Significance
Inorganic phosphate (Pi) is an essential constituent for nodule performance. Consequently, symbiotic efficiency in dinitrogen (N2)-fixing legumes, where nodules act as a sink for Pi, is affected by Pi availability. Low Pi availability results in major inhibition of symbiotic performance; thus, understanding the adaptive responses of nodule metabolism to Pi deficiency is crucial to improve symbiotic efficiency under Pi-limited conditions. This study reports the key mechanisms responsible for decreasing nodule activity under Pi deficiency and examines whether nodule responses to low Pi availability are mediated by changes in the metabolism of other organs of nodulated plants. These findings can be used to develop chickpea cultivars and perhaps, other leguminous crops with effective symbiotic efficiency under Pi-limited conditions through genetic engineering.
Keywords: phosphate deficiency, nitrogen fixation, metabolomics, carbon and nitrogen metabolism, phosphate homeostasis
Abstract
Low inorganic phosphate (Pi) availability is a major constraint for efficient nitrogen fixation in legumes, including chickpea. To elucidate the mechanisms involved in nodule acclimation to low Pi availability, two Mesorhizobium–chickpea associations exhibiting differential symbiotic performances, Mesorhizobium ciceri CP-31 (McCP-31)–chickpea and Mesorhizobium mediterranum SWRI9 (MmSWRI9)–chickpea, were comprehensively studied under both control and low Pi conditions. MmSWRI9–chickpea showed a lower symbiotic efficiency under low Pi availability than McCP-31–chickpea as evidenced by reduced growth parameters and down-regulation of nifD and nifK. These differences can be attributed to decline in Pi level in MmSWRI9-induced nodules under low Pi stress, which coincided with up-regulation of several key Pi starvation-responsive genes, and accumulation of asparagine in nodules and the levels of identified amino acids in Pi-deficient leaves of MmSWRI9-inoculated plants exceeding the shoot nitrogen requirement during Pi starvation, indicative of nitrogen feedback inhibition. Conversely, Pi levels increased in nodules of Pi-stressed McCP-31–inoculated plants, because these plants evolved various metabolic and biochemical strategies to maintain nodular Pi homeostasis under Pi deficiency. These adaptations involve the activation of alternative pathways of carbon metabolism, enhanced production and exudation of organic acids from roots into the rhizosphere, and the ability to protect nodule metabolism against Pi deficiency-induced oxidative stress. Collectively, the adaptation of symbiotic efficiency under Pi deficiency resulted from highly coordinated processes with an extensive reprogramming of whole-plant metabolism. The findings of this study will enable us to design effective breeding and genetic engineering strategies to enhance symbiotic efficiency in legume crops.
Phosphorus plays a critical role in numerous plant metabolic processes and contributes to the biosynthesis of cellular macromolecules, such as ATP, nucleic acids, phospholipids, and phosphorylated sugars (1). Thus, phosphorus has been established as one of the most important elements required for normal plant growth and development (1). Unfortunately, limited availability of inorganic phosphate (Pi), which is the only absorbable form of phosphorus for plants, in soils is nearly universal, because Pi readily forms various insoluble compounds with metals, such as calcium and iron in alkaline and acidic soils, respectively (1, 2). Pi deficiency can be overcome by the application of Pi fertilizers; however, the excessive use of chemical fertilizers can have serious environmental consequences, including the contamination of soil and water resources (1). Additionally, the global demand for and use of Pi fertilizers are projected to increase significantly with the explosive growth of the global population. Thus, it has been predicted that global Pi reserves will be depleted within 100 y or even sooner (3).
Symbiotic nitrogen fixation (SNF) in dinitrogen (N2)-fixing legumes is dramatically affected by numerous environmental limitations (4). Among the stressors, low Pi availability is a major restriction of SNF capacity, because this process depends on a series of energy-demanding metabolic steps. Reductions in the concentration of ATP and energy charge in Pi-deficient nodules result in significant declines in nitrogenase activity, SNF capacity, and ultimately, the growth and productivity of legume crops (5). Thus, an improvement in Pi levels in the nodules of plants grown in soils with low Pi availability can contribute to a more efficient atmospheric N2 fixing capacity and therefore, greater productivity of legumes (6). Although low Pi availability has been well-documented as a major restriction for SNF capacity, which imposes serious limitations on legume growth and crop production, little information is available pertaining to the mechanisms responsible for the decrease in SNF capacity under Pi starvation (7). A deep understanding of the complex strategies by which legume metabolism deals with nutritional Pi deficiency can pave the way to breeding crop plants with enhanced symbiotic efficiency or developing them through biotechnological strategies. The development of Pi-efficient crop plants that can grow and yield better under low Pi supply would have a significant beneficial impact on agricultural sustainability (8).
Being one of the major pulse crops in the world, chickpea (Cicer arietinum L.) is traditionally cultivated in many countries of different continents (9). It ranks third globally in total yield among grain legume crops (10). Chickpea has a high nutritive value and serves as an important source of protein in developing countries; also, it has the ability to increase soil fertility in terms of soil nitrogen (N) content because of the activity of SNF (11). Among legumes, chickpea has a superior ability to fix atmospheric N2 through its symbiotic relationship with compatible and effective Rhizobium strains and therefore, can reduce the need for chemical fertilizers, thus minimizing the adverse environmental effects of synthetic fertilizers. However, low soil fertility, particularly Pi deficiency, imposes an important constraint on chickpea crop production, especially in tropical and subtropical areas of Africa and Asia (12). Thus, the selection of Rhizobium–chickpea symbiotic associations with efficient N2 fixing capability under low Pi availability can help improve chickpea production in low Pi fields.
In this study, two Mesorhizobium strains inoculated on chickpea cultivar ILC482 were initially found to have differential symbiotic performances under both control and low Pi stress conditions. A comprehensive examination of the metabolic, biochemical, and molecular mechanisms underlying the differential SNF responses to Pi deficiency in these two Mesorhizobium–chickpea symbiotic associations allowed the identification of the key factors modulating SNF capacity in chickpea under Pi starvation and provided evidence that changes in the metabolism in plant organs other than nodules play an important role in determining the SNF capacity of chickpea plants.
Results
Chickpea Growth and Nodulation.
Under Pi-sufficient conditions, total shoot, root, and whole-plant dry matter (DM) accumulations in chickpea inoculated with Mesorhizobium mediterranum SWRI9 strain (MmSWRI9) were significantly greater than in plants inoculated with Mesorhizobium ciceri CP-31 strain (McCP-31) (Dataset S1). A reduction in Pi supply resulted in a significant decrease in DM accumulation in shoots, roots, and whole plants in both symbiotic combinations; however, the reduction in MmSWRI9-inoculated plants was significantly more severe than the reduction in McCP-31–inoculated plants (47.8% vs. 21.4% for shoots, 35.6% vs. 12.6% for roots, and 40.7% vs. 13.1% for whole plants) (Dataset S1). Shoot DM was more affected than root DM under low Pi conditions in both symbiotic systems. This observation was particularly evident in MmSWRI9-inoculated plants as evidenced by the increase in the root to shoot ratio when the Pi supply decreased (Dataset S1).
Under Pi-sufficient conditions, nodule number, total nodule DM, and individual nodule DM were similar in both symbiotic associations. Although nodule number increased under low Pi conditions in both symbiotic associations, MmSWRI9-inoculated plants displayed a 12.6% greater number of nodules per plant than McCP-31–inoculated plants. Individual nodule DM also decreased significantly in both symbiotic associations. This decline, however, was greater in MmSWRI9-inoculated plants than in McCP-31–inoculated plants (59.1% vs. 32.9%, respectively). The formation of more and smaller nodules in MmSWRI9-inoculated plants is expected to have a negative impact on its nodule energy expenditures (13). Such smaller nodules have been characterized to be less effective because of increased O2 diffusion caused by an increase in the surface to area ratio (14). In contrast, the lower increment of nodule number in McCP-31–inoculated plants would effectively help in saving the limited amount of available Pi with a positive impact on energy turnover of the existing nodules. Overall, McCP-31–inoculated plants displayed better intrinsic characteristics of nodulation under Pi starvation as measured by single and total nodule DM (Dataset S1).
Expression of nifD and nifK Genes in Chickpea Nodules.
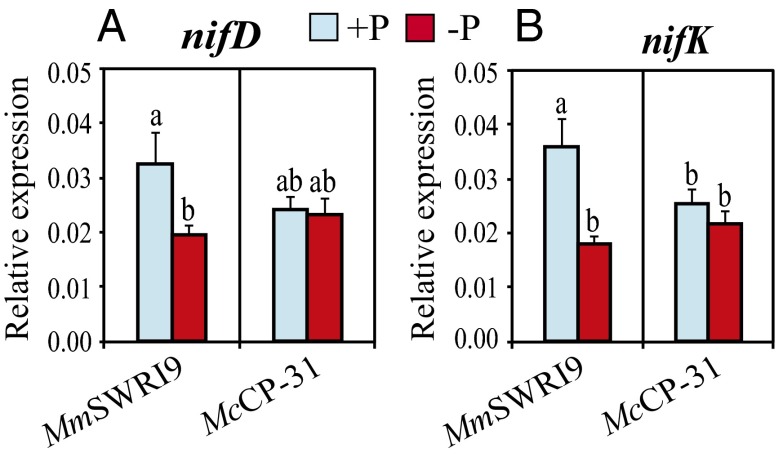
SNF capability of leguminous plants is controlled by the rhizobial nitrogenase complex, which catalyzes the reduction of N2 into ammonia (15). The nifDK genes are the structural genes encoding the MoFe protein subunits of the nitrogenase complex, and assessing their expression levels has been shown to be a reliable method for evaluating the contribution of the N2 fixing bacteria to plant N nutrition (9, 16–18). Thus, analysis of the expression levels of the nifDK genes will enable us to characterize the SNF capacity of the nodules induced by the tested strains under both normal and low Pi conditions (Fig. 1). Under sufficient Pi supply, the expression levels of nifD (encodes the α-subunit of the MoFe protein) and nifK (encodes the β-subunit of the MoFe protein) genes were ∼28.5% and ∼39.8%, respectively, higher in MmSWRI9-induced nodules than in McCP-31–induced nodules (Fig. 1). When chickpea plants were grown under low Pi conditions, the expression levels of both nifD and nifK were not significantly affected in McCP-31–induced nodules, whereas a significant reduction in the expression level of nifD and nifK genes (22.2% and 47.9%, respectively) was observed in MmSWRI9-induced nodules (Fig. 1).
Fig. 1.
Expression of (A) nifD and (B) nifK genes in nodules of chickpea plants. Chickpea seedlings were inoculated with either McCP-31 or MmSWR19 strain and grown over a period of 30 d after sowing under Pi-sufficient (+P) or -deficient (−P) conditions. Relative expression was computed based on the expression level of the target gene and the 16S rRNA reference gene. Data presented are the means ± SE of three independent biological replicates (with two technical repeats for each biological replicate). Data with different letters reveal significant differences as measured by a Duncan’s multiple range test (P ≤ 0.05).
Pi Levels in Chickpea Nodules, Roots, and Leaves.
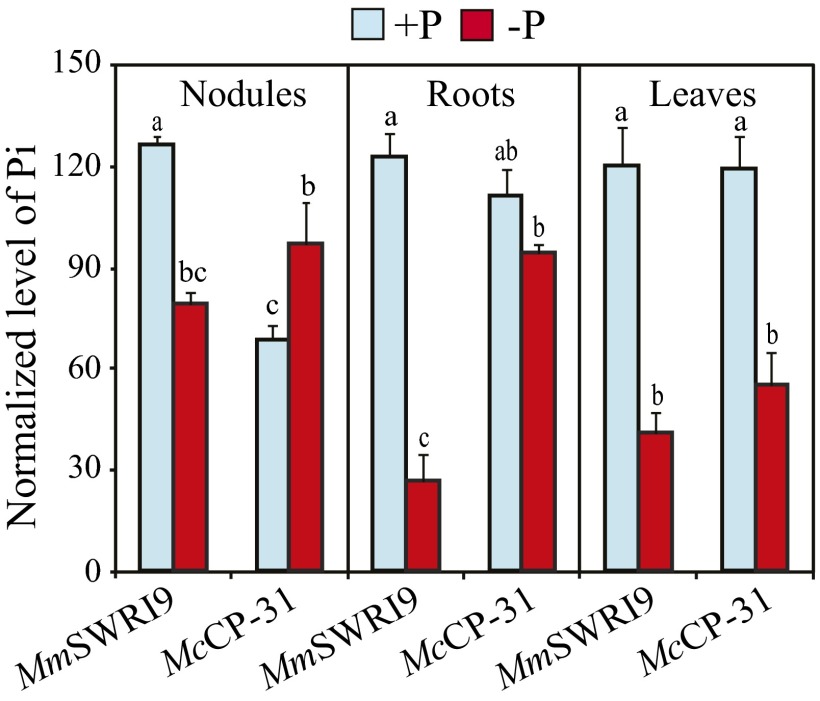
A comparison of Pi levels, as determined by metabolomic analysis (Datasets S2–S4), in nodules obtained from both symbiotic associations under Pi-sufficient levels revealed that MmSWRI9-induced nodules had an 84.4% higher Pi level than McCP-31–induced nodules (Fig. 2). Pi levels in nodules and roots of MmSWRI9-inoculated plants decreased by 37.5% and 78%, respectively, in response to Pi deficiency (Fig. 2). In contrast, the Pi level in McCP-31–induced nodules was almost 41.7% higher under Pi-deficient conditions than in Pi-sufficient conditions, whereas the Pi level in roots was not significantly affected. The Pi level in leaves significantly decreased in response to Pi limitation in both symbiotic associations. Pi deficiency, however, triggered a greater decline in the Pi level of leaves in MmSWRI9-inoculated plants (a 66% decline relative to the Pi level in Pi-sufficient control plants) than in McCP-31–inoculated plants (a 50% decline relative to the Pi level in Pi-sufficient control plants) (Fig. 2).
Fig. 2.
Pi levels in nodules, roots, and leaves of chickpea plants. Chickpea seedlings were inoculated with either McCP-31 or MmSWR19 strain and grown over a period of 30 d after sowing under Pi-sufficient (+P) or -deficient (−P) conditions. Relative Pi levels were determined by metabolite analysis. Data presented are the means ± SE of four independent biological replicates. Data with different letters reveal significant differences as measured by a Duncan’s multiple range test (P ≤ 0.05).
Genetic Responses of Chickpea Nodules to Pi Deficiency.
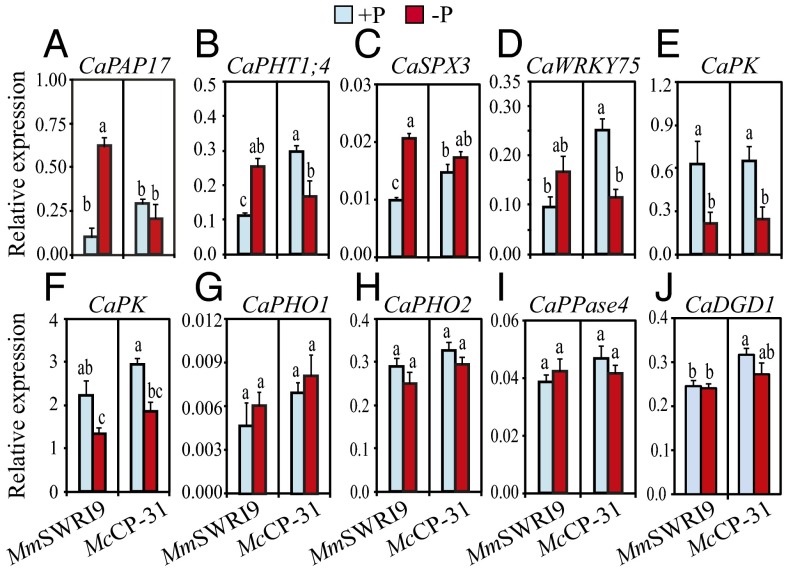
To determine whether the contrasting responses to Pi deficiency in the two symbiotic associations resulted from the differential expression of key Pi starvation responses-related genes, 10 chickpea genes were selected for comparative gene expression analysis using chickpea genome annotation information (19). The selected candidates included genes that are putatively involved in Pi uptake, transport, allocation, and mobilization/remobilization, such as C. arietinum phosphate 1 (CaPHO1), CaPHO2, phosphate transporter 1;4 (CaPHT1;4), purple acid phosphatase 17 (CaPAP17), pyrophosphatase 4 (CaPPase4), and digalactosyldiacylglycerol synthase 1 (CaDGD1); genes that are putatively involved in carbon (C) metabolism, such as pyruvate kinases (PKs; CaPK/LOC101509508 and CaPK/LOC101498888); and genes that are putatively involved in transcriptional regulation, such as CaWRKY75 and SYG1/Pho81/XPR1 domain containing protein 3 (CaSPX3) (Fig. 3 and Dataset S5).
Fig. 3.
Expression of Pi starvation response-related genes in chickpea nodules. Chickpea seedlings were inoculated with either McCP-31 or MmSWR19 strain and grown under Pi-sufficient (+P) or -deficient (−P) conditions. (A) CaPAP17, (B) CaPHT1;4, (C) CaSPX3, (D) CaWRKY75, (E) CaPK/LOC101509508, (F) CaPK/LOC101498888, (G) CaPHO1, (H) CaPHO2, (I) CaPPase4, and (J) CaDGD1. Relative expression was computed based on the expression level of the target gene and the CaIF4a reference gene. Data presented are the means ± SE of three independent biological replicates (with two technical repeats for each biological replicate). Data with different letters reveal significant differences as measured by a Duncan’s multiple range test (P ≤ 0.05).
Results indicated that CaPAP17, CaPHT1;4, and CaSPX3 were significantly up-regulated in MmSWRI9-induced nodules in response to low Pi supply, whereas the expression levels of CaPAP17 and CaSPX3 remained relatively unchanged and that of CaPHT1;4 was significantly repressed in McCP-31–induced nodules under low Pi conditions (Fig. 3 A–C). Furthermore, Pi deficiency reduced the expression of CaWRKY75 in McCP-31–induced nodules but not that in MmSWRI9-induced nodules (Fig. 3D). The expression levels of two CaPK genes significantly decreased in response to low Pi supply in both MmSWRI9- and McCP-31–induced nodules (Fig. 3 E and F). Transcript levels of CaPHO1, CaPHO2, CaPPase4, and CaDGD1 remained relatively unchanged under low Pi conditions in nodules of both symbiotic associations (Fig. 3 G–J).
Effects of Pi Deficiency on Total Soluble Protein Contents and C and N Metabolism-Related Enzyme Activities in Nodulated Chickpea.
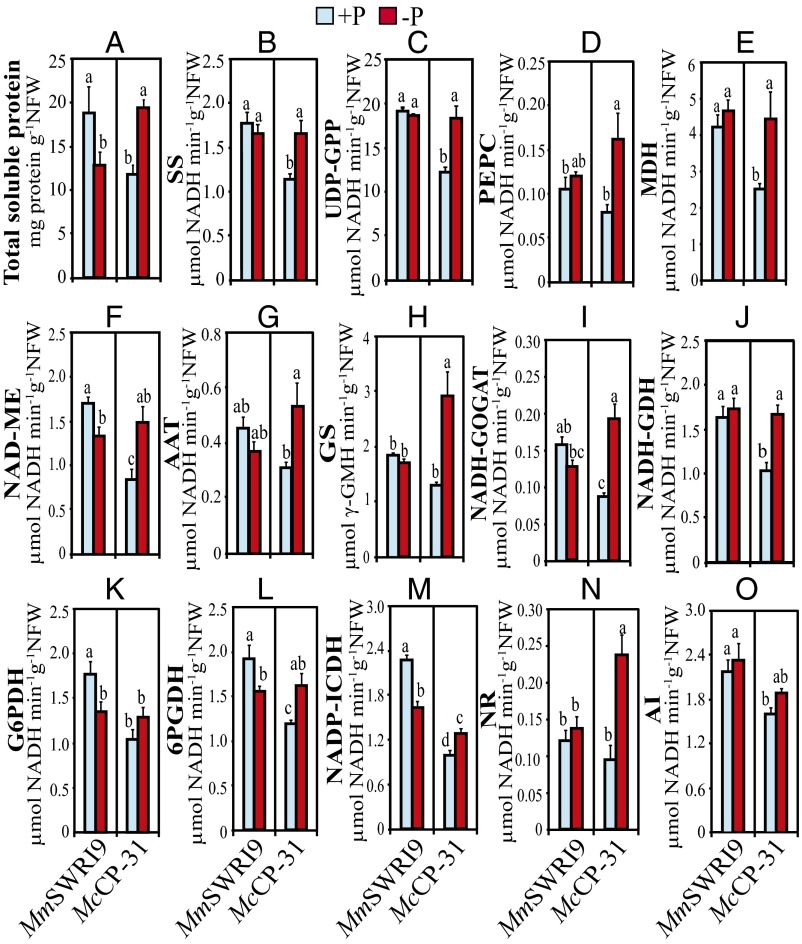
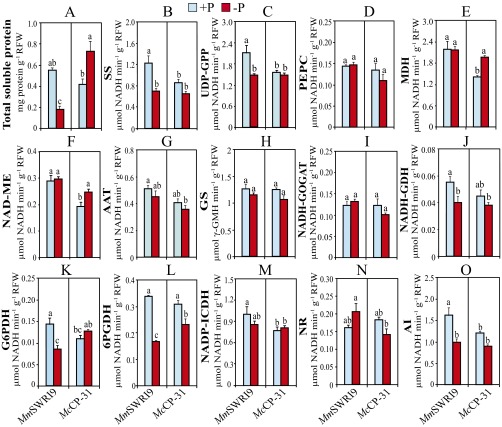
Measurements of total soluble protein contents and the activities of key enzymes involved in nodule C and N metabolism displayed a profound variation in response to Pi deficiency (Fig. 4). Under Pi-sufficient conditions, total soluble protein content (Fig. 4A) and the activities of the majority of the examined enzymes, including sucrose synthase (SS) (Fig. 4B), uridine diphosphate–glucose pyrophosphorylase (UDP-GPP) (Fig. 4C), malate dehydrogenase (MDH) (Fig. 4E), NAD-malic enzyme (NAD-ME) (Fig. 4F), NADH-glutamate synthase (NADH-GOGAT) (Fig. 4I), NADH-glutamate dehydrogenase (NADH-GDH) (Fig. 4J), glucose-6-phosphate dehydrogenase (G6PDH) (Fig. 4K), 6-phosphogluconate dehydrogenase (6PGDH) (Fig. 4L), NADP-isocitrate dehydrogenase (NADP-ICDH) (Fig. 4M), and alkaline invertase (AI) (Fig. 4O), were significantly higher in MmSWRI9-induced nodules than in McCP-31–induced nodules. Low Pi supply decreased the level of total soluble proteins in MmSWRI9-induced nodules to 68% of that observed in MmSWRI9-induced nodules under Pi-sufficient conditions. In contrast, the level of total soluble proteins in nodules of McCP-31–inoculated chickpea plants increased by 64.5% in response to low Pi supply compared with in Pi-sufficient conditions (Fig. 4A). Under Pi-deficient conditions, a significant increment in the activity of most C and N metabolism-related enzymes was observed in McCP-31–induced nodules, whereas the enzyme activity in MmSWRI9-induced nodules had a tendency to be relatively stable for most of the enzymes examined. For instance, in McCP-31–inoculated plants, a low Pi supply resulted in a significant increase in the activity of SS (46%), UDP-GPP (48%), phosphoenolpyruvate carboxylase (PEPC; 102%), MDH (70%), NAD-ME (75%), aspartate aminotransferase (AAT; 43%), glutamine synthetase (GS; 134%), and NADH-GOGAT (121%) (Fig. 4 B–I). In contrast, with the exception of a significant reduction in the specific activity of NAD-ME (21.7%) (Fig. 4F), G6PDH (23.5%) (Fig. 4K), 6PGDH (18.8%) (Fig. 4L), and NADP-ICDH (27.7%) (Fig. 4M), an insignificant variation was observed in the activity of the enzymes examined in MmSWRI9-inoculated plants in response to low Pi supply.
Fig. 4.
Total soluble protein contents and the activities of key enzymes involved in C and N metabolism in nodules of chickpea plants. Chickpea seedlings were inoculated with either McCP-31 or MmSWR19 strain and grown under Pi-sufficient (+P) or -deficient (−P) conditions. (A) Total soluble protein, (B) SS, (C) UDP-GPP, (D) PEPC, (E) MDH, (F) NAD-ME, (G) AAT, (H) GS, (I) NADH-GOGAT, (J) NADH-GDH, (K) G6PDH, (L) 6PGDH, (M) NADP-ICDH, (N) NR, and (O) AI. Data presented are the means ± SE of four independent biological replicates. Data with different letters reveal significant differences as measured by a Duncan’s multiple range test (P ≤ 0.05). NFW, nodule fresh weight.
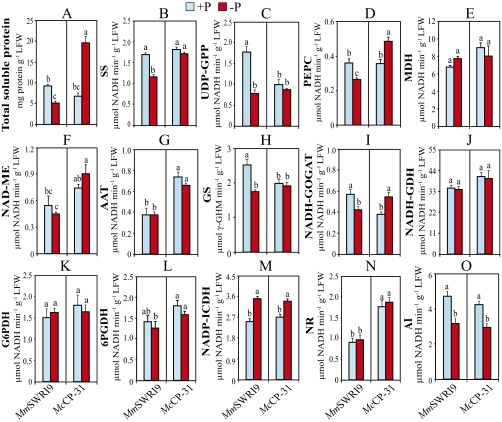
Significant differences in the level of total soluble proteins in the roots of the two symbiotic associations were not observed under Pi-sufficient conditions. In contrast, a reduction of 68% of total soluble protein content was observed in MmSWRI9-nodulated roots, and a 74% increase in total soluble protein content in McCP-31–nodulated roots was observed in response to low Pi (Fig. S1A). Furthermore, the activities of SS (Fig. S1B), UDP-GPP (Fig. S1C), MDH (Fig. S1E), NAD-ME (Fig. S1F), G6PDH (Fig. S1K), NADP-ICDH (Fig. S1M), and AI (Fig. S1O) were higher in MmSWRI9-nodulated roots than in McCP-31–nodulated roots under Pi-sufficient conditions. Differing from McCP-31–nodulated roots, where only the activities of MDH and NAD-ME significantly increased (39% and 28.5%, respectively) (Fig. S1 E and F) and those of 6PGDH and AI significantly decreased (24% and 26%, respectively) (Fig. S1 L and O) in response to Pi deficiency, low Pi treatment caused a significant decline in the activities of 6 of 15 of the examined enzymes in MmSWRI9-nodulated roots relative to the level of activity in Pi-sufficient conditions, including SS (43.5%) (Fig. S1B), UDP-GPP (31%) (Fig. S1C), NADH-GDH (27%), G6PDH (41%), 6PGDH (50%) (Fig. S1 J–L), and AI (40%) (Fig. S1O). The activities of the other enzymes remained unchanged.
Fig. S1.
The concentrations of total soluble proteins and the activities of key enzymes involved in C and N metabolism in roots of chickpea plants. Chickpea seedlings were inoculated with either the McCP-31 strain or the MmSWR19 strain and grown under Pi-sufficient (+P) or -deficient (−P) conditions. (A) Total soluble protein, (B) SS, (C) UDP-GPP, (D) PEPC, (E) MDH, (F) NAD-ME, (G) AAT, (H) GS, (I) NADH-GOGAT, (J) NADH-GDH, (K) G6PDH, (L) 6PGDH, (M) NADP-ICDH, (N) NR, and (O) AI. Data presented are the means ± SE of four independent biological replicates. Data with different letters reveal significant differences as measured by a Duncan’s multiple range test (P ≤ 0.05). RFW, root fresh weight.
Under Pi-sufficient conditions, no significant difference in total soluble protein levels was observed in chickpea leaves of plants with the two symbiotic associations (Fig. S2A). Among the enzymes assayed, however, the activities of UDP-GPP (Fig. S2C), GS (Fig. S2H), and NADH-GOGAT (Fig. S2I) were higher in leaves of MmSWRI9-inoculated plants than in leaves of McCP-31–inoculated plants. In contrast, AAT (Fig. S2G) and nitrate reductase (NR) (Fig. S2N) were lower in MmSWRI9-inoculated plants than in leaves of McCP-31–inoculated plants. A low Pi supply, however, led to a 189% increase in total soluble protein content in leaves of McCP-31–inoculated plants, whereas it resulted in a 45% decrease in leaves of MmSWRI9-inoculated plants relative to leaves of plants grown under Pi-sufficient conditions (Fig. S2A). The activities of C and N metabolism-related enzymes in the leaves of plants grown under Pi-deficient conditions differed in MmSWRI9- and McCP-31–inoculated plants. Low Pi supply decreased the activities of SS (31.7%), UDP-GPP (55.5%), PEPC (26.6%) (Fig. S2 B–D), GS (30.6%), and NADH-GOGAT (25%) (Fig. S2 H and I) in leaves of MmSWRI9-inoculated plants. Conversely, low Pi supply induced the activities of PEPC (35.7%) (Fig. S2D) and NAD-GOGAT (44.5%) (Fig. S2I) in leaves of McCP-31–inoculated plants compared with in leaves from plants grown under Pi-sufficient conditions.
Fig. S2.
The concentrations of total soluble proteins and the activities of key enzymes involved in C and N metabolism in leaves of chickpea plants. Chickpea seedlings were inoculated with either the McCP-31 strain or the MmSWR19 strain and grown under Pi-sufficient (+P) or -deficient (−P) conditions. (A) Total soluble protein, (B) SS, (C) UDP-GPP, (D) PEPC, (E) MDH, (F) NAD-ME, (G) AAT, (H) GS, (I) NADH-GOGAT, (J) NADH-GDH, (K) G6PDH, (L) 6PGDH, (M) NADP-ICDH, (N) NR, and (O) AI. Data presented are the means ± SE of four independent biological replicates. Data with different letters reveal significant differences as measured by a Duncan’s multiple range test (P ≤ 0.05). LFW, leaf fresh weight.
C and N Metabolism-Related Metabolite Profiles in Nodulated Chickpea Under Pi Deficiency.
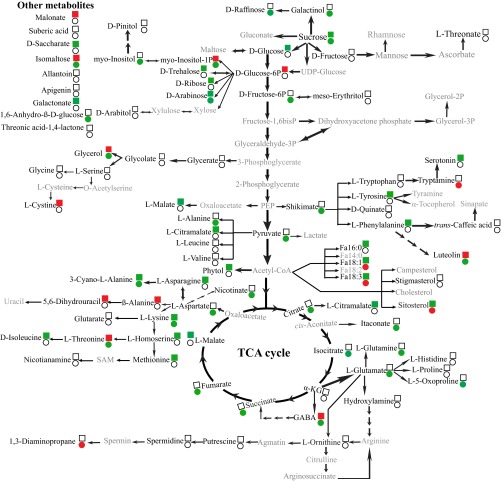
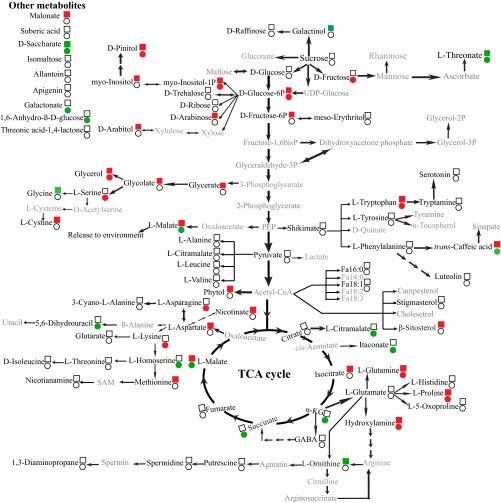
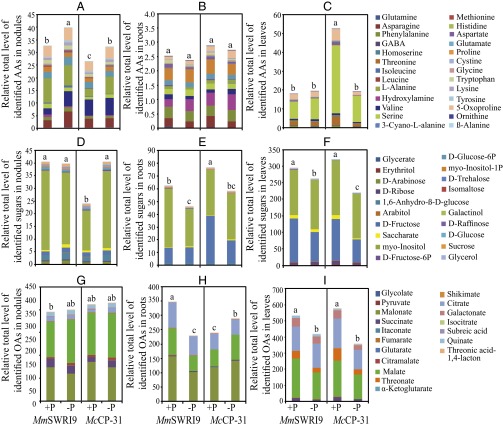
In total, 83, 80, and 79 metabolites were identified in nodules, roots, and leaves of both symbiotic associations, respectively, under sufficient and deficient Pi conditions, of which 35 and 35 metabolites in nodules, 29 and 25 metabolites in roots, and 31 and 53 metabolites in leaves were significantly affected by Pi deficiency in MmSWRI9- and McCP-31–inoculated plants, respectively (Datasets S2–S4). Differential responses in C and N metabolism-related metabolites were observed between MmSWRI9- and McCP-31–inoculated plants in response to Pi deficiency (Figs. S3, S4, and S5). Low Pi conditions increased the total level of detected amino acids in nodules of chickpea inoculated with either bacterial strain (Fig. 5A and Dataset S2). At the individual level, Pi deficiency resulted in a significant increase in the levels of asparagine, homoserine, isoleucine, 3-cyano-l-alanine, methionine, lysine, tyrosine, and phenylalanine in MmSWRI9-induced nodules and significantly increased the levels of glutamine, GABA, l-alanine, 3-cyano-l-alanine, aspartate, glutamate, threonine, lysine, and 5-oxoproline in McCP-31–induced nodules (Fig. S3 and Dataset S2). The total levels of identified amino acids in roots of the two symbiotic associations were not significantly different when plants were grown under Pi-deficient conditions (Fig. 5B). Nevertheless, the root metabolite profiles of several individual amino acids in plants grown under low Pi exhibited a significant decrease, including glutamine, hydroxylamine, methionine, aspartate, proline, cystine, and tryptophan in MmSWRI9-nodulated roots and glutamine, asparagine, serine, hydroxylamine, proline, tryptophan, and lysine in McCP-31–nodulated roots (Fig. S4 and Dataset S3). Moreover, a low Pi supply resulted in a strong reduction in the total level of identified amino acids in the leaves of McCP-31–inoculated plants, whereas no significant change was observed in leaves of MmSWRI9-nodulated plants, with the exception of a few individual amino acids (Fig. 5C and Dataset S4). Of all of the amino acids detected in the leaves of MmSWRI9-inoculated plants, only decreases in the levels of glutamine, phenylalanine, asparagine, and glycine were observed in response to low Pi. In contrast, however, a decrease in the levels of almost all of the detected amino acids was observed in leaves of McCP-31–inoculated plants in response to low Pi (Fig. S5 and Dataset S4).
Fig. S3.
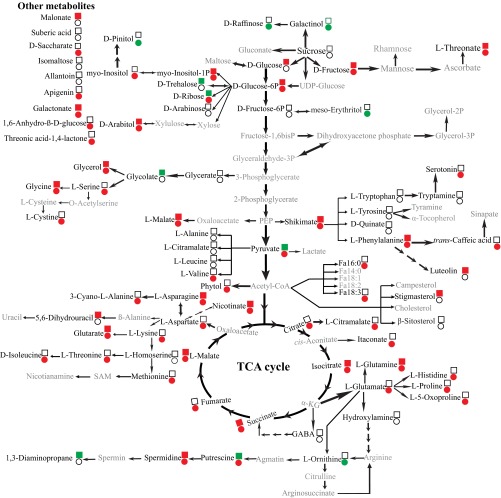
Metabolic changes in nodules of chickpea plants. Chickpea seedlings were inoculated with either the McCP-31 strain (●) or the MmSWR19 strain (■) and grown under Pi-sufficient (+P) or -deficient (−P) conditions. Significant differences (P ≤ 0.05) in the levels of metabolites between Pi-deficient and -sufficient nodules are indicated in green (increased levels) and red (reduced levels); whereas unchanged levels of metabolites are shown by no color. The metabolites with gray letters were unidentifiable. α-KG, α-ketoglutarate; PEP, phosphoenolpyruvate; SAM, S-adenosyl methionine.
Fig. S4.
Metabolic changes in roots of chickpea plants. Chickpea seedlings were inoculated with either the McCP-31 strain (●) or the MmSWR19 strain (■) and grown under Pi-sufficient (+P) or -deficient (−P) conditions. Significant differences (P ≤ 0.05) in the levels of metabolites between Pi-deficient and -sufficient nodules are indicated in green (increased levels) and red (reduced levels); whereas unchanged levels of metabolites are shown by no color. The metabolites with gray letters were unidentifiable. α-KG, α-ketoglutarate; PEP, phosphoenolpyruvate; SAM, S-adenosyl methionine.
Fig. S5.
Metabolic changes in leaves of chickpea plants. Chickpea seedlings were inoculated with either the McCP-31 strain (●) or the MmSWR19 strain (■) and grown under Pi-sufficient (+P) or -deficient (−P) conditions. Significant differences (P ≤ 0.05) in the levels of metabolites between Pi-deficient and -sufficient nodules are indicated in green (increased levels) and red (reduced levels); whereas unchanged levels of metabolites are shown by no color. The metabolites with gray letters were unidentifiable. α-KG, α-ketoglutarate; PEP, phosphoenolpyruvate; SAM, S-adenosyl methionine.
Fig. 5.
Total levels of identified amino acids (AAs) in nodules (A), roots (B), and leaves (C); total levels of identified sugars in nodules (D), roots (E), and leaves (F); and total levels of identified organic acids (OAs) in nodules (G), roots (H), and leaves (I) of chickpea plants. Chickpea seedlings were inoculated with either McCP-31 or MmSWR19 strain and grown under Pi-sufficient (+P) or -deficient (−P) conditions. Data presented are the means of four independent biological replicates. The SE values for each metabolite are shown in Datasets S2–S4. Data with different letters reveal significant differences as measured by a Duncan’s multiple range test (P ≤ 0.05).
Low Pi supply increased the total level of identified sugars by 68.8% in McCP-31–induced nodules, whereas the level of total detected sugars in MmSWRI9-induced nodules remained unchanged (Fig. 5D and Dataset S2). The individual levels of sugars in MmSWRI9-induced nodules in response to Pi deficiency, however, exhibited a remarkable increase in d-arabinose, d-glucose, sucrose, d-trehalose, d-ribose, and saccharate. In contrast, Pi deficiency in McCP-31–inoculated nodules significantly increased the accumulation of galactinol, d-arabinose, d-fructose-6P, sucrose, myo-inositol-1P, isomaltose, 1,6-anhydro-β-d-glucose, myo-inositol, glycerol, and d-raffinose (Fig. S3 and Dataset S2). The total levels of detected sugars significantly decreased in roots of both symbiotic associations when plants were grown under Pi-deficient conditions (Fig. 5E and Dataset S3). A remarkable decrease in the levels of several individual sugars was observed in MmSWRI9-nodulated roots of plants grown under low Pi, including myo-inositol, d-arabinose, d-glucose-6P, d-fructose-6P, myo-inositol-1P, glycerol, glycerate, and arabitol, whereas a significant increase was observed in the levels of saccharate, and galactinol. In contrast, low Pi conditions resulted in a significant decrease in the contents of several sugars in roots of McCP-31–nodulated plants, including d-fructose, d-glucose-6P, myo-inositol-1P, and glycerol, but a considerable increase in the levels of saccharate and 1,6-anhydro-β-d-glucose (Fig. S4 and Dataset S3). Pi deficiency resulted in a significant decrease in the levels of the majority of sugars in leaves of both McCP-31– and MmSWRI9-inoculated plants (Fig. 5F and Dataset S4). The only two sugars exhibiting a significant increase in MmSWRI9-inoculated plants in response to low Pi were d-trehalose and d-ribose. Conversely, d-fructose, d-glucose, myo-inositol-1P, d-glucose-6P, arabitol, and glycerol tended to show a decrease in leaves of MmSWRI9-inoculated plants subjected to Pi deficiency. Leaves of McCP-31–inoculated plants subjected to low Pi conditions exhibited an increase in the levels of erythritol, galactinol, and d-raffinose and a significant decrease in the contents of d-fructose, d-glucose-6P, myo-inositol-1P, d-ribose, arabitol, myo-inositol, saccharate, glycerol, and 1,6-anhydro-β-d-glucose (Fig. S5 and Dataset S4).
The total levels of detected organic acids in nodules of the two different symbiotic associations were similar under Pi-deficient conditions (Fig. 5G and Dataset S2). However, several individual compounds showed a remarkable change in their level in MmSWRI9- and McCP-31–induced nodules obtained from plants grown under Pi deficiency (Fig. 5G and Dataset S2). Among the organic acids detected, a notable accumulation of citrate, shikimate, fumarate, succinate, pyruvate, itaconate, and isocitrate was observed in response to Pi deficiency in McCP-31–induced nodules. Interestingly, when individual organic acids were examined in MmSWRI9-induced nodular tissues obtained from plants grown under Pi-deficient conditions, a significant increase in the levels of several metabolites was observed, including malate, citramalate, and galactonate (Fig. S3 and Dataset S2). Pi deficiency induced a notable accumulation of organic acids in McCP-31–nodulated roots, whereas it significantly decreased the total level of detected organic acids in MmSWRI9-nodulated roots (Fig. 5H and Dataset S3). More specifically, levels of malate, glycolate, malonate, and isocitrate exhibited a notable decrease in MmSWRI9-nodulated roots, whereas the levels of α-ketoglutarate, malate, succinate, citramalate, galactonate, threonate, itaconate, and threonic acid-1,4-lactone displayed significant accumulation in McCP-31–nodulated roots under Pi-deficient conditions (Fig. S4 and Dataset S3). Furthermore, the total levels of identified organic acids markedly reduced in leaves of McCP-31– and MmSWRI9-inoculated plants subjected to low Pi conditions (Fig. 5I and Dataset S4). Pi deficiency resulted in a significant reduction in the levels of eight individual organic acids, including shikimate, malate, glutarate, threonate, succinate, malonate, isocitrate, and galactonate, in the leaves of MmSWRI9-inoculated plants (Fig. S5 and Dataset S4). However, the contents of 13 individual organic acids significantly decreased in leaves of McCP-31–inoculated plants in response to Pi deficiency, including those of citrate, malate, citramalate, threonate, fumarate, succinate, pyruvate, itaconate, glutarate, threonic acid-1,4-lactone, shikimate, isocitrate, and galactonate (Fig. S5 and Dataset S4).
Discussion
The limitation of growth and productivity in N2 fixing leguminous plants induced by low Pi availability in the soil is mainly caused by the deleterious effects of Pi shortage on the formation, development, and activity of nodules and ultimately, SNF capacity (5, 7). Considering the final aim of designing more effective breeding and genetic engineering strategies for developing Pi-efficient legume crops for Pi-deficient soils, this study was undertaken to identify the mechanisms responsible for acclimation of SNF to low Pi availability. In this study, SNF capacity was analyzed by characterizing the expression levels of nifD and nifK genes and the number of key Pi starvation-related genes in nodules and measuring several growth parameters. Whole-plant metabolite profiles were also obtained, and the activities of C and N metabolism-related enzymes were measured in leaves, roots, and nodules of chickpea plants that were inoculated with specific strains of two different species of Mesorhizobium.
Previous reports have suggested that expression levels of nifD and nifK genes could be used as indicators of SNF capacity (9, 16–18). The higher levels of nifD and nifK expression under Pi-sufficient conditions observed in MmSWRI9-induced nodules relative to McCP-31–induced nodules suggest that the MmSWRI9-inoculated association may have higher SNF capacity than the McCP-31–inoculated association under Pi-sufficient conditions (Fig. 1). The greater accumulation of shoot, root, and total DM in MmSWRI9-inoculated chickpea plants than in McCP-31–inoculated plants grown under Pi-sufficient conditions provides supportive evidence for this premise (Dataset S1). Both nifD and nifK genes exhibited unaltered expression in McCP-31–induced nodules in response to low Pi conditions, whereas both genes were down-regulated in MmSWRI9-inoculated nodules under the same conditions (Fig. 1). These results suggest that N2 fixation performance in the nodules of McCP-31–inoculated plants is less affected by low Pi conditions than that in the nodules of MmSWRI9-inoculated plants. This premise is again supported by a better growth performance of McCP-31–inoculated chickpea plants under Pi-limiting conditions compared with that in MmSWRI9-inoculated plants (Dataset S1).
The differential responses of McCP-31– and MmSWRI9-inoculated plants to low Pi supply could be explained by several not mutually exclusive explanations. Because Pi deficiency-induced inhibition of SNF is caused by dramatic reductions in the cytosolic Pi pool in nodules, maintenance of an appropriate nodular Pi level represents a critical adaptive strategy for nodules to diminish the adverse effects of Pi deficiency on SNF capacity (6, 8, 20). In this study, there was an apparent difference between the two symbiotic systems regarding the maintenance of Pi homeostasis in their organs, particularly nodules and roots, for acclimation to low Pi stress. Although MmSWRI9-inoculated plants significantly reduced Pi level in the nodules and roots, McCP-31–inoculated plants tended to increase the Pi level in the nodules and maintain the high Pi level in the root fractions under Pi stress (Fig. 2). This result indicated that the changes in nodular Pi level in the two symbiotic systems showed opposite tendency under Pi deprivation. Furthermore, both symbiotic systems significantly reduced Pi levels in leaves under Pi deprivation, with MmSWRI9-inoculated plants exhibiting a greater decline (66%) than McCP-31–inoculated plants (50%) (Fig. 2). These data suggest an adaptive strategy for both symbiotic systems to tradeoff Pi allocation (i.e., plasticity) by the translocation of Pi from the aerial parts for supporting the SNF under Pi limitation (7, 13).
Apparently, MmSWRI9-inoculated plants tended to reallocate Pi from both leaves and roots to their nodules. Hence, a sharp reduction (78%) particularly observed in the Pi level in roots of MmSWRI9-inoculated plants subjected to low Pi might help prevent the complete depletion of nodular Pi level in MmSWRI9-inoculated plants (Fig. 2). These data indicate that Pi is preferentially allocated from other organs through the vascular network to MmSWRI9-induced nodules and imply that these nodular tissues are stronger sinks for Pi than roots or leaves. A competition in Pi is expected between the roots and nodules in MmSWRI9-inoculated plants, particularly under conditions of Pi limitation (21). The elevated levels of Pi mobilization from the roots and leaves to nodules in MmSWRI9-inoculated plants were particularly evidenced by the up-regulation of CaPAP17, CaPHT1;4, and CaSPX3 genes, involved in Pi remobilization and transport, in MmSWRI9-induced nodules in response to low Pi stress (Fig. 3 A–C) (22–24), which could be interpreted as an acclimation response to decreased nodular Pi levels under Pi deficiency. However, for a better adaptation, McCP-31–inoculated plants translocated Pi from the leaves and more importantly, stabilized the Pi level in the root fraction (Fig. 2). Pi concentrations need to be conserved in the roots to facilitate the acquisition of external Pi sources and upgrade the level of Pi in nodules during Pi starvation conditions (8, 21), as observed with the McCP-31-inoculated plants under Pi-deficient conditions (Fig. 2). In agreement with this finding, up-regulation of low Pi-induced CaPAP17, CaPHT1;4, and CaSPX3 was not observed in the McCP-31–induced nodules under low Pi conditions, and it could be further supported by the fact that CaWRKY75, which regulates the expression of low Pi-responsive genes (25), was not induced but rather repressed in McCP-31–induced nodules by Pi deficiency (Fig. 3 A–D). These results together indicate that McCP-31–inoculated plants possess a regulatory system that more efficiently maintains nodular Pi homeostasis under low Pi conditions than MmSWRI9-inoculated plants. Thus, McCP-31– and MmSWRI9-inoculated plants have evolved diverse mechanisms in response to low Pi availability that strictly control the level of nodular Pi level to fulfill the metabolic requirements of the nodules.
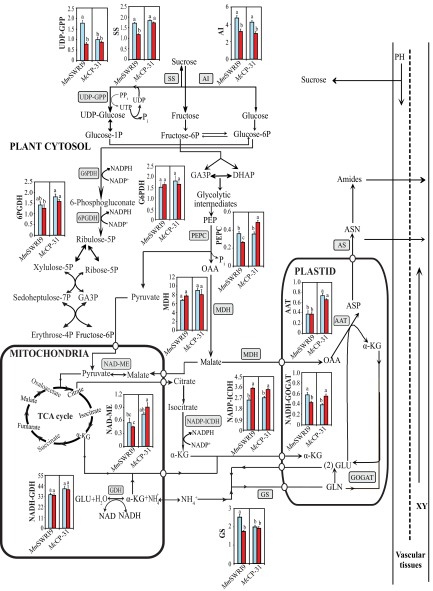
Nodules require sufficient photoassimilates in the form of sucrose to maintain optimum levels of SNF (26, 27). The results of this study showed that the relatively stable levels of sucrose in roots and leaves of plants subjected to low Pi were accompanied by an apparent accumulation of this photosynthate in the nodules of both symbiotic associations (Figs. S3, S4, and S5 and Datasets S2–S4). These data support the premise that legume nodules subjected to low Pi are not sucrose-limited (13, 28). Instead of the initial cleaving of sucrose with the aid of ATP-dependent AI, McCP-31–induced nodules subjected to low Pi significantly activated the energy-saving SS/UDP-GPP pathway that remained unaffected in MmSWRI9-induced nodules (Fig. 4 B and C and Fig. S6). The SS/UDP-GPP pathway can efficiently use pyrophosphates (PPis) as an autonomous energy donor during Pi deficiency (29, 30). Therefore, the induction of sucrose hydrolysis through the alternative PPi-dependent cytosolic pathway in McCP-31–induced nodules confers a considerable bioenergetic advantage, thereby improving nodule performance under Pi starvation. Furthermore, the Pi released in this pathway can be recycled for other metabolic processes under low Pi conditions (Fig. S6) (22).
Fig. S6.
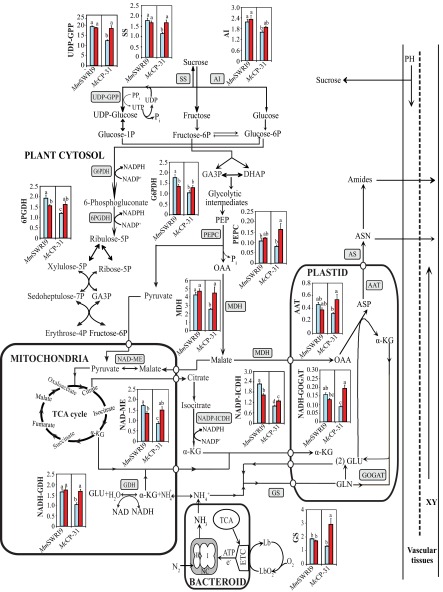
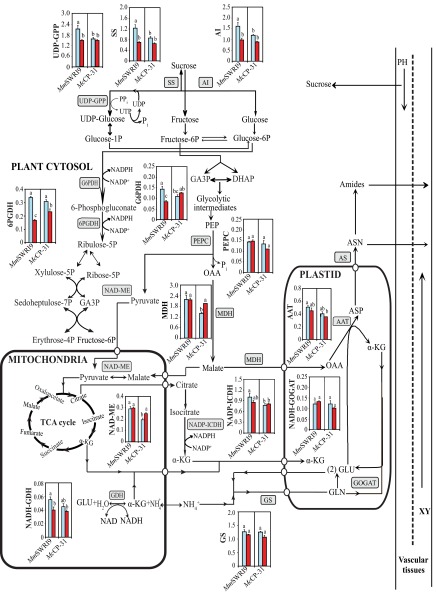
Schematic model representing C and N metabolism and key associated enzymes in nodules of chickpea plants inoculated with either the McCP-31 strain or the MmSWR19 strain under Pi-sufficient (blue columns) or -deficient (red columns) conditions. AS, asparagine synthetase; ASN, asparagine; ASP, aspartate; DHAP, dihydroxyaceton phosphate; GA3P, glyceraldehyde-3-phosphate; GLN, glutamine; GLU, glutamate; α-KG, α-ketoglutarate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PH, phloem; UTP, uridine triphosphate; XY, xylem.
The activities of PEPC, MDH, and NAD-ME increased in McCP-31–induced nodules in response to low Pi but not in MmSWRI9-induced nodules (Fig. 4 D–F and Fig. S6). These data suggest that the adenylate- and Pi-independent PEPC−MDH−ME glycolytic bypass is more active in McCP-31– than in MmSWRI9-induced nodules, resulting in greater conservation of cellular Pi (5). Additionally, pyruvate increased in McCP-31–induced nodules subjected to low Pi but not in MmSWRI9-induced nodules (Fig. S3 and Dataset S2). These observations support the hypothesis that the increased pyruvate might be synthesized from malate through the cytosolic PEPC–MDH–ME glycolytic bypass rather than through the Pi-dependent PK route to ensure the continuation of mitochondrial respiration under Pi-limited conditions (29). In support of this finding, the expression levels of CaPK-encoding genes, at least the two genes that were examined, were down-regulated in response to low Pi in nodules of both symbiotic associations (Fig. 3 E and F). It is plausible that this down-regulation occurs as a way to restrict pyruvate synthesis through the conventional Pi-requiring PK route in nodules under low Pi conditions. Because pyruvate can act as a scavenger of reactive oxygen species (ROS) (31), the higher level of pyruvate observed under low Pi conditions in McCP-31–induced nodules but not in MmSWRI9-induced nodules (Fig. S3 and Dataset S2) may help stabilize the cellular redox status of stressed McCP-31–induced nodules. In addition, utilization of pyruvate for l-alanine synthesis has also been suggested as an adaptation mechanism during energy-deficient conditions (32). Our results showed an increased level of l-alanine in parallel with pyruvate accumulation in McCP-31–induced nodules under low Pi availability (Fig. S3 and Dataset S2). However, metabolite profiling of MmSWRI9-induced nodules subjected to low Pi supply identified a significant accumulation of malate (23% greater than in Pi-sufficient nodules) (Fig. S3 and Dataset S2). The accumulation of malate in MmSWRI9-induced nodules under low Pi conditions was not the result of increased activity of enzymes involved in its synthesis (e.g., PEPC and MDH) (Fig. 4 D and E). Thus, the increase may be related to a decreased utilization of malate by bacteroid respiration. Alternatively, it may also be because of the lower provision of C skeletons for fixed N assimilation, which is likely caused by the altered activity of nodules and SNF under Pi-deficient conditions, as observed in earlier studies of N2 fixation capacity under drought conditions (33). Despite the significant increase of PEPC and MDH in McCP-31–induced nodules in response to low Pi availability (Fig. 4 D and E), the malate content remained at a basal level (Fig. S3 and Dataset S2). The stability of the malate content was most likely caused by its enhanced utilization during bacteroid respiration and/or as a source of C skeletons for fixed N assimilation (34). Furthermore, the conversion of malate to pyruvate in McCP-31–induced nodules might effectively prevent additional malate accumulation in this efficient symbiosis, because malate accumulation is inhibitory to N2 fixation (20).
Results of the study indicate that, in contrast to what was observed in the roots of MmSWRI9-inoculated plants subjected to low Pi conditions, roots in McCP-31–inoculated plants could accumulate a higher level of organic acids (Fig. 5H). It is plausible that this accumulation of organic acids could be potentially released into the rhizosphere to facilitate Pi solubilization in the soil (35, 36), thereby leading to a more efficient Pi uptake in McCP-31–inoculated plants. Remarkably, the increased production of organic acids, such as malate, succinate, citramalate, and α-ketoglutarate, through the TCA cycle in roots of McCP-31–inoculated plants subjected to Pi deficiency (Fig. S4 and Dataset S3) is likely associated with the increased activities of several C metabolism-related enzymes in the roots, such as MDH and NAD-ME (Figs. S1 E and F and S7). Moreover, the higher overall performance of McCP-31–chickpea plants when subjected to low Pi availability, relative to MmSWRI9–chickpea plants, is likely associated with the different levels of several enzymes involved in C metabolism in leaves, such as SS, UDP-GPP, and PEPC (Figs. S2 B–D and S8). These findings suggest that the adaptive mechanisms that modulate symbiotic performance under conditions of low Pi involve a complex suite of modifications in metabolic pathways in nodules and other plants organs (leaves and roots), which play an essential role in sustaining nodule metabolism when the availability of Pi is low or limiting. The activities of key enzymes connected with the oxidative pentose phosphate pathway also varied greatly in the nodules of both symbiotic associations, although the direction and degree of changes differed between the two (Fig. 4 K–M and Fig. S6). Pi deficiency increased the activities of 6PGDH and NADP-ICDH in McCP-31–induced nodules, whereas it decreased the activities of both enzymes as well as that of G6PDH in MmSWRI9-induced nodules (Fig. 4 K–M and Fig. S6). The response in McCP-31–induced nodules may be an adaptive mechanism to provide adequate antioxidant defense through NADPH, because NADPH supply is known to be critical in maintaining the cellular antioxidant system and adjusting the cellular redox status, both of which are important for efficient SNF (37).
Fig. S7.
Schematic model representing C and N metabolism and key associated enzymes in roots of chickpea plants inoculated with either the McCP-31 strain or the MmSWR19 strain under Pi-sufficient (blue columns) or -deficient (red columns) conditions. AS, asparagine synthetase; ASN, asparagine; ASP, aspartate; DHAP, dihydroxyaceton phosphate; ETC, electron transport chain; GA3P, glyceraldehyde-3-phosphate; GLN, glutamine; GLU, glutamate; α-KG, α-ketoglutarate; Lb, leghemoglobin; NC, nitrogenase enzyme complex; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PH, phloem; UTP, uridine triphosphate; XY, xylem.
Fig. S8.
Schematic model representing C and N metabolism and key associated enzymes in leaves of chickpea plants inoculated with either the McCP-31 strain or the MmSWR19 strain under Pi-sufficient (blue columns) or -deficient (red columns) conditions. AS, asparagine synthetase; ASN, asparagine; ASP, aspartate; DHAP, dihydroxyaceton phosphate; GA3P, glyceraldehyde-3-phosphate; GLN, glutamine; GLU, glutamate; α-KG, α-ketoglutarate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PH, phloem; UTP, uridine triphosphate; XY, xylem.
The important role of N metabolism in the adaptation of chickpea to low Pi stress was also evident in this study. In response to low Pi availability, a similar increase (22–23%) in the total level of identified amino acids was observed in the nodules of both symbiotic systems (Fig. 5A and Dataset S2). Therefore, the greater decrease in SNF capacity observed in MmSWRI9-induced nodules under low Pi conditions (Fig. 1 and Dataset S1) cannot be related to the accumulation of the identified amino acids, which has been reported in other studies (5, 38). The accumulation of specific amino acids in plants subjected to low Pi conditions, however, might be implicated in down-regulating SNF capacity in MmSWRI9-induced nodules relative to McCP-31–induced nodules. For example, an accumulation of asparagine was detected in MmSWRI9-induced nodules under Pi-deficient conditions, whereas the asparagine level was unaffected in McCP-31–inoculated nodules under the same conditions (Fig. S3 and Dataset S2). Several studies have reported that down-regulation of SNF capacity is associated with the accumulation of asparagine in nodules of Pi-stressed plants (28, 39). These reports further strengthen the premise that asparagine accumulation plays a role in N feedback regulation of SNF capacity in MmSWRI9-inoculated plants. However, the higher activities of the main enzymes associated with N metabolism, including AAT, GS, and GOGAT, in McCP-31–induced nodules (Fig. 4 G–I and Fig. S6) imply that the substantial accumulation of identified amino acids in McCP-31–induced nodules may be the result of the higher SNF capacity and N assimilation under low Pi conditions (Fig. 5A) (40). Indeed, the significant increase in total soluble protein content observed in McCP-31–induced nodules under low Pi treatment (Fig. 4A) supports this idea. Alternatively, the increase in the total level of the identified amino acids in Pi-stressed MmSWRI9-induced nodules might be attributed to protein degradation, which was confirmed by the observed decline in total soluble protein content in nodules (Fig. 4A) and the unaltered activities of N metabolism-related enzymes in these nodules (Fig. 4 G–I and Fig. S6). Increasing evidence has also suggested that SNF capacity is regulated by plant growth and shoot N requirement through a whole-plant N feedback mechanism (4, 41). In this study, a sharp decrease in shoot and total DM of Pi-deficient MmSWRI9-inoculated plants was observed (Dataset S1), although the level of identified amino acids was unchanged in leaves (Fig. 5C). These data suggest that the amino acid levels in Pi-deficient leaves of MmSWRI9-inoculated plants may exceed the N level required for plant growth. Thus, these excessive amino acids could signal the shoot N status to nodules, which would then lead to a whole-plant N feedback inhibition of SNF in MmSWRI9-inoculated plants under low Pi conditions (4, 41). In contrast, the identified amino acids in leaves of McCP-31–inoculated plants under Pi-deficient conditions were reduced to a very low level (Fig. 5C), perhaps because of their incorporation into proteins (Fig. 4A), thus preventing whole-plant N feedback inhibition. In this study, increased activity of NADH-GDH was observed in conjunction with an induction of the main enzymes of NH4+ assimilation in McCP-31–induced nodules in response to low Pi stress (Fig. 4 G–J and Fig. S6). It is possible that this increase may help prevent toxic accumulation of NH4+ resulted from a higher SNF capacity in McCP-31–induced nodules relative to MmSWRI9-induced nodules. In addition, glutamate produced by NADH-GDH in McCP-31–induced nodules in response to low Pi stress can be used to assimilate NH4+ derived from SNF through the GS activity. In agreement with this finding, we observed an increase in the levels of glutamate and glutamine in McCP-31–induced nodules under Pi deficiency (Fig. S3 and Dataset S2). Our results confirmed the role of NADH-GDH in detoxification of NH4+ and also, the supply of glutamate for glutamine synthesis (42).
A significant increase in GABA content was observed in McCP-31–induced nodules under low Pi conditions, which would lead to a higher SNF capacity, whereas GABA levels were reduced in MmSWRI9-induced nodules (Fig. S3 and Dataset S2). This finding is consistent with the physiological functions of GABA that include amino acid cycling, providing succinate for normal functioning of nodules, restriction of ROS, and protecting the TCA cycle against oxidative stress (43). GABA shunt, as an NAD+-independent mechanism, maintains normal energy metabolism through bypassing the conversion of α-ketoglutarate to succinate in the TCA cycle (44), and also, limits the accumulation of ROS, protecting certain enzymes of the TCA cycle against oxidative stress conditions (45). In addition, succinate as the end product of GABA shunt supplies the energy needed for SNF (43), because we also detected the increased succinate level in Pi-deficient McCP-31–induced nodules (Fig. S3 and Dataset S2). Moreover, because glutamate is the starting point for GABA synthesis, GABA production is strongly associated with glutamate content (45). In a recent study, a significant increase of glutamate content in McCP-31–induced nodules under Pi deficiency was mirrored in the increased amount of GABA (Fig. S3 and Dataset S2). Additionally, to better prevent oxidative stress under low Pi conditions, the specific activity of NR also increased in McCP-31–induced nodules (Fig. 4N). This enzyme is known to have a critical contribution to the maintenance of redox and cellular energy stability (46). Additionally, the comprehensive metabolite analysis conducted in this study indicated that several other metabolites were differentially synthesized in Pi-deficient McCP-31– and MmSWRI9-induced nodules. These differences in metabolite profiles may also contribute to the better SNF capacity observed in McCP-31–inoculated plants subjected to low Pi. Among the detected metabolites, d-raffinose and galactinol accumulated to higher levels in McCP-31–induced nodules than in MmSWRI9-induced nodules subjected to low Pi (Fig. S3 and Dataset S2). Both of these sugars could significantly help protect cellular metabolism against oxidative damage (47). In addition, an apparent reduction in the levels of certain phosphorylated metabolites, including d-glucose-6P and myo-inositol-1P, was observed in MmSWRI9-induced nodules under low Pi, whereas these same metabolites as well as d-fructose-6P were unchanged or increased in McCP-31–induced nodules under low Pi conditions (Fig. S3 and Dataset S2). It is possible that reduced levels of these metabolites could potentially have a negative impact on several vital biochemical processes, such as glycolysis and phospholipids (48).
In summary, we present, to our knowledge, the first comprehensive metabolic, biochemical, and molecular analyses of contrasting Mesorhizobium–chickpea associations under Pi limitation. Comparative analyses of symbiotic performance conducted in this study showed that the higher SNF capacity of McCP-31–nodulated chickpea under Pi-deficient conditions, relative to that of MmSWRI9-induced nodules, results from a highly coordinated process that includes an extensive reprogramming of various metabolic and biochemical pathways in nodules, roots, and leaves. Thus, the findings of this study fundamentally advance our understanding of how contrasting microsymbionts facilitate acclimation to Pi deficiency. Furthermore, our analyses support the idea that legume adaptability to Pi stress depends on intricate signaling pathways, including those of Pi, sugars, and amino acids. These data revealed the existence of cross-talk among numerous signaling pathways in regulation of Mesorhizobium–chickpea adaptation to Pi limitation. Nevertheless, it cannot be disregarded that other master players, such as small RNAs, might also contribute to regulation of SNF capacity in chickpea under low Pi availability. Additional investigations are, thus, warranted to identify previously unidentified crucial regulators and further clarify the contribution of different Rhizobium strains in host plant acclimation to Pi deficiency. A careful selection of Rhizobium strains with efficient SNF capacity under Pi-deficient conditions may help improve legume crop productivity in low Pi soils. Additionally, the results of this study also provide information that can be used to genetically engineer chickpea cultivars and perhaps, other leguminous crops to have effective symbiotic efficiency under Pi-limiting conditions by targeting the key biochemical responses observed in the presented data.
Materials and Methods
Biological Materials and Growth Conditions.
Chickpea (C. arietinum L. cultivar ILC482) was used throughout the study combined with two symbionts (McCP-31 and MmSWRI9) of the genus Mesorhizobium. ILC482 is a high-yielding elite Kabuli variety with relatively high adaptability to abiotic stress, particularly water scarcity (49). Seeds of ILC482 were germinated in 1.5-L pots containing autoclaved vermiculite as a rooting substrate. Pots were placed in a greenhouse under controlled conditions (continuous 30 °C temperature, 60% relative humidity, 12-h/12-h photoperiod, and 150 μmol m−2 s−1 photon flux density). Seeds were inoculated with a suspension of ∼109 cells mL−1 of either McCP-31 or MmSWRI9 as previously described (50). Plant irrigation was done three times per week with N-free nutrient solution (pH 6.8) (50). KH2PO4 was supplemented to the nutrient solution to a final concentration of either 0.5 mM or 5 μM to establish Pi-sufficient or -deficient conditions, respectively. An appropriate concentration of K2SO4 was also added to the Pi-deficient solution to ensure an equal supply of potassium. Thirty days after sowing, shoots, roots, and nodules were separately collected and dried at 60 °C to a constant weight for DM determination. In addition, leaf, root, and nodule fractions used in assays of enzyme activity, gene expression, and metabolite profiling were also collected, frozen immediately in liquid N2, and stored at −80 °C until the analyses were conducted.
Gene Expression Analysis.
Total RNA purification from chickpea nodules of three biological replicates, DNase I treatment, and cDNA synthesis were carried out as previously described (51).
Chickpea genome annotation (Bioproject PRJNA175619) and key genes identified as Pi starvation response-related genes in Arabidopsis were used in a search for putative Pi starvation response-related genes in chickpea (19, 23, 52–55). The identified chickpea genes and the specific primers used to amplify these genes using real-time quantitative RT-PCR (qRT-PCR) are listed in Dataset S5; 16S rRNA (NR_025953) (9) and IF4a (XM_004513380) (56) were used as reference genes for expression analysis of the nifDK and selected Pi starvation response-related chickpea genes. Analyses of the qRT-PCR data were performed as previously described (57).
Assays for C and N Metabolism-Related Enzymes.
Leaf, root, and nodule samples of four biological replicates were extracted as previously described (9). Total soluble protein content was estimated using the Bradford reagent (58). The activities of SS, AI, PEPC, GS, and AAT were assayed using standard methods (59). UDP-GPP and NAD-MDH activities were assayed according to described protocols (60). The activities of NADH-GOGAT and NADH-GDH were measured as described earlier (61), whereas G6PDH and 6PGDH were assayed using published protocols (62). NADP-ICDH was measured according to previously described methods (63). NR was determined based on a standard protocol (64), and the activity of NAD-ME was assayed as previously described (20). All enzymes were evaluated on a GENios Microplate Reader (Tecan) in a reaction mixture at 25 °C.
Metabolite Profiling.
Samples of nodules, leaves, and roots (25 mg fresh weight of each sample and four biological replicates per tissue type) were extracted in 1 mL extraction mixture comprised of chloroform:MeOH:H2O (2:6:2 vol/vol/vol) and 10 stable isotope reference compounds (65). After extraction, 200 μL supernatant was evaporated to dryness and then, derivatized as previously described (66). An equivalent of 55.6 µg fresh weight derivatized samples were subjected to GC–time of flight MS according to a published method (65). All raw data of detected metabolites were transferred from the ChromaTOF software in NetCDF format to MATLAB software 6.5 (MathWorks). The chromatograms were preprocessed using the high-throughput data analysis (HDA) method (67) and normalized using the cross-contribution compensating multiple standard normalization (CCMN) algorithm (68).
Statistical Analysis.
Three biological replicates from each Pi treatment and tissue type were used to evaluate the level of gene expression. Each biological replicate was run in duplicate using qRT-PCR. For growth parameters, assays of C and N metabolism-related enzymes, and metabolite profiling analyses, four biological replicates from each Pi treatment were used. Data were subjected to a two-way ANOVA, and significant differences between treatments (Pi-sufficient and -deficient treatments) and the two symbiotic associations were measured by Duncan’s multiple range test (P ≤ 0.05). All statistical analyses were determined using SPSS software package 16.0.
Supplementary Material
Acknowledgments
We thank Mr. Makoto Kobayashi for technical assistance. M.N.E. thanks the Lorestan University Research Council for financial support.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609440113/-/DCSupplemental.
References
- 1.López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu Rev Plant Biol. 2014;65(1):95–123. doi: 10.1146/annurev-arplant-050213-035949. [DOI] [PubMed] [Google Scholar]
- 2.Richardson AE, Hocking PJ, Simpson RJ, George TS. Plant mechanisms to optimise access to soil phosphorus. Crop Pasture Sci. 2009;60(2):124–143. [Google Scholar]
- 3.Herrera-Estrella L, López-Arredondo D. Phosphorus: The underrated element for feeding the world. Trends Plant Sci. 2016;21(6):461–463. doi: 10.1016/j.tplants.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Aranjuelo I, Arrese-Igor C, Molero G. Nodule performance within a changing environmental context. J Plant Physiol. 2014;171(12):1076–1090. doi: 10.1016/j.jplph.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Valentine AJ, Benedito VA, Kang Y. Legume nitrogen fixation and soil abiotic stress: From physiology to genomics and beyond. In: Foyer CH, Zhang H, editors. Annual Plant Reviews: Nitrogen Metabolism in Plants in the Post-Genomic Era. vol 42. Wiley-Blackwell; Chichester, UK: 2011. pp. 207–248. [Google Scholar]
- 6.Kleinert A, Venter M, Kossmann J, Valentine A. The reallocation of carbon in P deficient lupins affects biological nitrogen fixation. J Plant Physiol. 2014;171(17):1619–1624. doi: 10.1016/j.jplph.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Sulieman S, Tran L-SP. Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 2015;239:36–43. doi: 10.1016/j.plantsci.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Qin L, et al. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 2012;159(4):1634–1643. doi: 10.1104/pp.112.199786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasr Esfahani M, et al. Approaches for enhancement of N₂ fixation efficiency of chickpea (Cicer arietinum L.) under limiting nitrogen conditions. Plant Biotechnol J. 2014;12(3):387–397. doi: 10.1111/pbi.12146. [DOI] [PubMed] [Google Scholar]
- 10.Jain M, et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.) Plant J. 2013;74(5):715–729. doi: 10.1111/tpj.12173. [DOI] [PubMed] [Google Scholar]
- 11.Gaur PM, Jukanti AK, Varshney RK. Impact of genomic technologies on chickpea breeding strategies. Agronomy. 2012;2(3):199–221. [Google Scholar]
- 12.Srinivasarao C, Ganeshamurthy AN, Ali M, Venkateswarlu B. Phosphorus and micronutrient nutrition of chickpea genotypes in a multi-nutrient-deficient typic ustochrept. J Plant Nutr. 2006;29(4):747–763. [Google Scholar]
- 13.Cabeza RA, et al. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N₂ fixation before emerging P deficiency reaches the nodules. J Exp Bot. 2014;65(20):6035–6048. doi: 10.1093/jxb/eru341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thuynsma R, Valentine A, Kleinert A. Phosphorus deficiency affects the allocation of below-ground resources to combined cluster roots and nodules in Lupinus albus. J Plant Physiol. 2014;171(3-4):285–291. doi: 10.1016/j.jplph.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Fox JE, Gulledge J, Engelhaupt E, Burow ME, McLachlan JA. Pesticides reduce symbiotic efficiency of nitrogen-fixing rhizobia and host plants. Proc Natl Acad Sci USA. 2007;104(24):10282–10287. doi: 10.1073/pnas.0611710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curatti L, Brown CS, Ludden PW, Rubio LM. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc Natl Acad Sci USA. 2005;102(18):6291–6296. doi: 10.1073/pnas.0501216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, et al. A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet. 2013;9(10):e1003865. doi: 10.1371/journal.pgen.1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasr Esfahani M, et al. Mechanisms of physiological adjustment of N2 fixation in Cicer arietinum L. (chickpea) during early stages of water deficit: Single or multi-factor controls. Plant J. 2014;79(6):964–980. doi: 10.1111/tpj.12599. [DOI] [PubMed] [Google Scholar]
- 19.Varshney RK, et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol. 2013;31(3):240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- 20.Le Roux MR, Khan S, Valentine AJ. Organic acid accumulation may inhibit N2 fixation in phosphorus-stressed lupin nodules. New Phytol. 2008;177(4):956–964. doi: 10.1111/j.1469-8137.2007.02305.x. [DOI] [PubMed] [Google Scholar]
- 21.Cabeza RA, et al. The activity of nodules of the supernodulating mutant Mtsunn is not limited by photosynthesis under optimal growth conditions. Int J Mol Sci. 2014;15(4):6031–6045. doi: 10.3390/ijms15046031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang C, Wang J, Zhao J, Tian J, Liao H. Control of phosphate homeostasis through gene regulation in crops. Curr Opin Plant Biol. 2014;21:59–66. doi: 10.1016/j.pbi.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Misson J, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102(33):11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Secco D, et al. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012;193(4):842–851. doi: 10.1111/j.1469-8137.2011.04002.x. [DOI] [PubMed] [Google Scholar]
- 25.Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;143(4):1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larrainzar E, Gil-Quintana E, Seminario A, Arrese-Igor C, González EM. Nodule carbohydrate catabolism is enhanced in the Medicago truncatula A17-Sinorhizobium medicae WSM419 symbiosis. Front Microbiol. 2014;5:447. doi: 10.3389/fmicb.2014.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsikou D, et al. Cessation of photosynthesis in Lotus japonicus leaves leads to reprogramming of nodule metabolism. J Exp Bot. 2013;64(5):1317–1332. doi: 10.1093/jxb/ert015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sulieman S, Ha CV, Schulze J, Tran L-SP. Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J Exp Bot. 2013;64(10):2701–2712. doi: 10.1093/jxb/ert122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plaxton WC, Tran HT. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011;156(3):1006–1015. doi: 10.1104/pp.111.175281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plaxton WC, Podesta FE. The functional organization and control of plant respiration. CRC Crit Rev Plant Sci. 2006;25(2):159–198. [Google Scholar]
- 31.Le Roux MR, Ward CL, Botha FC, Valentine AJ. Routes of pyruvate synthesis in phosphorus-deficient lupin roots and nodules. New Phytol. 2006;169(2):399–408. doi: 10.1111/j.1469-8137.2005.01594.x. [DOI] [PubMed] [Google Scholar]
- 32.Juszczuk IM, Rychter AM. Pyruvate accumulation during phosphate deficiency stress of bean roots. Plant Physiol Biochem. 2002;40(9):783–788. [Google Scholar]
- 33.Aranjuelo I, et al. Concerted changes in N and C primary metabolism in alfalfa (Medicago sativa) under water restriction. J Exp Bot. 2013;64(4):885–897. doi: 10.1093/jxb/ers367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, et al. Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics. 2011;11(24):4648–4659. doi: 10.1002/pmic.201100231. [DOI] [PubMed] [Google Scholar]
- 35.Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157(3):423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Liao H, Lucas WJ. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol. 2014;56(3):192–220. doi: 10.1111/jipb.12163. [DOI] [PubMed] [Google Scholar]
- 37.Corpas FJ, Barroso JB. NADPH-generating dehydrogenases: Their role in the mechanism of protection against nitro-oxidative stress induced by adverse environmental conditions. Front Environ Sci. 2014;2:55. [Google Scholar]
- 38.Almeida JP, Hartwig UA, Frehner M, Nösberger J, Lüscher A. Evidence that P deficiency induces N feedback regulation of symbiotic N2 fixation in white clover (Trifolium repens L.) J Exp Bot. 2000;51(348):1289–1297. [PubMed] [Google Scholar]
- 39.Sulieman S, Tran L-SP. Asparagine: An amide of particular distinction in the regulation of symbiotic nitrogen fixation of legumes. Crit Rev Biotechnol. 2013;33(3):309–327. doi: 10.3109/07388551.2012.695770. [DOI] [PubMed] [Google Scholar]
- 40.Sulieman S, Schulze J, Tran L-SP. Comparative analysis of the symbiotic efficiency of Medicago truncatula and Medicago sativa under phosphorus deficiency. Int J Mol Sci. 2013;14(3):5198–5213. doi: 10.3390/ijms14035198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze J. Source-sink manipulations suggest an N-feedback mechanism for the drop in N2 fixation during pod-filling in pea and broad bean. J Plant Physiol. 2003;160(5):531–537. doi: 10.1078/0176-1617-00709. [DOI] [PubMed] [Google Scholar]
- 42.Magadlela A, Vardien W, Kleinert A, Steenkamp ET, Valentine AJ. Variable P supply affects N metabolism in a legume tree, Virgilia divaricata, from nutrient-poor Mediterranean-type ecosystems. Funct Plant Biol. 2016;43(3):287–297. doi: 10.1071/FP15262. [DOI] [PubMed] [Google Scholar]
- 43.Sulieman S. Does GABA increase the efficiency of symbiotic N2 fixation in legumes? Plant Signal Behav. 2011;6(1):32–36. doi: 10.4161/psb.6.1.14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michaeli S, Fromm H. Closing the loop on the GABA shunt in plants: Are GABA metabolism and signaling entwined? Front Plant Sci. 2015;6:419. doi: 10.3389/fpls.2015.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouché N, Fromm H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004;9(3):110–115. doi: 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Bianucci E, Fullana C, Furlan A, Castro S. Antioxidant defense system responses and role of nitrate reductase in the redox balance maintenance in Bradyrhizobium japonicum strains exposed to cadmium. Enzyme Microb Technol. 2013;53(5):345–350. doi: 10.1016/j.enzmictec.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Nishizawa A, Yabuta Y, Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147(3):1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang CY, et al. Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.) Plant Cell Physiol. 2008;49(5):691–703. doi: 10.1093/pcp/pcn044. [DOI] [PubMed] [Google Scholar]
- 49.Rozrokh M, Sabaghpour SH, Armin M, Asgharipour M. The effects of drought stress on some biochemical traits in twenty genotypes of chickpea. Eur J Exp Biol. 2012;2(6):1980–1987. [Google Scholar]
- 50.Nasr Esfahani M, Mostajeran A. Rhizobial strain involvement in symbiosis efficiency of chickpea–rhizobia under drought stress: Plant growth, nitrogen fixation and antioxidant enzyme activities. Acta Physiol Plant. 2011;33(4):1075–1083. [Google Scholar]
- 51.Nguyen KH, et al. Correlation between differential drought tolerability of two contrasting drought-responsive chickpea cultivars and differential expression of a subset of CaNAC genes under normal and dehydration conditions. Front Plant Sci. 2015;6:449. doi: 10.3389/fpls.2015.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morcuende R, et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30(1):85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- 53.Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 2007;143(1):156–171. doi: 10.1104/pp.106.090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin W-Y, Lin S-I, Chiou T-J. Molecular regulators of phosphate homeostasis in plants. J Exp Bot. 2009;60(5):1427–1438. doi: 10.1093/jxb/ern303. [DOI] [PubMed] [Google Scholar]
- 55.Ha S, Tran LS. Understanding plant responses to phosphorus starvation for improvement of plant tolerance to phosphorus deficiency by biotechnological approaches. Crit Rev Biotechnol. 2014;34(1):16–30. doi: 10.3109/07388551.2013.783549. [DOI] [PubMed] [Google Scholar]
- 56.Garg R, Sahoo A, Tyagi AK, Jain M. Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.) Biochem Biophys Res Commun. 2010;396(2):283–288. doi: 10.1016/j.bbrc.2010.04.079. [DOI] [PubMed] [Google Scholar]
- 57.Le DT, et al. Differential gene expression in soybean leaf tissues at late developmental stages under drought stress revealed by genome-wide transcriptome analysis. PLoS One. 2012;7(11):e49522. doi: 10.1371/journal.pone.0049522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 59.González EM, Gordon AJ, James CL, Arrese-lgor C. The role of sucrose synthase in the response of soybean nodules to drought. J Exp Bot. 1995;46(10):1515–1523. [Google Scholar]
- 60.Gordon AJ, Kessler W. Defoliation-induced stress in nodules of white clover: II. Immunological and enzymic measurmens of key proteins. J Exp Bot. 1990;41(10):1255–1262. [Google Scholar]
- 61.Groat RG, Vance CP. Root nodule enzymes of ammonia assimilation in alfalfa (Medicago sativa L.): Development patterns and response to applied nitrogen. Plant Physiol. 1981;67(6):1198–1203. doi: 10.1104/pp.67.6.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Copeland L, Vella J, Hong Z. Enzymes of carbohydrate metabolism in soybean nodules. Phytochemistry. 1989;28(1):57–61. [Google Scholar]
- 63.Canino S, Nieri B, Pistelli L, Alpi A, De Bellis L. NADP+-isocitrate dehydrogenase in germinating cucumber cotyledons: Purification and characterization of a cytosolic isoenzyme. Physiol Plant. 1996;98(1):13–19. [Google Scholar]
- 64.Hall N, Tomsett AB. Structure-function analysis of NADPH:nitrate reductase from Aspergillus nidulans: Analysis of altered pyridine nucleotide specificity in vivo. Microbiology. 2000;146(Pt 6):1399–1406. doi: 10.1099/00221287-146-6-1399. [DOI] [PubMed] [Google Scholar]
- 65.Kusano M, et al. Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855(1):71–79. doi: 10.1016/j.jchromb.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 66.Kusano M, et al. Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst Biol. 2007;1(1):53. doi: 10.1186/1752-0509-1-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jonsson P, et al. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal Chem. 2005;77(17):5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- 68.Redestig H, et al. Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal Chem. 2009;81(19):7974–7980. doi: 10.1021/ac901143w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.