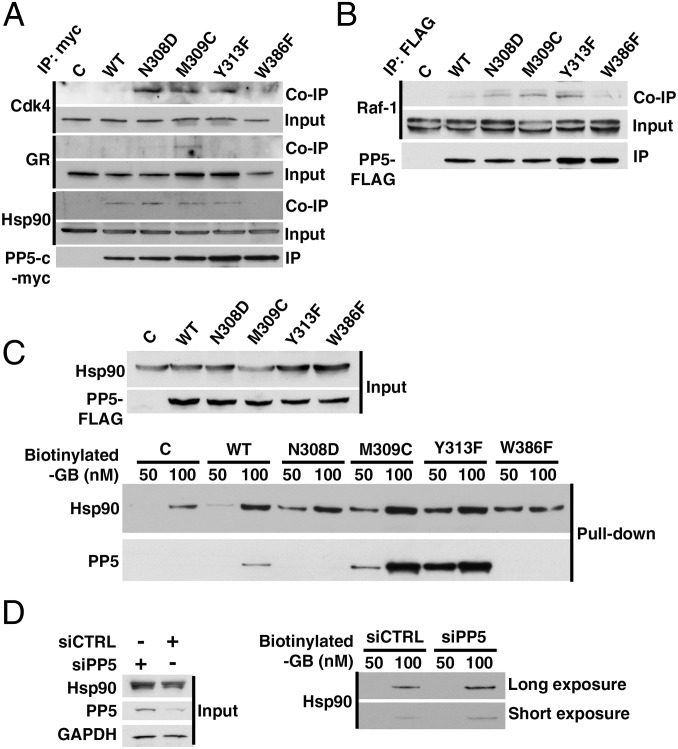
PP5-mediated dephosphorylation of Cdc37 is an essential step in the activation of Hsp90-dependent kinases. Dysregulation of this step by overexpression of PP5 with concomitant reduction in phospho–Cdc37-Ser13 results in reduced Raf-1 activity in HCT116 cells. In yeast, both the overexpression and deletion of the ortholog, Ppt1, causes reduced v-Src activity (8). To understand how Cdc37 dephosphorylation affects the activation mechanism of Hsp90-dependent kinases, we investigated the impact of PP5 mutants on chaperoning of the kinase clients Cdk4 and Raf-1. The PP5 mutants were immunoprecipitated, and their interaction with Cdk4 and Hsp90 was assessed by immunoblotting. Cdk4 did not coimmunoprecipitate with WT PP5, presumably because of its dynamic interaction within the Hsp90 chaperone cycle. However, the N308D, M309C, and Y313F PP5 mutations that prevent the dephosphorylation of Cdc37 in vitro and in vivo trap the kinase in the Hsp90-PP5 complex (Fig. 4A). We also observed similar results with PP5 mutants’ interaction with Raf-1 (Fig. 4B). Coimmunoprecipitation of Hsp90 with PP5 mutants at steady state shows that they bind to Hsp90 with similar affinity, with the exception of the W386F mutant, which is hyperactive, resulting in a transient interaction (Fig. 4A). These results provide mechanistic details that elaborate our previous work (8), showing that kinase release from the chaperone complex is critically dependent on Cdc37 dephosphorylation by PP5.
Fig. 4.
Inactive mutants of PP5 cause client proteins to be stalled in the Hsp90 chaperone complex in vivo. (A) Empty plasmid (C), WT PP5–c-myc, or indicated mutants were transiently transfected in HEK293 cells. PP5 proteins were subject to immunoprecipitation (IP). Co-IP of Hsp90, CDK4, and GR was examined by immunoblotting. (B) IP of C, PP5-FLAG WT, or mutants from transiently transfected HEK293 cells was immunoblotted for co-IP of Raf-1. (C) C, PP5-FLAG WT, or mutants were transiently expressed in HEK293 cells. Lysates were incubated with 50 or 100 nM biotinylated ganetespib (GB). Using streptavidin beads, Hsp90 and PP5 bound to the drug were isolated and immunoblotted. (D) HEK293 were transiently transfected with control and PP5 siRNA. Lysates were incubated with indicated amounts of biotinylated GB. Using streptavidin beads, Hsp90 bound to the drug was isolated and immunoblotted.
Hyper- or Hypoactivity of PP5 Mutants Enhances Hsp90 Binding to Drug.
It is well-established that increased binding of Hsp90 to its inhibitors is associated with sensitivity of cells to Hsp90 drugs (24, 25). We have previously shown that overexpression of the yeast PP5 ortholog (Ppt1) sensitizes cells to the Hsp90 inhibitor geldanamycin (8). We reasoned that PP5 mutants might influence Hsp90 inhibitor association; therefore, we expressed FLAG-PP5 WT and mutants in HEK293 cells followed by biotinylated ganetespib (GB) pulldown experiments. Our data showed that the hypoactive (N308D, M309C, and Y313F) and hyperactive (W386F) PP5 mutants significantly enhanced Hsp90 binding to biotinylated GB (Fig. 4C). Although PP5 mutants bound with similar affinity as the WT PP5 to Hsp90 at steady state (Fig. 4A), these mutants differentially impacted Hsp90 binding to its inhibitor GB. Additionally, after Hsp90 was bound to biotinylated GB, it formed different complexes with the PP5 mutants (Fig. 4C). We obtained further evidence to support our hypothesis by down-regulating PP5 using siRNA (Fig. 4D and Fig. S4). These data suggest that the activity of PP5 is a determining factor for Hsp90 affinity to its inhibitor.
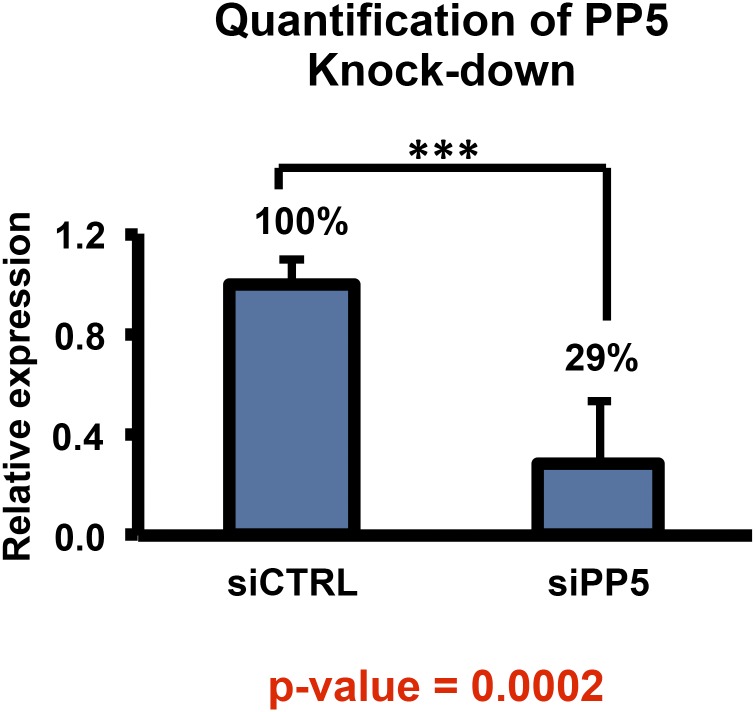
Fig. S4.
Related to Fig. 4. HEK293 cells were transiently transfected with control or PP5 siRNA. RNA was isolated from cells, reverse-transcribed, and DNA quantitated. Quantification is reported as relative expression of mean ± SE. ***P value < 0.0005.
Implications for PP5 Specificity.
Having established that particular residues in the catalytic cleft are necessary for Cdc37 recognition, the question arises whether these residues are able to confer specificity of phosphatase activity (i.e., does their mutation have differential effects on different substrates?). PP5 also dephosphorylates GR, another Hsp90 client, at Ser203, Ser211, and Ser226 and subsequently, controls its activity and nucleocytoplasmic translocation (2). To examine specificity, we, therefore, investigated the consequences of mutations in the PP5 catalytic cleft on its ability to dephosphorylate GR-Ser211. Mutations of PP5-M309C and Y313F prevent dephosphorylation of GR-Ser211 as observed with Cdc37 (Fig. 3D). In addition, an equivalent trapping of GR on Hsp90-containing PP5 complexes is observed for the M309C mutation (Fig. 4A). The Y313F mutation does not result in a trapped GR complex, but because this mutant has an activity profile that is distinct from the other mutants (Fig. S3B), this difference may be a result of unpredicted structural changes in the vicinity of the substrate binding site. However, unlike the case for Cdc37, the PP5-N308D mutant had no effect on dephosphorylation of GR-Ser211. In the PP5-Cdc37 structure, the side chain of Asn308 provides interactions with the backbone carbonyl of Cdc37-Ile10 and the carboxylate of Glu11 as well as van der Waals contacts to Glu11. The equivalent position for GR phospho-Ser211 substrate is Asn209 (sequence TNEpSPWR); therefore, the PP5-N308D mutant may provide similar electrostatic interactions to WT PP5. In agreement with our structural analysis, the contrasting effect of this mutation on different substrates confirms that the binding groove on PP5 for the substrate −2 position provides a degree of specificity, while providing an environment that allows a variety of side chains to bind (Fig. 1D and Fig. S2D).
Discussion
The structure presented here defines the interaction of the Cdc37 substrate with the PPP Ser/Thr phosphatase PP5. The conformation of the substrate is primarily dictated by interactions between its backbone and residues of PP5 that line the active site. Interactions between the side chains of the Cdc37 substrate and PP5 are largely water-mediated and within spacious pockets that form part of the Y-shaped channel that surrounds the catalytic site. This channel is highly conserved in sequence and structure throughout the PPP superfamily, and together, the use of the main chain of the substrate and the versatility provided by water-mediated interactions to accommodate a range of side chains reveal how different members of this superfamily can accommodate a multitude of unrelated substrates with highly divergent sequences. In addition, these two factors suggest that the substrate conformation observed here may provide a template for many substrates that occupy the hydrophobic and C-terminal grooves of the PPP superfamily.
Our structure illustrates the fundamental difference in the mechanism of substrate binding at the catalytic sites of kinases and phosphatases and shows how this is achieved on an atomic level. For kinases, the sequence of the substrate is critical in ensuring specificity through highly conserved interactions dictated by the substrate side chains (26, 27), whereas Ser/Thr PPPs allow sequence plasticity by providing an environment in which a variety of side chains can be accommodated within a conserved substrate backbone conformation.
Although this sequence plasticity is a key feature of PPP substrate recognition, our biochemical and cell biology data indicate that, contrary to the current widespread assumption in the field, the catalytic subunit can confer some degree of local specificity. Nonetheless, regulatory subunits are still fundamental in controlling the primary specificity of a Ser/Thr PPP through their role in recruiting substrates and increasing their local concentration; thus, phosphatases other than PP5 cannot dephosphorylate Cdc37, because they are unable to form the complete tripartite interaction required for efficient dephosphorylation (8).
Our results delineate mechanistic details of one step in the Hsp90-dependent kinase activation cycle for Cdk4 and Raf-1. Although the requirement for PP5-mediated dephosphorylation of Cdc37 was previously known to be essential for activation of several Hsp90 client kinases (8), its role at a molecular level was not understood. The results presented here identify this dephosphorylation step as a requirement for release of these clients from the Hsp90 chaperone. Previous results suggest that association of PP5 with Hsp90 chaperone complexes is accompanied by structural changes, because within a purified Hsp90-Cdc37-Cdk4 complex, the phospho-Ser13 Cdc37 substrate is inaccessible to dephosphorylation by the nonspecific λ-phosphatase but readily accessible to dephosphorylation by PP5 (8). This conclusion is further supported by recent results indicating that residues in the vicinity of the Ser13 are responsible for binding client kinases (9, 22, 23) where it is likely to be buried. A conformational change would, therefore, be required to allow PP5 access to its substrate. It may be these conformational changes rather than the absence of the phosphorylation itself that are the trigger for client release, because eventual dephosphorylation by λ-phosphatase, which occurs after prolonged incubation with the complex, does not cause release of the Cdk4 client from the Hsp90-Cdc37 complex (8).
Although phosphorylation of Cdc37 Ser13 is essential for client kinase activation in vivo (11, 12) and in vitro (28), its mechanistic role remains enigmatic. In the cell, the interaction of client kinase with Cdc37 and its subsequent recruitment to Hsp90 are likely to always occur with the phospho-Ser13 Cdc37, because CK2 is constitutively active. Phospho-Ser13 Cdc37 has a more stable and compact conformation than the WT (29), in which the phospho-Ser13 is accessible to dephosphorylation by both calf intestine alkaline phosphatase (29) and λ-phosphatase (8). However, at least for the kinase B-Raf, this conformation is not a requirement for either client vs. nonclient recognition (9) or subsequent association with Hsp90 as a stable complex between Hsp90, Cdc37, and B-Raf can be formed from the individual proteins in vitro in the absence of phosphorylation (22, 23), although phospho-Ser13 enhances its stability (22). These observations are consistent with recent data indicating that the primary interaction of Cdc37 with B-Raf is through a distinct C-terminal domain of Cdc37 and remote from this site of phosphorylation (9, 23). Nonetheless, our results augment the set of known, nonoverlapping residues residing in a relatively short stretch of the Cdc37 polypeptide that encompasses Ser13 responsible for interactions that regulate Cdc37 chaperone function, namely CK2 phosphorylation (11, 12), client kinase recognition in vitro (9), client kinase complex formation in vivo (20), and PP5 dephosphorylation.
PP5-mediated dephosphorylation of Cdc37 is one of a number of modifications that controls progression of the chaperone cycle by affecting either cochaperone or client association. Phosphorylation of Y197 on Hsp90 dissociates Cdc37 from Hsp90 immune complexes, whereas phosphorylation of Y313 stimulates association with Aha1 (21), the cochaperone required for the enhancement of Hsp90’s ATPase activity and completion of one round of the chaperone cycle (30). Indeed, these two cochaperones are not found in the same immune complexes (21), highlighting the degree of control imposed by these posttranslational modifications (PTMs).
We have identified the trapping of GR, a Cdc37-independent Hsp90 client, on the Hsp90 chaperone when PP5 activity is abrogated, showing that PP5 activity exerts influence on the chaperone cycle regulation beyond kinases alone. Serine/threonine phosphorylation of Hsp90 itself is, in general, found to result in reduced affinity with clients (31–33). Client kinase release is also triggered by tyrosine phosphorylation of Cdc37 on Y298 by the Src family kinase YES, and tyrosine phosphorylation of Hsp90 on Y627 dissociates Cdk4 (21). The latter modification also dissociates Aha1 and PP5 from Hsp90 (21) and as such, may be a mechanism for terminating one cycle of chaperone activity. Together, these events emphasize the intricacy of the client activation process and the many layers of control provided by serine/threonine and tyrosine phosphorylation and its reversal.
Finally, this study shows that both hyper- and hypoactivity of PP5 mutants enhances Hsp90 binding to its inhibitor GB. Previous works have shown that PTM of cochaperones influences their activity. It remains to be seen whether PP5 activity is also regulated by PTMs. Targeting the potential enzymes that catalyze those PTMs may influence cell sensitivity to Hsp90 inhibitors.
Methods
Details are in SI Methods. Briefly, the chimera protein, comprising the catalytic domain of PP5 (residues 175–499) and a peptide comprising residues 5–20 of Cdc37, with the mutation S13E joined by a nine-residue flexible linker was generated by PCR (Table S3). The protein was purified to homogeneity using standard chromatographic techniques and crystallized using hanging drop vapor diffusion. Data were processed using XDS (34) and Scala (35). The structure was solved by molecular replacement of PDB ID code 1S95 using Phaser (36) and refined using Phenix Refine (37) with manual rebuilding in Coot (38). Data collection and refinement statistics are summarized in Table S1. In vitro dephosphorylation was assessed using antiphospho-Serine 13 antibody (Sigma). Immunoprecipitation assays in transiently transfected HEK293 cells were carried out using anti-FLAG (Sigma) or anti-myc (ThermoScientific) antibody-conjugated beads. Biotinylated GB (Synta Pharmaceuticals) and streptavidin agarose (ThermoScientific) were used for drug binding assays. siRNA knockdown of PP5 (Origene) was carried out using standard methods.
Table S3.
Primer sequences
 |
For the PP5-Cdc37 chimera construct (primers 1–3), PP5 sequence is in bold, the linker sequence is in green, and Cdc37 sequence is italic. Restriction sites are underlined. For immunoprecipitation experiments (primers 4–22), mutated sequences are in red. Epitope sequences are highlighted in blue.
Note Added in Proof.
While this article was in print the cryo-EM structure of the Hsp90-Cdc37-Cdk4 complex was published showing that Cdc37-phospho-Ser13 contributes towards the coordination of a specific conformation of the Cdc37 N-terminal domain and its position relative to Hsp90 (39). This structure further supports our conclusion that a structural change is required for PP5-mediated dephosphorylation of Cdc37 in the context of the client-loaded Hsp90 chaperone complex and that this may be the trigger for client kinase release from the Hsp90-chaperone complex.
SI Methods
Protein Expression and Purification.
The human PP5-Cdc37 chimera was cloned in two steps. A linker [sequence (GS)3ASR] was added 3′ to the PP5 catalytic domain, comprising residues 175–499, using PCR and subsequently, used as a template for the further addition of residues 5–20 of Cdc37 including the S13→E mutation, 3′ to the linker. The chimera gene was subcloned as a BamHI–XhoI fragment into a modified pET-44 vector (Novagen) with an N-terminal Tobacco Etch Virus-cleavable His6-NusA tag. The PP5-Cdc37 chimera was purified using a DEAE-Sepharose Anion Exchange Column (GE Healthcare) followed by cleavage of the tag with TEV protease. The protein was reloaded onto the anion exchange column to separate the cleaved protein from the tag and further purified using a Superdex 200 HR16/60 Gel Filtration Column (GE Healthcare). Full-length human Cdc37 was cloned into the pET-28a vector (Novagen) with a C-terminal His6 tag and purified using Talon Metal Affinity Resin (Clontech) followed by PreScission Protease Cleavage, Glutathione Sepharose Resin (GE Healthcare), and finally, Superdex 200 HR16/60 Gel Filtration (GE Healthcare). Phospho–Cdc37-Ser13 was generated by coexpression of Cdc37 with GST-tagged Zea mays CK2α in pGEX 6P-1 and purified as described for WT Cdc37. Phosphorylation was confirmed by MS and Western blot using antiphospho-Ser13 Cdc37 (Sigma). Human PP5 (residues 16–499) was purified as previously described (6). Full-length WT Hsp90 with an N-terminal His6 tag in a pRset-A vector was purified as previously described (40). All constructs were expressed in Escherichia coli BL21 star (DE3) cells (Invitrogen).
PP5 and Cdc37 mutants were created using the QuikChangeII site-directed mutagenesis method (Stratagene) and confirmed by DNA sequencing. Mutant proteins were expressed and purified using the same method as in WT proteins.
Crystallization, Data Collection, and Structure Determination.
The PP5-Cdc37 chimera crystals were grown by hanging drop vapor diffusion at 4 °C by mixing the protein at 12 mg/mL in a buffer containing 100 mM NaCl, 20 mM Tris, pH 8, and 2% (vol/vol) glycerol with an equal volume of well buffer containing 18% (vol/vol) PEG 10,000, 8% (vol/vol) ethylene glycol, 0.1 M Hepes, pH 7.5, and 20 mM hexamine cobalt chloride. Crystals were cryoprotected in well buffer supplemented with 15% (vol/vol) ethylene glycol. Data were collected at the Diamond light source (beamline I24; wavelength = 0.968600 Å). Data were processed using XDS (34) and Scala (35). The chimera structure was solved by molecular replacement using Phaser (36), with the PP5 catalytic domain (PDB ID code 1S95) as a search model. Clear difference density was observed for the phosphomimetic Glu13 of Cdc37. Additional rounds of refinement using Phenix Refine (37) allowed the remainder of the peptide to be built manually into difference density using Coot (38). There is clear electron density for the main chain of residues 175–490 of PP5 and residues 7–15 of the Cdc37 peptide. The αJ helix of PP5 (residues 491–499), the linker, and the N and C termini of Cdc37 (residues 5–6 and 16–20) are not seen in the maps; 95.3% of residues are in the most favored region of the Ramachandran plot (only two residues, PP5-Phe272 in the core of the protein and Cdc37-His9, are outliers; density for both of these residues is clearly defined).
PP5 Activity Assays.
The phosphatase activity of PP5 WT and mutants was assayed by monitoring hydrolysis of the artificial substrate para-nitrophenyl phosphate (pNPP). Samples were assayed in 384-well plates in triplicate. Each well contained 0.015 μM PP5 and 1.5 μM Hsp90 in a buffer containing 0.5 mM MnCl2, 50 mM NaCl, and 100 mM Tris, pH 8.0. The reaction was started by addition of pNPP, and the change in absorbance at 405 nm was measured over 10 min at 30 °C. Data were fit to the Michaelis–Menten equation using nonlinear least squares regression in MATLAB.
Cdc37 Dephosphorylation Assays.
To monitor dephosphorylation of Cdc37-Ser13 by PP5, 5 μM purified phospho-Ser13 Cdc37 (WT or mutant) was mixed with 2.5 μM Hsp90 in a buffer containing 100 mM NaCl, 50 mM Tris, pH 7.5, 2 mM DTT, and 1 mM MnCl2. The reaction was started by adding 0.25 μM PP5 (WT or mutant), and the samples were incubated at 30 °C. Samples were taken every 45 min for SDS/PAGE analysis. The phosphorylation state of Cdc37 Ser13 was probed by Western blot using a phospho-Ser13–specific antibody (Sigma). Figures are representative of results from a minimum of two independent assays.
Conservation Analysis.
ConSurf (41) was used to map the relative conservation of residues between PP5 homologs and within the PPP family. The input alignments were generated in ClustalW2 (42). For PP5 subfamily analysis, greater than 70 PP5 homologs were identified using Blast (43). For the PPP family, all common homologs were identified for each family member (typically 10 homologs per family member isoform, except for PP7, for which only Arabidopsis thaliana was included).
Plasmids for Cellular Assays.
N-terminal FLAG or c-myc–tagged human PP5 and N-terminal FLAG-tagged human Cdc37 in mammalian expression plasmid pCDNA3 and their mutants were derived using a QuikChangeII Site-Directed Mutagenesis Kit (Stratagene) with the primers listed in Table S3. Mutations were checked by DNA sequencing.
Immunoprecipitation and Immunoblotting.
HEK293 cells were transiently transfected with each construct using TransIT-2020 (Mirus) transfection reagent and incubated at 37 °C for 24 h. Cells were washed with cold 1× PBS (Dulbecco’s PBS without calcium or magnesium; Sigma) on ice. Total cell lysate was collected in cold lysis buffer [20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM MgCl2, 0.1% Nonidet P-40, protease inhibitor mixture (Roche), PhosSTOP (Roche)] on ice. Lysates were incubated with anti-FLAG (Sigma) or anti-myc antibody-conjugated beads (ThermoScientific) for 2 h at 4 °C with gentle rotation. After incubation, beads were washed four times with cold lysis buffer, eluted with 5× Laemmli buffer, and boiled for 5 min. Western blotting was carried out as previously described (44). Coimmunoprecipitated proteins were detected by immunoblotting with indicated antibodies recognizing FLAG (Sigma-Aldrich); Raf-1 and CDK4 (Santa Cruz); Cdc37, Ulk1, myc-tag, GR, phospho-GR (S211), CKIIα, and PP5 (Cell Signaling Technology); Hsp90 (Enzo); or PP5 and phospho-Cdc37 (S13; Abcam). The data are representative of three biological replicates.
Drug Binding Assay.
Total cell lysates prepared as described above were incubated with 50 or 100 nM biotinylated GB (Synta Pharmaceuticals) at 4 °C for 1 h followed by streptavidin agarose beads (ThermoScientific) for an additional 2 h with gentle rotation. Bound Hsp90 and PP5-FLAG were detected by immunoblotting as described above. The data are representative of three biological replicates.
PP5 Knockdown by siRNA.
siRNA scramble control and PP5 targeting duplexes were purchased from OriGene (SKU: SR303702) and suspended in provided resuspension buffer. In a six-well plate, HEK293 cells were transiently transfected with the siRNA using TransIT-2020 (Mirus) with either 30 nM control siRNA (SKU: SR30004) or 10 nM of each PP5 siRNA duplex mixed (SKUs: SR303702A, SR303702B, and SR303702C). Transfections were incubated at 37 °C for 72 h. Then, cells were harvested, and protein was extracted as described above; alternately, cells were scraped for RNA isolation (see below). The data presented are representative of three biological replicates.
RNA Extraction, cDNA, and Quantitative RT-PCR Quantification.
Cells from one six-well plate of control siRNA and PP5 siRNA knockdown in HEK293 cells were scraped and washed in cold 1× PBS (Dulbecco’s PBS without calcium or magnesium; Sigma). RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s protocol. cDNA was synthesized using the iScript cDNA Synthesis Kit (BioRad) according to the manufacturer’s protocol. Quantitative RT-PCR was carried out using PP5 primers forward: 5′-AAGACTCAGGCCAATGACTAC-3′ and reverse: 5′-CGCGTAGCCATAGCACTCAG-3′ (PrimerBank ID 324021714c1) and GAPDH primers forward: 5′-GGAAGGTGAAGGTCGGAGTCA-3′ and reverse: 5′-GCAACAATATCCACTTTACCA-GAGTTAA-3′. PCR product was detected using iTaq Universal SYBR Green Supermix (BioRad) and a CFX96 Thermocycler (BioRad). The data presented are representative of three biological replicates.
Statistical Analysis.
Statistics for RNA relative quantitation are presented as means ± SE. Data were analyzed with an unpaired t test. Asterisks represent significance (***P value < 0.0005).
Acknowledgments
We thank David Barford for the human PP5 clone, Olaf-Georg Issinger for the Zea mays CK2 α-clone, Ambrose Cole for data collection, Synta Pharmaceuticals for the Hsp90 inhibitor GB, and David Barford and Mark Williams for critical discussions. This work was supported with funds from the State University of New York Upstate Medical University, the Foundation for Upstate Medical University, the One Square Mile of Hope Foundation (D.B. and M.M.), the Medical Research Council (J.O. and C.K.V.), the Biochemical Society (L.M.), and the Carol M. Baldwin Breast Cancer Fund (D.B. and M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5HPE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603059113/-/DCSupplemental.
References
- 1.Golden T, Swingle M, Honkanen RE. The role of serine/threonine protein phosphatase type 5 (PP5) in the regulation of stress-induced signaling networks and cancer. Cancer Metastasis Rev. 2008;27(2):169–178. doi: 10.1007/s10555-008-9125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez ER. Chaperoning steroidal physiology: Lessons from mouse genetic models of Hsp90 and its cochaperones. Biochim Biophys Acta. 2012;1823(3):722–729. doi: 10.1016/j.bbamcr.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y. Serine/threonine phosphatases: Mechanism through structure. Cell. 2009;139(3):468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Kang H, Sayner SL, Gross KL, Russell LC, Chinkers M. Identification of amino acids in the tetratricopeptide repeat and C-terminal domains of protein phosphatase 5 involved in autoinhibition and lipid activation. Biochemistry. 2001;40(35):10485–10490. doi: 10.1021/bi010999i. [DOI] [PubMed] [Google Scholar]
- 5.Ramsey AJ, Chinkers M. Identification of potential physiological activators of protein phosphatase 5. Biochemistry. 2002;41(17):5625–5632. doi: 10.1021/bi016090h. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, et al. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 2005;24(1):1–10. doi: 10.1038/sj.emboj.7600496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cliff MJ, Harris R, Barford D, Ladbury JE, Williams MA. Conformational diversity in the TPR domain-mediated interaction of protein phosphatase 5 with Hsp90. Structure. 2006;14(3):415–426. doi: 10.1016/j.str.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Vaughan CK, et al. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol Cell. 2008;31(6):886–895. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keramisanou D, et al. Molecular mechanism of protein kinase recognition and sorting by the Hsp90 kinome-specific cochaperone Cdc37. Mol Cell. 2016;62(2):260–271. doi: 10.1016/j.molcel.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearl LH. Hsp90 and Cdc37 -- a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15(1):55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Shao J, Prince T, Hartson SD, Matts RL. Phosphorylation of serine 13 is required for the proper function of the Hsp90 co-chaperone, Cdc37. J Biol Chem. 2003;278(40):38117–38120. doi: 10.1074/jbc.C300330200. [DOI] [PubMed] [Google Scholar]
- 12.Miyata Y, Nishida E. CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone, Cdc37. Mol Cell Biol. 2004;24(9):4065–4074. doi: 10.1128/MCB.24.9.4065-4074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg J, et al. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature. 1995;376(6543):745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 14.Maynes JT, et al. Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1. J Biol Chem. 2001;276(47):44078–44082. doi: 10.1074/jbc.M107656200. [DOI] [PubMed] [Google Scholar]
- 15.Kelker MS, Page R, Peti W. Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: A novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J Mol Biol. 2009;385(1):11–21. doi: 10.1016/j.jmb.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peti W, Nairn AC, Page R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2013;280(2):596–611. doi: 10.1111/j.1742-4658.2012.08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swingle MR, Honkanen RE, Ciszak EM. Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5. J Biol Chem. 2004;279(32):33992–33999. doi: 10.1074/jbc.M402855200. [DOI] [PubMed] [Google Scholar]
- 18.Miyata Y, Nishida E. CK2 binds, phosphorylates, and regulates its pivotal substrate Cdc37, an Hsp90-cochaperone. Mol Cell Biochem. 2005;274(1-2):171–179. doi: 10.1007/s11010-005-2949-8. [DOI] [PubMed] [Google Scholar]
- 19.Bandhakavi S, McCann RO, Hanna DE, Glover CVC. A positive feedback loop between protein kinase CKII and Cdc37 promotes the activity of multiple protein kinases. J Biol Chem. 2003;278(5):2829–2836. doi: 10.1074/jbc.M206662200. [DOI] [PubMed] [Google Scholar]
- 20.Shao J, Irwin A, Hartson SD, Matts RL. Functional dissection of cdc37: Characterization of domain structure and amino acid residues critical for protein kinase binding. Biochemistry. 2003;42(43):12577–12588. doi: 10.1021/bi035138j. [DOI] [PubMed] [Google Scholar]
- 21.Xu W, et al. Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50CDC37-AHA1 chaperone machine. Mol Cell. 2012;47(3):434–443. doi: 10.1016/j.molcel.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polier S, et al. ATP-competitive inhibitors block protein kinase recruitment to the Hsp90-Cdc37 system. Nat Chem Biol. 2013;9(5):307–312. doi: 10.1038/nchembio.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckl JM, et al. Hsp90·Cdc37 complexes with protein kinases form cooperatively with multiple distinct interaction sites. J Biol Chem. 2015;290(52):30843–30854. doi: 10.1074/jbc.M115.693150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beebe K, et al. Posttranslational modification and conformational state of heat shock protein 90 differentially affect binding of chemically diverse small molecule inhibitors. Oncotarget. 2013;4(7):1065–1074. doi: 10.18632/oncotarget.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodford MR, et al. Mps1 mediated phosphorylation of Hsp90 confers renal cell carcinoma sensitivity and selectivity to Hsp90 inhibitors. Cell Reports. 2016;14(4):872–884. doi: 10.1016/j.celrep.2015.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 27.Endicott JA, Noble MEM, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81(1):587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 28.Arlander SJH, et al. Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J Biol Chem. 2006;281(5):2989–2998. doi: 10.1074/jbc.M508687200. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Landgraf R. Phosphorylated and unphosphorylated serine 13 of CDC37 stabilize distinct interactions between its client and HSP90 binding domains. Biochemistry. 2015;54(7):1493–1504. doi: 10.1021/bi501129g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 31.Lu X-A, et al. The regulatory mechanism of a client kinase controlling its own release from Hsp90 chaperone machinery through phosphorylation. Biochem J. 2014;457(1):171–183. doi: 10.1042/BJ20130963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollapour M, et al. Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol Cell. 2011;41(6):672–681. doi: 10.1016/j.molcel.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soroka J, et al. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell. 2012;45(4):517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verba KA, et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 2016;352(6293):1542–1547. doi: 10.1126/science.aaf5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ali MMU, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440(7087):1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Web Server issue):W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mollapour M, et al. Asymmetric Hsp90 N domain SUMOylation recruits Aha1 and ATP-competitive inhibitors. Mol Cell. 2014;53(2):317–329. doi: 10.1016/j.molcel.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]