Significance
The recently identified rhinovirus C (RV-C) species of picornaviruses might be associated with more severe respiratory infections than other rhinoviruses in children. The RV-C have been linked to 50–85% of hospital-level childhood asthma exacerbations that can lead to significant adult respiratory problems. However, there are currently no effective antiviral treatments or vaccines available. Using cryoelectron microscopy, we have determined the atomic structures of the full virion and native empty particle (NEP) of a cell-adapted RV-C strain. The structures highlight novel immunogenic sites on the virus surface, a probable binding site for the RV-C receptor, molecular determinants of antiviral resistance, and the rationale of utilizing NEPs as vaccine candidates. These results provide the basis for designing antiviral therapeutics and vaccines.
Keywords: rhinovirus C, cryoelectron microscopy, atomic structure, asthma
Abstract
Isolates of rhinovirus C (RV-C), a recently identified Enterovirus (EV) species, are the causative agents of severe respiratory infections among children and are linked to childhood asthma exacerbations. The RV-C have been refractory to structure determination because they are difficult to propagate in vitro. Here, we report the cryo-EM atomic structures of the full virion and native empty particle (NEP) of RV-C15a. The virus has 60 “fingers” on the virus outer surface that probably function as dominant immunogens. Because the NEPs also display these fingers, they may have utility as vaccine candidates. A sequence-conserved surface depression adjacent to each finger forms a likely binding site for the sialic acid on its receptor. The RV-C, unlike other EVs, are resistant to capsid-binding antiviral compounds because the hydrophobic pocket in VP1 is filled with multiple bulky residues. These results define potential molecular determinants for designing antiviral therapeutics and vaccines.
The Picornaviridae family includes a variety of small, nonenveloped, icosahedral viruses with positive-strand RNA genomes (1). Many picornaviruses (e.g., rhinoviruses, polioviruses, coxsackieviruses, enterovirus A71, and enterovirus D68) that infect humans and cause high morbidity belong to the Enterovirus genus (EV) (1). A number of these viruses have been structurally characterized by X-ray crystallography (2–5), establishing the general mechanisms for virus infection and for the development of effective anti-EV therapeutics. Nevertheless, rhinovirus C (RV-C), a newly discovered species among the EVs, remains enigmatic.
RV-C viruses (55 types), together with RV-A and RV-B viruses (∼100 types), are the leading cause of common colds. However, the RV-C might be associated with more severe respiratory infections among children than any other known rhinoviruses (6, 7). In contrast to other RVs, RV-Cs use cadherin-related family member 3 (CDHR3) as a cellular receptor (8). This childhood asthma susceptibility gene product is expressed in the human lower respiratory tract (9). In line with this etiology, RV-Cs cause a significantly higher rate of lower respiratory tract infections in children than in adults (10) and are directly associated with childhood asthma exacerbations (7). Similar to influenza, RV-C infections peak in winter months. Currently, there are no vaccines or effective antiviral treatments available.
RV-C isolates have been refractory to structural characterization since their discovery in 2006 (11) because of an inability to infect standard tissue culture (e.g., HeLa) (12). Only modeled structures, based on amino acid sequence comparisons, have been available to aid biological investigations (12–15) However, with recent advances in direct electron detection (16) and image processing approaches (17, 18), single-particle cryoelectron microscopy (cryo-EM) has now emerged as a powerful method for determining near atomic resolution (better than 4 Å) structures of macromolecular assemblies (19). Cryo-EM requires only a limited amount of sample without intensive purification, offering advantages over X-ray crystallography in structural studies of samples that are difficult to produce.
Picornavirus capsids are assembled from 60 copies of biological protomers, each composed of four proteins: VP1, VP2, VP3, and VP4 (2). The three large surface polypeptides, VP1, VP2, and VP3, are folded into eight-stranded antiparallel “jelly rolls.” During the assembly process, autocatalytic cleavage of precursor VP0 into VP2 and VP4 in the presence of viral RNA results in the formation of full infectious virions (20). The arrangement of jelly rolls in the virions exhibits pseudo T = 3 icosahedral symmetry with an outer diameter of ∼300 Å (2, 3). The internal surface of the capsid is lined by the 60 copies of VP4. A surface depression or canyon (2), encircling each fivefold axis, is frequently the receptor binding site for many EVs (21). Amino acid residues located on the outer surface of the virus but not specifically within this canyon are typically involved in forming immunogenic sites recognized by neutralizing antibodies. The canyon allows only limited access to these antibodies (22). In many EVs, a hydrophobic pocket within the VP1 jelly roll and situated underneath the canyon floor is occupied by a fatty-acid like molecule, or “pocket factor” (23, 24), that regulates the conformational states of the virus during cell entry (25). Capsid-binding reagents that replace the pocket factor within VP1 are effective antiviral therapeutics against many EVs (26), but not RV-Cs (14).
Here we report atomic resolution cryo-EM structures of the full and native empty particles (NEP) of the cell-adapted RV-C15a strain. These structures highlight novel immunogenic surfaces, a probable binding site for the glycosylated CDHR3 receptor molecule, and the requirements for antiviral compound resistance. The external surface of both types of particles is almost identical. Thus, the RV-C NEPs represent potential vaccine candidates. The present findings also identify targets for designing anti–RV-C therapeutics.
Results
Production of RV-C15a Viruses.
Recently, a recombinant RV-C15 virus (12), adapted for tissue culture growth by serial passage in HeLa-E8 cells (8) (a transduced HeLa cell line expressing CDHR3) led to new protocols for enhanced virus yields. The derivative, RV-C15a, represents a cell-adapted, uncloned population. The consensus sequence of this population differs in the capsid region from that of RV-C15, primarily by a single, high-frequency, nucleotide polymorphism. The substitution converts residue 1125 from Thr to Lys. (Numbering convention adds 1,000 to VP1 residues, 2,000 to VP2 residues, 3,000 to VP3 residues, and 4,000 to VP4 residues.) In this study, an RV-C15a sample, purified only by sucrose cushion sedimentation, was used for cryo-EM structure analysis. To obtain a minimum of 5–6 particles per micrograph, given the high dilution of the sample, data collection was carried out at a low magnification. Specifically, movies of frozen RV-C15a particles within a thin layer of vitreous ice were recorded at a nominal magnification of 14,000× using a Gatan K2 Summit direct electron detector. However, the tradeoff was a low signal-to-noise ratio and a high anisotropic magnification distortion compared with what would be the case were high magnifications used for data collection. The primary data were collected in less than 1 wk.
Biochemical Characterization of Two Forms of Particles.
Cryo-EM micrographs of RV-C15a showed the presence of two major forms of particles. One form lacked density at their centers and another form had density at their centers (Fig. S1). When fractionated on sucrose gradients, these types of particles separated from each other. One form was full, infectious virions that contained VP1, VP2, VP3, and VP4, whereas the other form (∼30% of all particles) was NEP that had VP1, VP3, and uncleaved VP0, as shown by Western blot analyses using an antibody against VP2 (Fig. S2A). Unlike the full virions, NEPs were devoid of viral RNA and had no infectivity to HeLa-E8 cells (Fig. S2B).
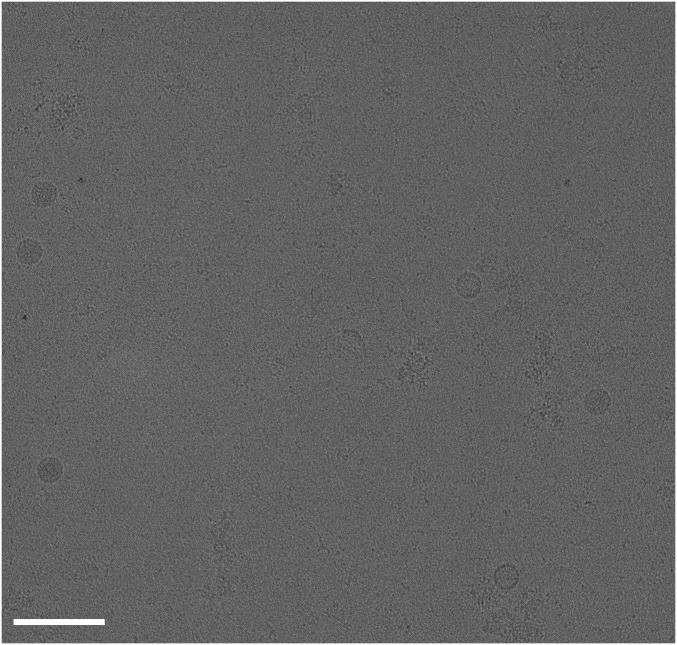
Fig. S1.
A cryo-EM micrograph shows the presence of full and empty RV-C15a particles. This micrograph was recorded at a defocus of 3.1 µm. (Scale bar, 100 nm.)
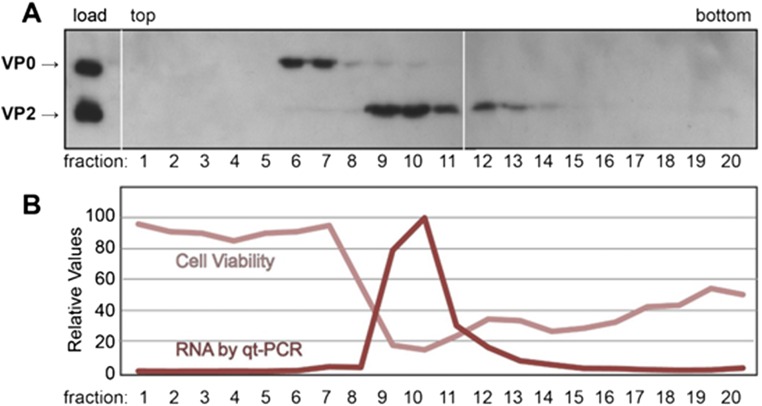
Fig. S2.
Characterization of two forms of RV-C15a particles. A sample of RV-C15a was sedimented through a sucrose gradient. Fractions (1 mL) were collected (from the top) and then probed for VP2/VP0 content by Western blot analyses (A) using mouse anti-RV-C15-VP2. These fractions were also tested for infectivity according to cytopathic effect and for RNA content by qRT-PCR (B).
Cryo-EM Structure Determination.
Images of full and empty particles were separated by reference-free 2D classification using the program Relion (17). A “truly independent” procedure of 3D reconstruction was used to avoid overfitting to noise (18). Essentially, initial model calculations, low-resolution refinements and high-resolution refinements were performed independently for each of the two half-data subsets. Parameters of anisotropic magnification distortion, a major resolution limiting factor for large assemblies (e.g., viruses), were estimated using powder diffraction patterns of polycrystalline gold particles (27). The resultant parameters were used in the program jspr (18) for correcting anisotropic magnification distortion on individual particles. Refinements of particle center, orientation, defocus, astigmatism, scale, and beam tilt resulted in icosahedral reconstructions of 8,973 full particles and 3,614 empty particles at 2.8 Å and 3.2 Å resolution, respectively (Fig. S3 and Table S1). The resolution of the maps was estimated by calculating the Fourier shell correlation between the two half maps, using 0.143 as a cutoff (28) (Fig. S4).
Fig. S3.
Typical densities of the full (A–C) and empty (D–F) particle EM maps.
Table S1.
Cryo-EM data statistics
| RV-C15a full particle | RV-C15a empty particle | |
| EM data collection and processing | ||
| Microscope | Titan Krios | Titan Krios |
| Accelerating voltage, kV | 300 | 300 |
| Camera | Gatan K2 Summit | Gatan K2 Summit |
| Pixel size, Å/pixel* | 1.04 | 1.04 |
| Dose rate, e−/(pixel⋅s) | 8 | 8 |
| Total dose, e−/Å2 | 25.7 | 25.7 |
| No. of particles for the final reconstruction | 8,973 | 3,614 |
| Resolution, ņ | 2.79 | 3.16 |
| Map sharpening B factor, Å2 | −108.6 | −122.2 |
| Model statistics | ||
| Correlation coefficient (around atoms)‡ | 0.885 | 0.880 |
| No. of atoms | ||
| Protein | 6,221 | 6,007 |
| Water | 60 | 0 |
| Average B factor, Å2 | 24.1 | 31.7 |
| rmsd§ | ||
| Bond lengths, Å | 0.010 | 0.010 |
| Bond angles, ° | 0.960 | 0.944 |
| Ramachandran plot§ | ||
| Favored, % | 93.4 | 92.0 |
| Allowed, % | 6.5 | 7.6 |
| Outliers, % | 0.1 | 0.4 |
Pixel size in superresolution mode. The physical pixel size is 2.08 Å per pixel.
Resolution was determined by the Fourier shell correlation between two half-maps using 0.143 as a cutoff value.
Real-space correlation coefficient (around atoms) between the final EM map and a density map calculated based on the coordinates.
Based on the criteria of MolProbity.
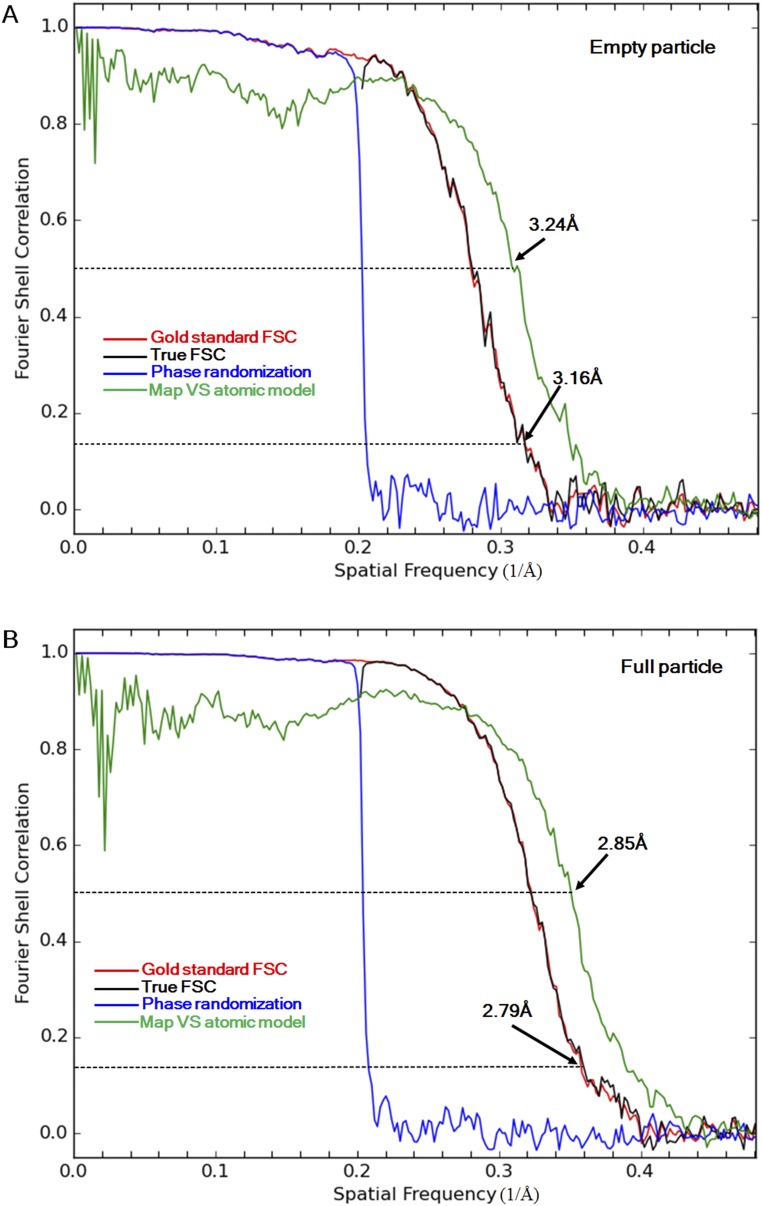
Fig. S4.
The resolution of the empty (A) and full (B) particle maps is estimated using FSC curves. The FSC curves between two half-subset maps calculated using the original images (gold standard FSC) and using phase randomized (beyond 5 Å) images (phase randomization) are colored red and blue. “True” FSC curves are colored black. The FSC curves between the final EM map and a density map computed based on the modeled atomic coordinates are colored green.
RV-C15a Has a Spiky Structure.
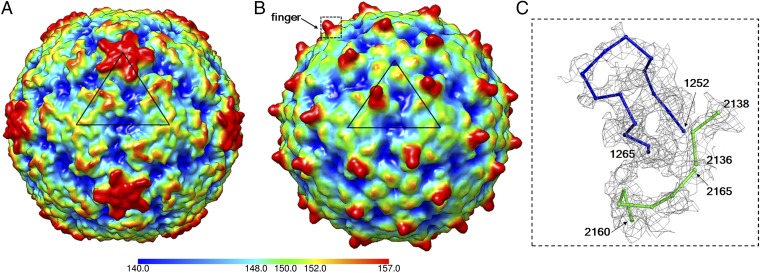
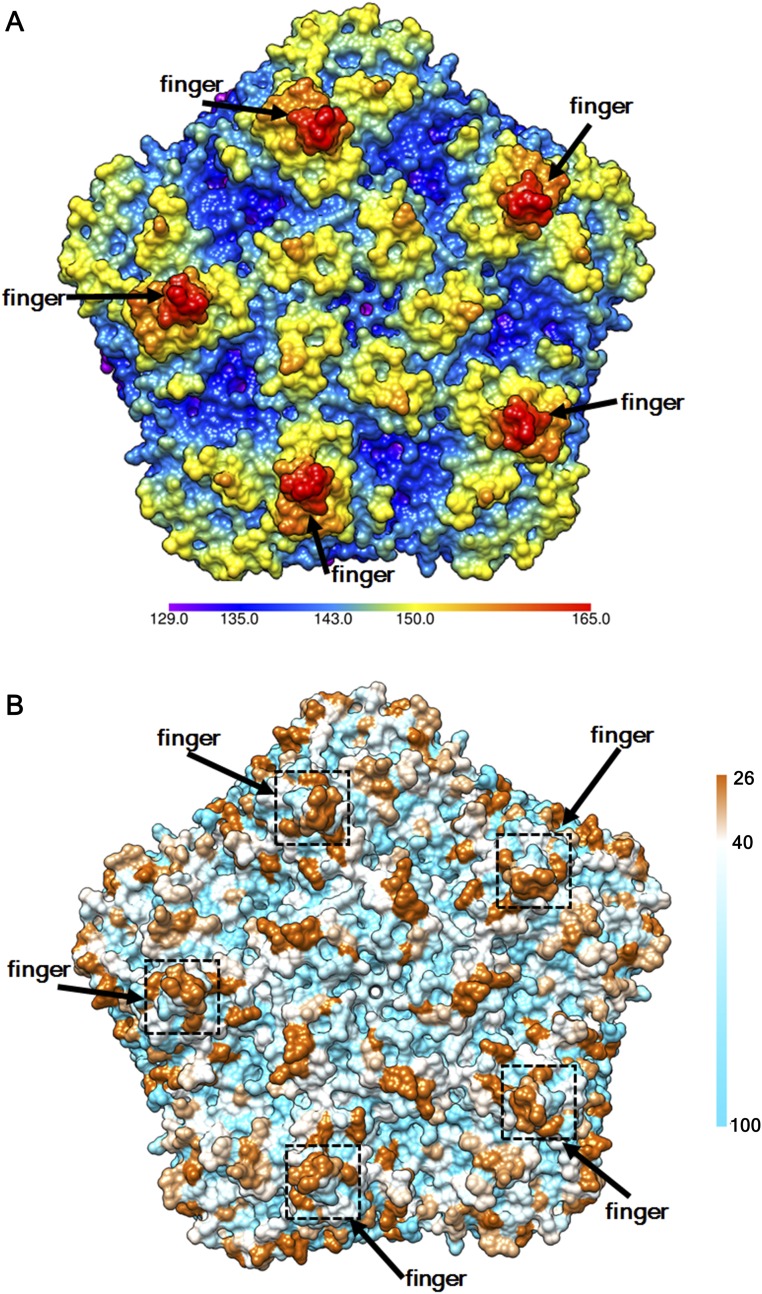
The structure of the RV-C15a full particle has 60 dominant spike-like protrusions, or “fingers,” on the outer surface of the virion (Fig. 1). In contrast, all other EV structures have smoother, spherical surfaces (Fig. 1). Each RV-C15a finger, located at the juncture between VP1, VP2, and VP3 that form a protomer, is formed by the VP1 C-terminal residues 1252–1265 as well as residues 2136–2138 and 2160–2165 that form part of the VP2 EF loop (the loop that connects β strand E and β strand F in the jelly roll; Fig. 1). The quality of the density for the finger is not as good as is typical for the rest of the virus, presumably because this density is on the periphery of the virus. It is noteworthy that residues 2160–2165 are highly variable among alignments of RV-C sequences (13). This segment corresponds to the neutralizing immunogenic site NIm-II on the RV-B14 structure (2, 13). The VP1 contribution to the finger, residues 1252–1265, is an RV-C-specific insertion. This region is also conserved in length but not in sequence among all members of the RV-C (Fig. S5).
Fig. 1.
The spiky structure of RV-C15a. A 10-Å resolution density map of RV-B14 (PDB ID code 4RHV) (A) and RV-C15a (B) calculated based on the respective coordinates is colored by radial distance (Å) to the virus center. A black triangle indicates an icosahedral asymmetric unit on each of the two viruses. A rectangle (black dashed line) outlines the limit of a close up view of a “finger” in C. Residues that form the finger region, which are fitted into the EM map densities (gray), are shown as Cα backbones and colored blue (VP1 residues) and green (VP2 residues).
Fig. S5.
The finger region on the RV-C15a structure is variable in amino acid sequence among RV-C viruses. A surface representation of a pentamer of the RV-C15a capsid is colored by radial distance (Å) to the virus center (A) or by conservation of amino acid sequences (B) among 33 types of RV-C virus. A total of 67 sequences of RV-C viruses for which the complete sequence of the P1 region is available are used in sequence alignment. Shown in the color key is the occurrence (%) of the most popular residue at a given alignment position among the 67 sequences.
Because of relatively large deletions (21–35 residues) in parts of the VP1 BC, DE, and HI loops, the RV-C15a structure lacks a protruding “plateau” around each of the fivefold vertices, a characteristic feature of other EV (Fig. 1 A and B). Thus, the RV-C do not have the analogous surface mass near the fivefold vertices to form immunogenic sites equivalent to NIm-IA (VP1 BC loop) and NIm-IB (VP1 DE loop) on RV-B14 (2). Instead, the finger regions, as mentioned above, probably function as the dominant antigenic sites (13). As another consequence of these finger regions, the RV-C15a particles have narrow, noncontinuous canyons, much like the surface of EV-D68, a virus that also causes respiratory illnesses (5). In each icosahedral asymmetric unit, the C-proximal, RV-C15a VP1 insertion helps create a wall-like feature blocking the eastern end of the canyon (defined with respect to the usual orientation of picornaviruses used in most figures; Fig. 1 A and B).
A Sequence-Conserved Depression Could Bind Glycosylated CDHR3.
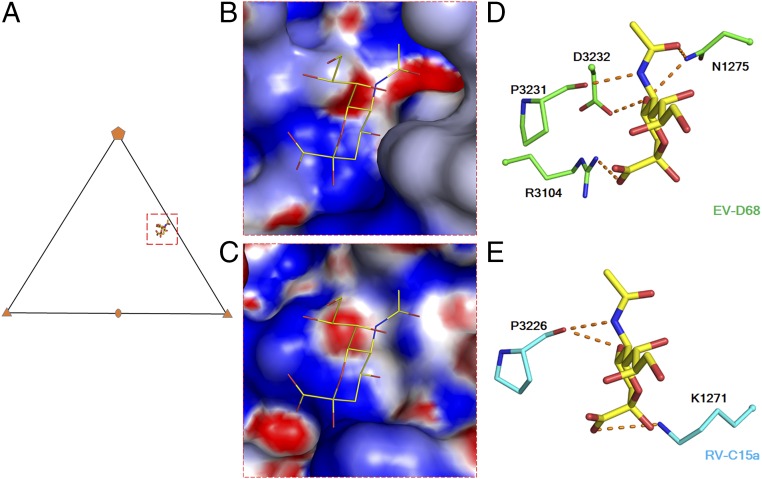
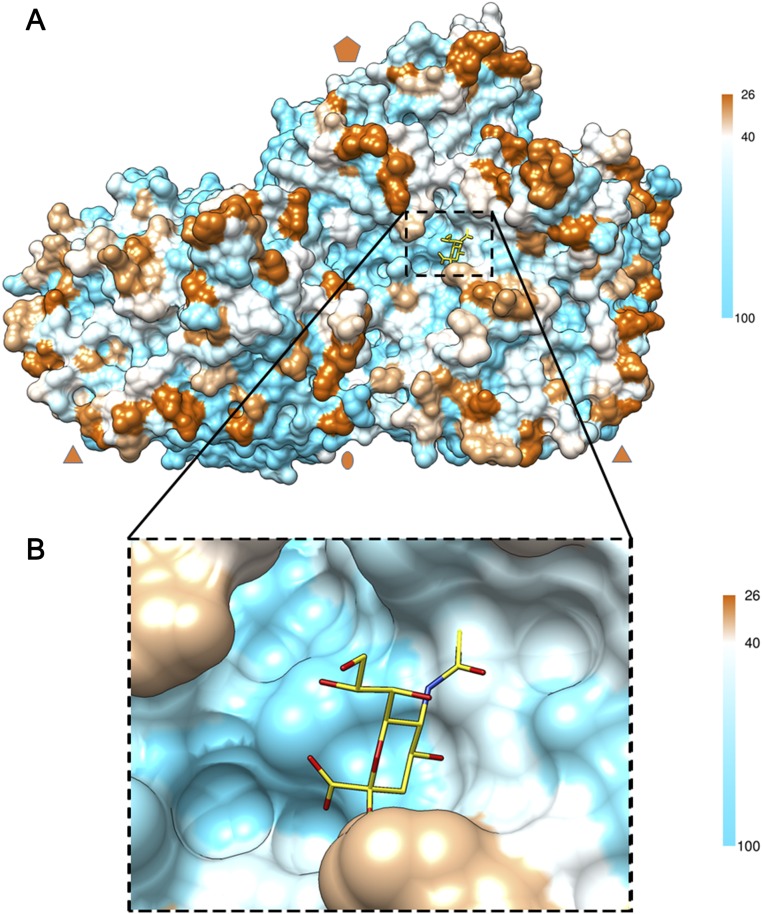
Sialic acid is the glycan moiety recognized by EV-D68 when it interacts with its cellular receptor (25). Superposition of EV-D68 structure complexed with sialic acid (Fig. 2A) onto the structure of RV-C15a showed that the region near the eastern end of the RV-C15a canyon has a similar surface electrostatic potential as the sialic acid binding site on EV-D68 (Fig. 2 B and C). In the EV-D68, sialic acid can be bound mainly by the Pro3231 carbonyl group and by the Arg3104 guanidinium group (Fig. 2D). In RV-C15a, potentially those interactions would be replaced by the structurally equivalent carbonyl group of Pro3226 and by the side chain amino group of Lys1271, respectively (Fig. 2E). Some of the nearby surface residues contributing to this region are conserved among all RV-C (Fig. S6), and it is clear that the overall topography could readily accommodate a sialic acid ligand. Therefore, this region, close to the base of each finger in the RV-C15a structure, is a likely binding site for a CDHR3 glycan. Consistent with this prediction, mutation of Asn186, a key glycosylation site on CDHR3, impairs RV-C15 binding to receptor-expressing cells (8). Therefore, glycans must play an important role in RV-C receptor interactions, as they do also for EV-D68.
Fig. 2.
A potential binding site for glycans on the RV-C receptor. (A) A triangle indicates an icosahedral asymmetric unit. A red rectangle (dashed line) outlines the limit of the sialic acid binding site shown in B and C. Surface electrostatic potential of EV-D68 (PDB ID code 5BNO) (B) and RV-C15a (C) is represented with a scale of −8kT/e (red) to 8kT/e (blue). (D and E) The sialic acid (yellow) interacts with surrounding residues on EV-D68 (green) and as anticipated on RV-C15a (cyan). Red dashed lines indicate (potential) polar interactions. Oxygen and nitrogen atoms are colored red and blue, respectively.
Fig. S6.
A sequenced-conserved region near the base of the finger on RV-C15a forms a potential binding site for sialic acid. (A) A surface representation of two protomers of the RV-C15a capsid is colored by conservation of amino acid sequences among 33 types of RV-C virus as in Fig. S5B. A black rectangle (dash line) outlines the limit of a close-up view of the potential sialic acid (yellow) binding site shown in B.
The VP1 Hydrophobic Pocket Is Unsuitable for Antiviral Capsid Binding Agents.
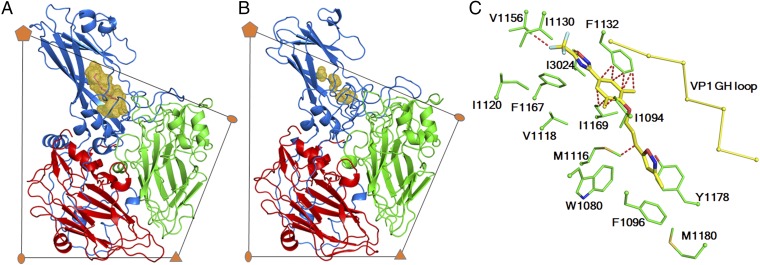
Unlike many EV structures, the hydrophobic pocket within the VP1 jelly roll fold, where a pocket factor is typically bound (4, 5, 23), is collapsed in RV-C15a (Fig. 3 A and B). The collapsed structure is similar to the empty pockets found in purified RV-B14 (2) and RV-B3 (29). None of these three structures have sufficient space to accommodate a fatty-acid pocket factor, because for each, the VP1 GH loop, located at the boundary between the canyon and the entrance to the VP1 pocket, is in a conformation that squeezes the pocket. Nevertheless, in RV-B3 and RV-B14, the flexibility of the VP1 GH loop allows enlargement of the pocket that then can bind antiviral reagents. The RV-B14 pocket is lined with multiple small residues (e.g., Ala, Ser, Val, etc.) that can accommodate such compounds. In contrast, the collapsed RV-C15a VP1 pocket is filled with bulky, hydrophobic residues (in particular, Trp1080, Phe1096, Met1116, and Met1180; Fig. 3C and Table S2). These amino acids are conserved in almost all RV-Cs (14). Additionally, Ile1198 and Tyr1246 partially block the entrance to the VP1 pocket. Therefore, as has been observed experimentally (14), no RV-Cs are likely to be responsive to antiviral therapies based on pocket-binding compounds.
Fig. 3.
RV-C15a has a collapsed VP1 hydrophobic pocket. RV-A16 (A) and RV-C15a (B) are colored by polypeptides: VP1 (blue), VP2 (green), and VP3 (red). The volume of the VP1 hydrophobic pocket, calculated using PyMOL, is colored gold. (C) Residues lining the VP1 pocket of RV-C15a clash with pleconaril (a capsid-binding inhibitor against many EV but not RV-C) when superimposing the structures of RV-C15a and RV-B14 complexed with pleconaril. The VP1 GH loop (yellow) of RV-B14 is shown as Cα backbones, which adopts a conformation that can accommodate pleconaril. Red dashed lines indicate a distance of closer than 2.5 Å between a given atom of a RV-C15 residue (green) and a given atom of pleconaril (yellow). Oxygen, nitrogen, and sulfur atoms are colored red, dark blue, and dark yellow, respectively.
Table S2.
Comparison of amino acid residues lining the VP1 hydrophobic pocket among enteroviruses
| Residues | RV-C15a | RV-A16 (1AYM) | RV-B14 (4RHV) | EV-A71 (3ZFE) | CVB3 (1COV) | PV1 (1ASJ) | EV-D68 (4WM8) |
| Head | |||||||
| 1096 | Phe | Leu | Leu | Ile | Pro | Tyr | Thr |
| 1106 | Phe | Phe | Leu | Val | Leu | Leu | Leu |
| 1114 | Phe | Phe | Phe | Phe | Phe | Phe | Phe |
| 1178 | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr |
| 1180 | Met | Met | Cys | Trp | Asn | His | Val |
| 1198 | Ile | Thr | Ile | Ala | Ile | Ala | Ile |
| 1246 | Tyr | His | Gly | Ala | Lys | — | Ala |
| Middle part | |||||||
| 1080 | Trp | Ile | Val | Val | Val | Val | Val |
| 1092 | Trp | Trp | Trp | Trp | Trp | Trp | Trp |
| 1094 | Ile | Ile | Ile | Ile | Ile | Ile | Ile |
| 1116 | Met | Ser | Ser | Ala | Leu | Met | Ala |
| 1169 | Ile | Leu | Val | Val | Val | Val | Ile |
| 1204 | Met | Met | Met | Met | Met | Phe | Ile |
| 1224 | Val | His | His | Met | Phe | Leu | Met |
| End of the pocket factor | |||||||
| 1118 | Val | Ile | Tyr | Phe | Leu | Leu | Ile |
| 1120 | Ile | Met | Ile | Phe | Phe | Phe | Ile |
| 1130 | Ile | Tyr | Ala | Tyr | Ile | Ile | Ala |
| 1132 | Phe | Tyr | Tyr | Phe | Tyr | Tyr | Phe |
| 1154 | Pro | Ala | Pro | Pro | Pro | Pro | Ala |
| 1156 | Val | Val | Val | Val | Val | Ile | Val |
| 1167 | Phe | Phe | Phe | Val | Met | Ile | Met |
| 1207 | Leu | Leu | Met | Phe | Leu | Leu | Leu |
| 3024 | Ile | Ala | Ala | Ile | Ala | Ala | Val |
Residues are numbered based on RV-C15a numbering. In analogy with EV that have a pocket factor bound in the VP1 pocket, RV-C15a residues that are close to the head, the middle part, and the end of the pocket factor are indicated.
Comparison of the Full and Empty Particle Structures.
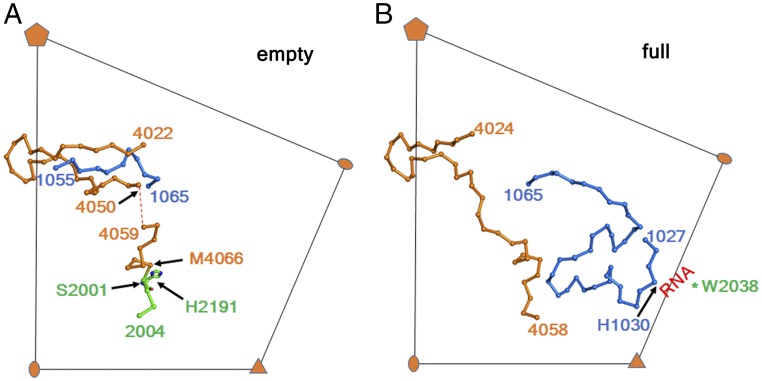
RV-C15a full and empty particles differed mainly in regions on the inner surfaces of their capsid shells (Fig. 4). In particular, the VP1 N-terminal residues 1017–1053 are well-ordered in the full particle map, but disordered in the empty particle map; this is consistent with other EV structures where the VP1 N terminus is involved in binding to viral RNA (23) and is externalized before ejecting the genome during infection (30). Thus, the specific configuration of this internal region is RNA-dependent and is likely to exert strong influence on VP0 cleavage when the RNA is packaged. In the empty particles, VP0 residues 4024–4050 form a hairpin loop positioning the VP0 cleavage site in close proximity to His2191 (Fig. 4A), a crucial residue in the cleavage mechanism (31). Nearby VP1 residues 1054–1064 interact with the VP0 hairpin within the same protomer, presumably helping to set up the pending cleavage reaction. However, in the full particle structure, the VP1 N-terminal residues 1027–1053 (disordered in the empty particles) interact with the C terminus of VP4 within the same protomer and participates in viral RNA binding (Fig. 4B).
Fig. 4.
The full and empty RV-C15a particle structures differ in regions at the capsid interior. The VP1 N terminus and VP4 undergo structural rearrangements when the empty particle (A) and full particle (B) structures are compared. Amino acid residues are shown as Cα backbones. VP4, VP2, and VP1 are colored orange, green, and blue, respectively. In the RV-C15a full particle structure, His1030, together with a conserved residue Trp2038 shared by many EV, are involved in forming an RNA binding site.
Discussion
The cryo-EM structure of RV-C15a showed a collapsed hydrophobic pocket in VP1 that is filled with multiple bulky residues that inhibit the entrance of compounds, which inhibit other EV by binding into the VP1 pocket; this is reminiscent of the collapsed pockets of non-EV picornaviruses, such as foot-and-mouth disease virus (genus Aphthovirus) (32) and mengovirus (genus Cardiovirus) (33). Those pockets are similarly occupied by multiple bulky, hydrophobic side chains and are unable to serve as drug targets.
Inclusion of large hydrophobic residues into the VP1 pocket of RV-Cs and incorporation of a fatty acid-like pocket factor into the VP1 pocket of many other EVs produce similar hydrophobic effects that favor the folding of the VP1 jelly roll β barrel. On one hand, large hydrophobic residues (e.g., Phe, Trp, and Met) are more effective than small residues (e.g., Val and Ala) at reducing solvent-accessible surface areas in the pocket. Likewise, a pocket factor with a long hydrophobic tail (more than eight carbon atoms) plays a crucial role in expelling water out of the VP1 pocket of many EVs that contain multiple small residues. On the other hand, the presence of large hydrophobic side chains or a pocket factor in a VP1 pocket keeps the two β sheets (one with β strands C, H, E, and F, and the other with β strands B, I, D, and G) in a favorable distance for forming a β barrel and offers hydrophobic interactions that stabilize the jelly roll fold.
Concerning enteroviruses (e.g., EV-A71 and RV-A2) that have a pocket factor bound in the VP1 pocket in the native form, a collapsed VP1 pocket is formed when these viruses are converted into uncoating intermediates, including altered (A)-particle (34) and emptied particle (35). In this process, the VP1 GH loop changes its conformation, whereas other residues lining the pocket remain nearly unchanged. Specifically, the VP1 GH loop moves into and squeezes the pocket, adopting a conformation that resembles that in the RV-C15a, RV-B3, and RV-B14 structures. It is thus probable that the collapsed pocket in RV-C15a, RV-B3, and RV-B14 might facilitate virus uncoating. These viruses might undergo particle expansion to become A-particles where pores are formed in the capsid, as was previously described for other EVs (34, 36).
The large interior rearrangements characterizing the full and the native empty particles of RV-C15a contrast with the conserved external surfaces. Both particle types have the same diameter and display the same finger protrusions, truncated fivefold vertices, and putative glycan binding regions. Presently, it is not clear whether the observed high proportion (∼30%) of empty particles is a property of all RV-C, or unique only to RV-C15a. Possibly the RV-C species use these native empty particles as immunogenic molecular decoys during infections, or they are merely by-products of the assembly process.
The published atomic models of RV-C (13, 15) predicted the loss of VP1 mass at the fivefold vertices, relative to the crystal structures of other RV virions. The more rigorous one of these models (14) also predicted steric impediments in the VP1 pockets as the cause of drug-binding failures. However, the overall rmsd between equivalent Cα atoms of this predicted model and the present cryo-EM structure was ∼1.3 Å, and none of the bioinformatics-derived information correctly projected the finger-like features that dominate the particle surface. The relative VP1 insertion contributing to each protrusion is unique to the RV-C.
The atomic structures of an RV-C virus, as reported here, show novel molecular targets for designing anti-RV-C therapeutics. Furthermore the noninfectious empty particles may have potential as vaccine candidates. These possibilities have clinical relevance because many RV-C, including RV-C15, are associated with severe, hospitalization-category infections in young children (7), especially those with asthma, and can also lead to significant adult respiratory problems, including chronic obstructive pulmonary disease.
Materials and Methods
Growth and Purification of RV-C15a.
RV-C15a (adapted) is a virus preparation derived by serial passage (13×) of recombinant C15 virus (12), in HeLa-E8 cells, a lentivirus-transduced line expressing the full-length human CDHR3 gene (Tyr529) linked to a GFP reporter sequence (8). To produce viruses for structure determination, HeLa-E8 cells were infected with RV-C15a at 34 °C. At 40 h postinfection after complete cytopathic effect (CPE) was observed, infected cells (2.4 × 108) were supplemented with Hepes (to 50 mM, pH 7.2) and then subject to multiple freeze/thaw cycles (3×). Clarified supernatants were treated with RNase A, then concentrated by pelleting through 30% sucrose before resuspension and being assayed for titer (8, 37). The procedure gave a sample of RV-C15a at ∼1.2 × 1010 PFUe [by quantitative reverse transcription PCR (qRT-PCR) using HeLa-E8 cells] equivalent to ∼125 μg RNA-containing infectious particles, assuming a particle-to-PFU ratio of ∼200. PFUe is the “PFU-equivalent” calculated based on calibrated standard curves for RV-A16/RV-B14 viruses.
Characterization of Two Forms of RV-C15a Particles.
A sample of RV-C15a, as prepared for structure determination, was sedimented through a 10–40% sucrose gradient [103,600 × g (Beckman SW41 Ti rotor) for 3 h at 4 °C]. Fractions (1 mL) were collected (from the top) and then probed for VP2/VP0 content by Western blot analyses using anti-RV-C15-VP2 mouse monoclonal antibody, clone no. 517 (kindly provided by MedImmune Inc.). The fractions were also tested for infectivity according to CPE (38), and for RNA content by qRT-PCR (8).
Cryo-EM.
Aliquots of 2.8 μL of purified RV-C15a sample were applied onto glow-discharged holey carbon EM grids (400 mesh; Ted Pella Inc.). Grids were blotted for ∼8 s at a relative humidity of 80% and then plunge-frozen in liquid ethane cooled down by liquid nitrogen using a Cryoplunge 3 system (Gatan). Movies of frozen RV-C15a particles embedded in vitreous ice were collected at liquid nitrogen temperature using a Titan Krios transmission electron microscope (FEI) operated at 300 kV and equipped with a Gatan K2 Summit direct electron detector (3,838 × 3,710 in physical pixels). All of the movies were automatically recorded in superresolution mode using Leginon (39) at a nominal magnification of 14,000× and with a defocus range of 0.7–3.5 µm, which resulted in a superresolution pixel size of 1.04 Å per pixel. The dose rate was ∼8 e−/(pixel⋅s). For each movie, the total electron dose was ∼25.7 e/Å2, which was fractionated into 70 frames with an exposure time of 200 ms per frame.
Image Processing.
Cryo-EM data were collected of the RV-15a particles. A total of 2,979 movies were subjected to whole-frame motion correction using a modified version of MotionCorr (16) as modified by Wen Jiang (Purdue University). This process was integrated into the Appion data processing pipeline (40). Aligned frames were subsequently summed to obtain individual micrographs. Micrographs that had ice contamination or severe drift were discarded. The remaining micrographs were used for estimating of the contrast transfer function (CTF) parameters using CTFFIND3 (41). For a part of the data set particles were selected semiautomatically. In this process the program e2boxer.py in the EMAN2 package (42) picked particles based on several templates (derived from manually selected particles). The selection was then confirmed by visual inspection. For the rest of the data set, automatic particle selection was accomplished by using DoG picker (43). The final number of selected particles was 24,882. Individual particle images were boxed, extracted, and subjected to reference-free 2D classification into 156 classes using the program Relion (17). Some of these classes were clearly composed of full particles, some of empty particles, and some were just junk. This procedure yielded 13,390 full particles and 5,324 empty particles. Particles were reboxed and reextracted from the micrographs using jspr (18). CTF parameters of the particles from each micrograph were estimated using fitctf2.py (44).
Images of the full particles were divided into two half-data subsets. A truly independent 3D reconstruction strategy, using the program jspr (18), was applied to each of the two subsets assuming icosahedral symmetry. For each subset, eightfold binned particle images (squares of 8 × 8 original pixels separated by 1.04 Å were averaged to represent one pixel with a spacing of 8.32 Å) were used to compute 10 initial 3D reconstructions by assigning random initial angles to each of 150 particle images. Refinements were performed by searching for the best orientation and particle center of each particle image relative to the 10 current 3D reconstructions. Three of these structures were selected for further refinement with all of the available particle images in the half-subset. After multiple iterations, these structures converged to roughly the same reconstruction. One of these structures was randomly chosen to extend the refinement with four-binned, then two-binned data, and, finally, unbinned data. At this point, anisotropic magnification distortion remained the major resolution limiting factor. Ten images of polycrystalline gold particles were taken at a nominal magnification of 14,000× in superresolution mode. Fourier transform of these gold particle images gave powder diffraction like patterns that were used to estimate parameters of anisotropic magnification distortion (27). The estimated degree of distortion and angle were 2.87% and 31.3°, respectively. These parameters were then used to correct anisotropic magnification distortion for individual particle images using jspr (18). Subsequent refinement of particle center, orientation, defocus, astigmatism, scale, and beam tilt using jspr led to the final optimal reconstructions in terms of resolution. Fourier shell correlation (FSC) between the two subsets was used to monitor convergence. The same procedures were used for determining the 3D structure of the empty particles. A 2.79-Å resolution map of the full particle was reconstructed using 8,973 particles, and a 3.16-Å resolution map of the empty particle was reconstructed using 3,614 particles. The map resolution was determined based on the FSC between the two half-maps (masked with a soft mask) independently calculated using the two half-data subsets following the 0.143 cutoff criterion (28, 45). To further validate the map resolution, phase-randomized (beyond 5 Å) data were refined using the same procedures as were used for the original data that were not phase randomized. A “true FSC” curve (46) was calculated using the FSC curve based on the original data and the FSC curve based on the phase-randomized data. The full and empty particle maps were sharpened (28) using a B factor of −108.6 Å2 and −122.2 Å2, respectively.
Model Building and Refinement.
For the full particle structure, a predicted atomic structure of the RV-C15 (13) (including coordinates for a protomer, VP1–VP4) was manually fitted into a region of the final EM map that corresponds to one protomer of the capsid using Chimera (47). Atomic positions were refined using Phenix (48) in real space to maximize the correlation coefficient between the final EM map and a map calculated based on the coordinates. Model statistics, including bond lengths, bond angles, and all-atom clash; rotamer statistics; and Ramachandran plot statistics, were monitored; this was followed by model rebuilding with the program Coot (49). The combination of real-space refinement in Phenix and model rebuilding in Coot were repeated multiple times to achieve an optimized fit between the coordinates and the final EM map. At this point, the coordinates fit well into the densities by visual inspection.
Next, a mask, which included all grid points within a radius of 5 Å around each atom, was used to cut out densities from the final EM map using the CCP4 (50) program suite. The resultant segment of the final EM map was placed into a pseudo crystallographic unit cell (P1 space group) and was back-transformed into pseudo structure factors (including both amplitudes and phases). The coordinates were then subjected to refinement of individual B factors, atom positions, and occupancy against the pseudo structure factors using standard reciprocal space refinement procedures in Phenix (48). R factors were monitored during the refinement cycles. Only the coordinates were refined, whereas the map was kept constant. Subsequently, the coordinates were refined in real space against the final EM map by applying noncrystallographic symmetry (60-fold) constraints using Phenix (48). Validation of the final coordinates was based on the criteria of MolProbity (51). The full particle atomic model (excluding VP4 and the VP1 N-terminal residues 1101–1160) was used as a starting atomic model for model building and refinement of the empty particle structure.
As a further validation of the EM map resolution and the fitting between the atomic model and the final EM map, a density map was calculated based on the atomic model specifying a resolution of 2.79 Å for the full particle structure and 3.16 Å for the empty particle structure. An FSC was computed between the resultant density map and the final EM map. The resolution determined using 0.5 FSC as a cutoff was 2.85 Å (full particle) and 3.24 Å (empty particle). Oligomers were generated using VIPERdb (52). Figures were made using Chimera (47) and PyMOL (https://www.pymol.org).
Acknowledgments
We thank Roland Rueckert for critically reading the manuscript, Guimei Yu for helpful discussions, Valorie Bowman of the Purdue University Cryo-EM Facility and Ken Hibler of the FEI Company for technical assistance, and Sheryl Kelly for help preparing this manuscript. This work was supported by NIH Grants R37 AI011219 (to M.G.R.) and U19 AI104317 (to A.C.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of the RV-C15a full particle and of the RV-C15a empty particle have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5K0U and 5JZG). The cryo-EM maps for the RV-C15a full particle and empty particles have been deposited in the Electron Microscopy Data Bank (EMDB ID codes EMD-8189 and EMD-8184).
See Commentary on page 8903.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606595113/-/DCSupplemental.
References
- 1.Knowles NJ, et al. Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego: 2012. pp. 855–880. [Google Scholar]
- 2.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 3.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 4.Hogle JM. A 3D framework for understanding enterovirus 71. Nat Struct Mol Biol. 2012;19(4):367–368. doi: 10.1038/nsmb.2276. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, et al. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science. 2015;347(6217):71–74. doi: 10.1126/science.1261962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller EK, et al. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol. 2009;46(1):85–89. doi: 10.1016/j.jcv.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bizzintino J, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37(5):1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkov YA, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112(17):5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bønnelykke K, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46(1):51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 10.Piralla A, et al. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol. 2009;45(4):311–317. doi: 10.1016/j.jcv.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78(9):1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bochkov YA, et al. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17(5):627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basta HA, Sgro JY, Palmenberg AC. Modeling of the human rhinovirus C capsid suggests a novel topography with insights on receptor preference and immunogenicity. Virology. 2014;448:176–184. doi: 10.1016/j.virol.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basta HA, et al. Modeling of the human rhinovirus C capsid suggests possible causes for antiviral drug resistance. Virology. 2014;448:82–90. doi: 10.1016/j.virol.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McErlean P, et al. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS One. 2008;3(4):e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo F, Jiang W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol. 2014;1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartesaghi A, et al. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348(6239):1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basavappa R, et al. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: Structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci. 1994;3(10):1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossmann MG, He Y, Kuhn RJ. Picornavirus-receptor interactions. Trends Microbiol. 2002;10(7):324–331. doi: 10.1016/s0966-842x(02)02383-1. [DOI] [PubMed] [Google Scholar]
- 22.Rossmann MG. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem. 1989;264(25):14587–14590. [PubMed] [Google Scholar]
- 23.Filman DJ, et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8(5):1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth M, Pettitt T, Symonds A, Martin J. Identification of the pocket factors in a picornavirus. Arch Virol. 2003;148(6):1225–1233. doi: 10.1007/s00705-002-0974-4. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, et al. Sialic acid-dependent cell entry of human enterovirus D68. Nat Commun. 2015;6:8865. doi: 10.1038/ncomms9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers JM, Diana GD, McKinlay MA. Pleconaril. A broad spectrum antipicornaviral agent. Adv Exp Med Biol. 1999;458:69–76. [PubMed] [Google Scholar]
- 27.Grant T, Grigorieff N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J Struct Biol. 2015;192(2):204–208. doi: 10.1016/j.jsb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Zhao R, et al. Human rhinovirus 3 at 3.0 A resolution. Structure. 1996;4(10):1205–1220. doi: 10.1016/s0969-2126(96)00128-1. [DOI] [PubMed] [Google Scholar]
- 30.Fricks CE, Hogle JM. Cell-induced conformational change in poliovirus: Externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64(5):1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hindiyeh M, Li QH, Basavappa R, Hogle JM, Chow M. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J Virol. 1999;73(11):9072–9079. doi: 10.1128/jvi.73.11.9072-9079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acharya R, et al. The three-dimensional structure of foot-and-mouth disease virus at 2.9 A resolution. Nature. 1989;337(6209):709–716. doi: 10.1038/337709a0. [DOI] [PubMed] [Google Scholar]
- 33.Luo M, et al. The atomic structure of Mengo virus at 3.0 A resolution. Science. 1987;235(4785):182–191. doi: 10.1126/science.3026048. [DOI] [PubMed] [Google Scholar]
- 34.Ren J, et al. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun. 2013;4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garriga D, et al. Insights into minor group rhinovirus uncoating: The X-ray structure of the HRV2 empty capsid. PLoS Pathog. 2012;8(1):e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butan C, Filman DJ, Hogle JM. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol. 2014;88(3):1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griggs TF, Bochkov YA, Nakagome K, Palmenberg AC, Gern JE. Production, purification, and capsid stability of rhinovirus C types. J Virol Methods. 2015;217:18–23. doi: 10.1016/j.jviromet.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ledford RM, Collett MS, Pevear DC. Insights into the genetic basis for natural phenotypic resistance of human rhinoviruses to pleconaril. Antiviral Res. 2005;68(3):135–138. doi: 10.1016/j.antiviral.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 42.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166(2):205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang W, Guo F, Liu Z. A graph theory method for determination of cryo-EM image focuses. J Struct Biol. 2012;180(2):343–351. doi: 10.1016/j.jsb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 51.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrillo-Tripp M, et al. VIPERdb2: An enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37(Database issue):D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]