Significance
Parasitic interactions can result in changes to the host’s behavior in a way that promotes the distribution or life cycle of the parasite. Inteins are molecular parasites found in all three domains of life. Here we look at the influence of an intein in the DNA polymerase on a population of halophilic archaea in simulations, in experiments, and in the wild. This intein has a fitness cost that is higher than expected for a self-splicing genetic element. In these populations, where mating is independent of host replication, the intein increases the recombination rate between cells with and without inteins. This modification may contribute to the long-term persistence of these genetic parasites, despite the fitness burden they impart on their host.
Keywords: homing cycle, intein, homing endonuclease, horizontal gene transfer, selfish genetic elements
Abstract
Inteins are parasitic genetic elements that excise themselves at the protein level by self-splicing, allowing the formation of functional, nondisrupted proteins. Many inteins contain a homing endonuclease (HEN) domain and rely on its activity for horizontal propagation. However, successful invasion of an entire population will make this activity redundant, and the HEN domain is expected to degenerate quickly under these conditions. Several theories have been proposed for the continued existence of the both active HEN and noninvaded alleles within a population. However, to date, these models were not directly tested experimentally. Using the natural cell fusion ability of the halophilic archaeon Haloferax volcanii we were able to examine this question in vivo, by mating polB intein-positive [insertion site c in the gene encoding DNA polymerase B (polB-c)] and intein-negative cells and examining the dispersal efficiency of this intein in a natural, polyploid population. Through competition between otherwise isogenic intein-positive and intein-negative strains we determined a surprisingly high fitness cost of over 7% for the polB-c intein. Our laboratory culture experiments and samples taken from Israel’s Mediterranean coastline show that the polB-c inteins do not efficiently take over an inteinless population through mating, even under ideal conditions. The presence of the HEN/intein promoted recombination when intein-positive and intein-negative cells were mated. Increased recombination due to HEN activity contributes not only to intein dissemination but also to variation at the population level because recombination tracts during repair extend substantially from the homing site.
Homing endonucleases (HENs) are selfish genetic elements found in all domains of cellular life, as well as many viruses. HENs recognize and cleave highly specific target sequences, up to 40 bp long, usually corresponding to a singular site within the genome (1, 2). HENs can reside within conserved genes because they are nearly always located within self-splicing selfish elements: either group I introns (ref. 3; for review, see ref. 4), which excise themselves at the mRNA level, or inteins, which splice out of the protein product (ref. 5; for a recent review, see ref. 6). HENs contribute to the horizontal transmission of these selfish elements into intronless or inteinless alleles, by cleaving the vacant allele to induce homologous recombination or reverse transcription, where the allele containing the intron or intein serves as template. Thus, the intein or intron effectively invades the vacant site and can later be passed on to daughter cells vertically.
Paradoxically, if a HEN is highly successful in invading cells, it will saturate all target cells, and then its activity will no longer be under purifying selection. This may result in degeneration of the HEN domain due to accumulation of mutations and will prevent the future horizontal propagation of the intein in question. This phenomenon has been observed in group I introns of bacteriophages (7, 8), eukaryotes (9), and archaea (10). However, active HENs are often observed, and in most natural populations surveyed to date, there are in fact strains with inteins (or introns) containing degenerate HENs, whereas other isolates maintain a fully intact HEN (9). The homing cycle model (11, 12) resolves this paradox by hypothesizing that after the intein-/intron-containing allele has been fixed in the population and after the HEN has completely decayed due to the lack of selection for function of HEN activity, the splicing element (either intron or intein) can be deleted, thus returning the homing cycle to its beginning. In the case of introns, a precise deletion is somewhat likely and can occur via a processed mRNA intermediate (13, 14). In contrast, a precise deletion of an intein by a random event is expected to be very rare (15). In both introns and inteins, an imprecise deletion in a critical gene would often yield a dysfunctional product that may be highly deleterious or even lethal (16); we suggest that a precise deletion mainly occurs through homologous recombination with an intein-free allele that is transferred into the population or that survived within the population in case of incomplete invasion.
Recently, two models based on incomplete invasion were described. Both are based on the coexistence of intein-free and intein-containing alleles with and without functioning HEN in homogeneous populations. It was demonstrated computationally that if the cost of an inactive HEN is lower than that of the active HEN, the two forms can coexist with alleles carrying the empty target site even in a homogeneous population (15, 17). In many instances, the rock–paper–scissors dynamic between the three allele types (empty target site, intein with HEN, and intein without an active HEN) results in oscillations typical for predator–prey systems (15). In this scenario, both the intein (or intron) and the HEN activity have separate and cumulative fitness costs. Intein-negative cells are slightly more fit than their intein-containing counterparts and therefore outcompete intein-positive cells while becoming invaded. Cells with an intein but a defective HEN are slightly fitter than those with an intact HEN because they do not suffer the fitness cost from the large size of the HEN or from HEN activity such as off-target cleavage but are still immune to invasion. Under these assumptions a tripartite equilibrium between these three cell populations can result that may undergo periodical oscillations (15, 17).
Although these models provided fresh insights into the HEN enigma, they assumed homogeneous conditions throughout the population and used parameter estimates that were never tested, such as fitness costs associated with HEN activity and/or intein presence. Here we use the advanced genetic tools available for halophilic archaea to test the validity of these assumptions and gain an in vivo perspective into HEN/intein dynamics. Halophilic archaea were shown to undergo a unique mechanism of cell fusion (18–20), in which two or more cells can become fused into one cell, containing all of the genetic material of the parental cells. This heterodiploid (heterozygous) state, where two different chromosome types are present in the same cell, allows contact between HEN-positive and HEN-negative alleles. Thus, the efficiency of the spread of an active HEN can be assessed in a realistic natural system. We have previously shown that the HEN that resides in the intein in the polB gene, encoding DNA polymerase B, at insertion site c (henceforth abbreviated as polB-c) in Haloferax volcanii is highly active in vivo and converts close to 100% of engineered plasmids that contain its homing site upon transformation. Deleting the entire intein did not significantly alter the growth rate of H. volcanii cells (21). An additional aspect of this experimental system is that H. volcanii was shown to be polyploid, the number of chromosome copies per cell ranging between 10 and 20 (22). Here we explore through simulations the consequence of the invading HEN having to convert multiple chromosomes, running the risk of being back-converted into a vacant allele through homologous recombination. We demonstrate experimentally that intein invasion is an inefficient process in H. volcanii and that having an intein results in a substantial fitness cost. These genetic experiments were followed up by a survey of H. volcanii strains isolated from various sampling locations along the eastern Mediterranean shore, facilitating a comparison between laboratory findings and ecogenetic observations.
Results
Intein Presence Incurs a Fitness Cost.
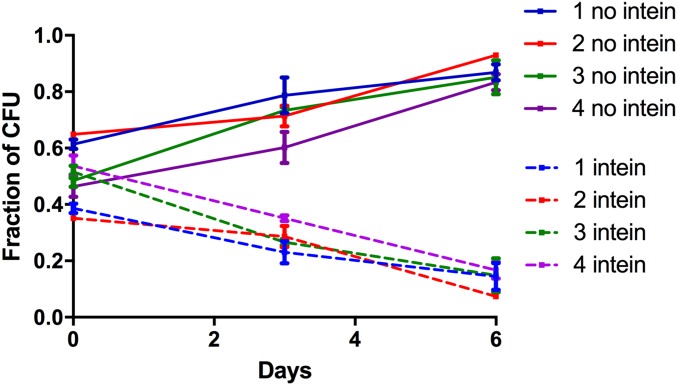
We had previously observed that the growth rate of the intein deletion strain is highly similar to its parental strain (21); however, growth rates were compared using growth curve analysis, a method that cannot detect small differences in fitness or ones associated with the size of the lag phase when growth is resumed after cells from stationary phase are transferred into fresh medium. We therefore performed direct competition assays between a strain containing the intein (H26; Table 1) and its intein-deletion isogenic strain (HAN12). Fig. 1 shows the relative abundance of intein-containing and intein-deletion cells in the mixed cultures, at time 0, and after 3 and 6 days of cogrowth. It is evident that cells containing the intein grew slower and were outcompeted by the intein-negative cells. Following the approach described by Lenski et al. (23) we used the change in average growth rate to quantify the intein’s effect on host fitness. Calculating the growth rates from three time points in each of eight parallel experiments, we calculated the relative fitness of the intein harboring cells to be 92.8% (SEM: 0.4%); that is, the fitness cost of the intein is 7.2% (SEM: 0.4%). This corresponds to an increase in the average doubling time from 4.17 to 4.49 h averaged over the repeated culture cycles (see spreadsheet in Dataset S1 for the calculation).
Table 1.
Strains used in this study
Fig. 1.
Relative abundance of intein-containing and intein-free cells in a direct competition assay. Haloferax volcanii with and without the polB-c intein (otherwise isogenic) were grown in coculture, aliquots were sampled, and the fraction of intein-containing and intein-free colony forming units was determined at different time points. Colors indicate independent parallel experiments.
Intein Spread Is Not Completely Efficient During Cell Fusion Events.
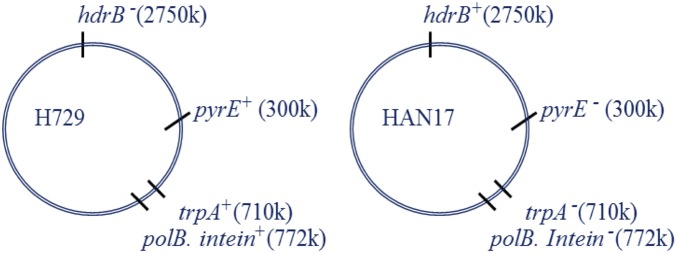
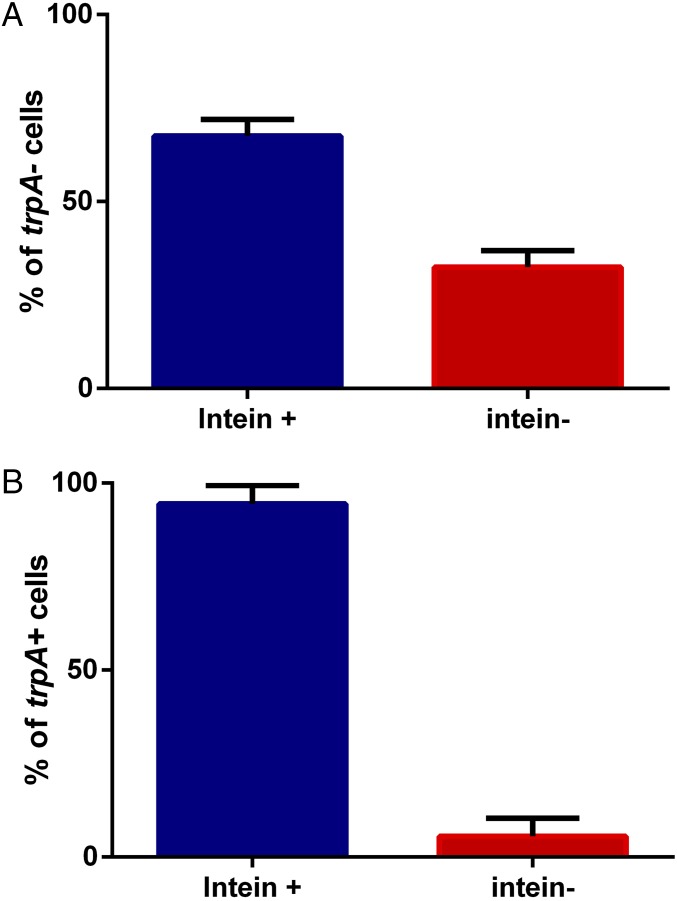
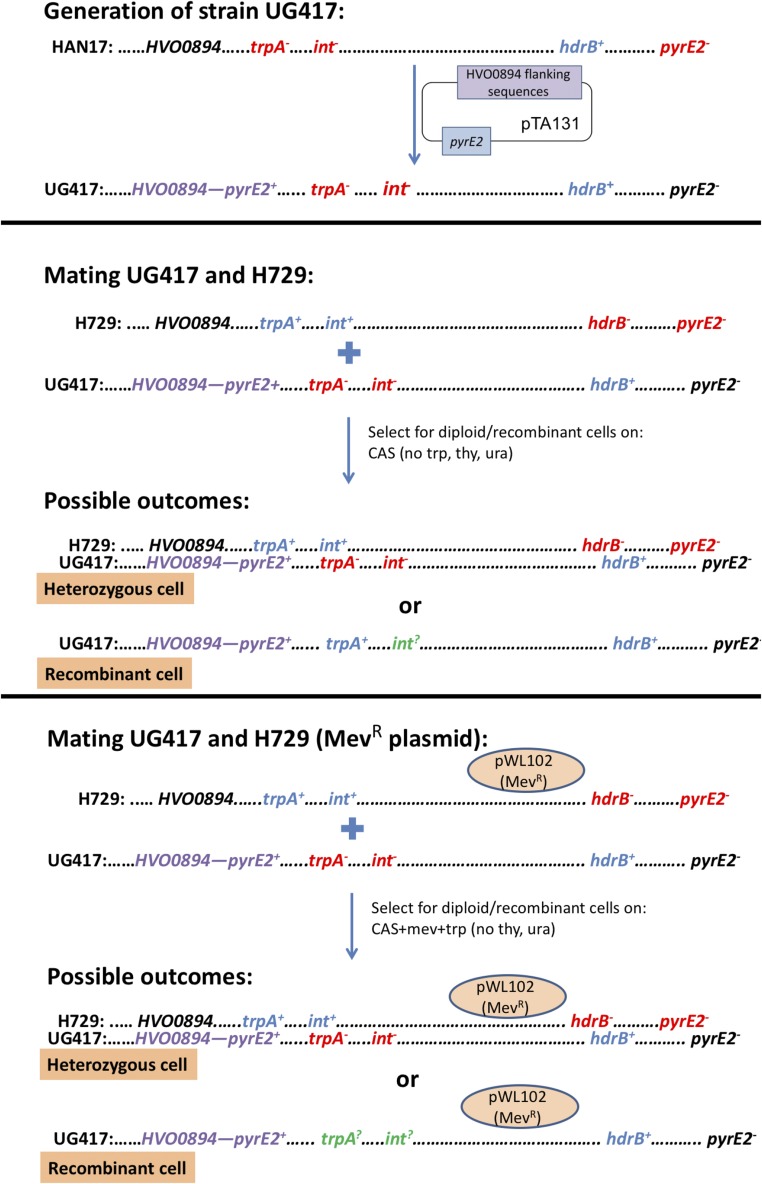
Examination of the natural invasion capacity of an intein/HEN in archaea requires a system where intein/HEN-positive cells can come into contact and fuse with cells carrying vacant alleles of the same gene that can be invaded. Presumably, invasion requires contact between alleles as well as the expression of the HEN protein domain, and thus, in this work, only cells that undergo fusion can be invaded. We therefore selected for mated cells so that invasion frequency can be simply assessed, by mating two strains that carry different gene deletions, serving as selectable markers (Materials and Methods). We used the following strains: H729, which is auxotrophic for thymidine (ΔhdrB, deficient in the gene encoding dihydrofolate reductase) and contains an intact intein with a HEN, and HAN17 (Fig. S1), which is auxotrophic for both uracil and tryptophan (ΔpyrE, deleted for the gene encoding orotate phosphoribosyl transferase; and ΔtrpA, deleted for the gene encoding tryptophan synthase) and intein-negative, thus containing a vacant HEN cleavage site. Following mating, we selected for mated cells by plating on media lacking thymidine and uracil, such that only cells that underwent mating could grow. The cells that underwent mating were then grown for four to five generations in rich media and subsequently plated, and each colony had its genotype determined. After establishing the original genotype, either H729 or HAN17 using the trpA locus, we examined intein presence using PCR for each colony (see spreadsheet in Dataset S2). As shown in Fig. 2, 68.5% of the cells that had the trpA locus from the intein-deleted parent (i.e., a trpA− allele—HAN17) became intein-positive, and 31.5% of the cells that underwent mating remained intein-negative. We also examined the cells that retained the trpA+ allele from the intein-positive parent H729, and as expected, most (92.5%) of the cells remained intein-positive; however, over all four biological replicates, 7.5% of the cells were now intein-negative (mean fraction of 5.5%; Fig. 2). This is probably due to random recombination and gene conversion events, not involving endonuclease activity that resulted in the elimination of the intein-occupied allele. Such events can mechanistically explain how such vacant alleles are formed without resorting to additional molecular mechanisms, such as precise intein deletion. Because even under conditions where all intein-negative cells are forced to make contact with intein-containing alleles, homing efficiency was less than 70%, i.e., nowhere near saturation, inteins are unlikely to rapidly invade all cells in a natural population.
Fig. S1.
Schematic representation of the H. volcanii genotypes used for measuring intein invasion during mating. Numbers in parentheses give the coordinates of the loci in base pairs. hdrB, dihydrofolate reductase gene B; pyrE, pyrimidine biosynthesis gene E; trpA, tryptophan synthase subunit A gene; polB, DNA polymerase B gene.
Fig. 2.
Percent of intein-containing and intein-free cells following mating between intein-containing and intein-free cells. (A) The percent of cells that had the genotype of the intein-free parent (trpA−). (B) The percent of cells that had the genotype of the intein-containing parent (trpA+).
Simulation of Intein Invasion.
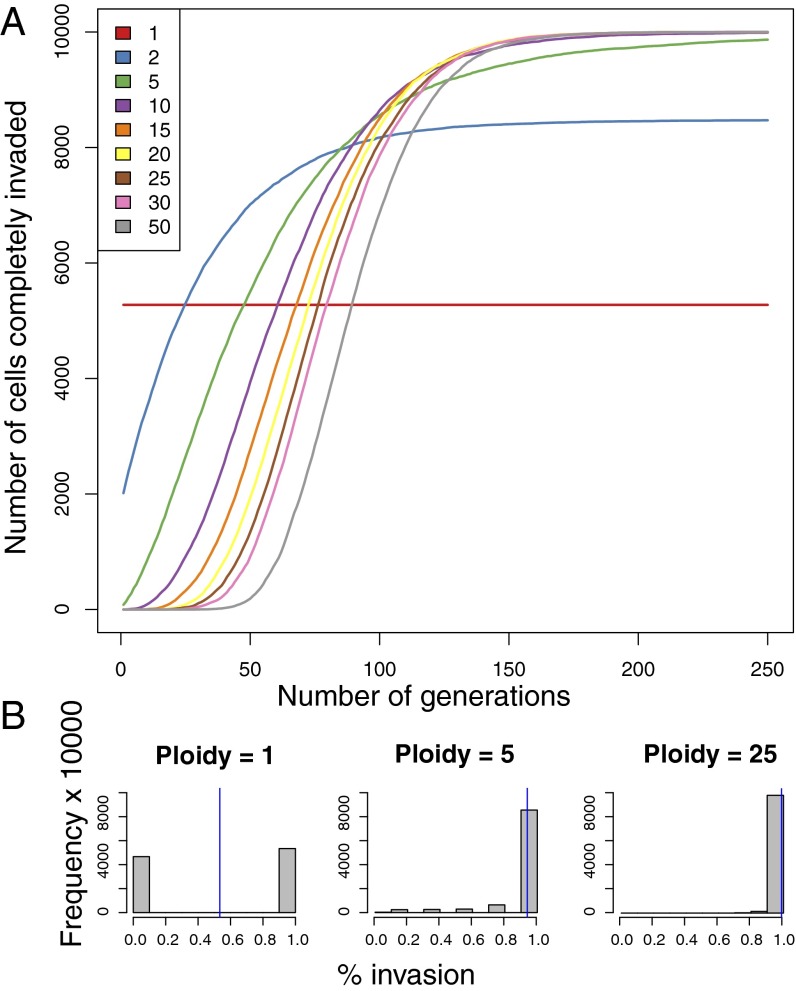
H. volcanii, like other haloarchaea and methanogens, are polyploid, with the number of genomes per cell varying between about 20 (during exponential growth) and about 10 during stationary phase (22). To assess the effect of polyploidy on intein invasion of a lineage, we performed several simulations. In the simulations depicted in Fig. 3, chromosomes were assumed to segregate randomly after mating. Under this condition a higher ploidy level increases likelihood of complete invasion of the lineage, but the rate of invasion is delayed with higher levels of ploidy.
Fig. 3.
Simulations to assess the effect of ploidy on intein invasion dynamics after fusion. Two cells, one completely invaded and one uninvaded, fuse, and chromosomes are assumed to assort randomly onto the daughter cells. Ten thousand cells are followed for 250 generations. (A) The number of cells completely invaded after each generation is recorded with the ploidy number as parameter, which is indicated by the colors in the legend. (B) Ten thousand cells are followed over 100 generations, and the percent of intein invasion in each cell is recorded. The blue line indicates the mean percent invasion for each ploidy number simulated.
To illustrate the effect of a high fitness cost on intein invasion of a population, we performed simulations of invasion of a population that over time approaches the carrying capacity of the environment (the maximum population size that the environment can support long term) (Fig. 4 and Fig. S2). A recent study of gene flow and recombination in a deep lake (24) suggests that gene transfer and homologous recombination continue in the absence of population growth in halophilic archaea. In the simulation in Fig. 4, the intein-free genotype outcompetes the intein-containing genotype during the initial growth phase, but the intein-containing allele spreads in the population, once the overall growth rate declines. Incorporating the fitness effect of the intein into the contribution of intein-containing cells on the carrying capacity of the population does not change the overall outcome, except that the resulting population size decreases as the intein-free cells are invaded by the intein (Fig. S2).
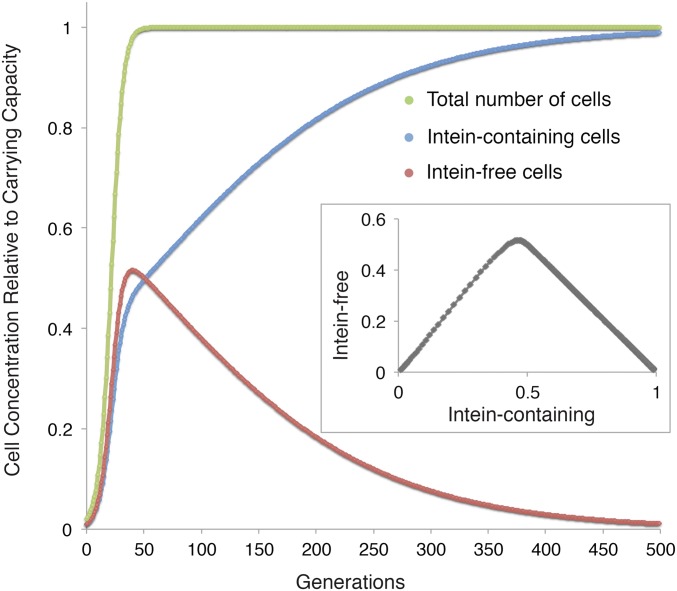
Fig. 4.
Simulation of intein invasion dynamics in a population with carrying capacity. Cell number is plotted relative to the carrying capacity over 500 generations. The inset gives concentration of intein-free alleles plotted against the concentration of intein-containing alleles. Note that initially, due to the fitness cost of the intein, the intein-free cells outgrow the intein-containing ones; however, even with the low homing efficiency of 0.01 assumed in this simulation, the intein spreads throughout the population as the growth rate declines closer to carrying capacity. The results remain qualitatively the same even when the carrying capacity is assumed to be equally affected by the fitness cost (Fig. S2). Parameters are as follows: homing efficiency = 0.01, fitness cost of the intein = 0.075, carrying capacity = 1, growth rate = 0.2 per generation, and starting concentration = 0.01 each for the intein-containing and intein-free cells.
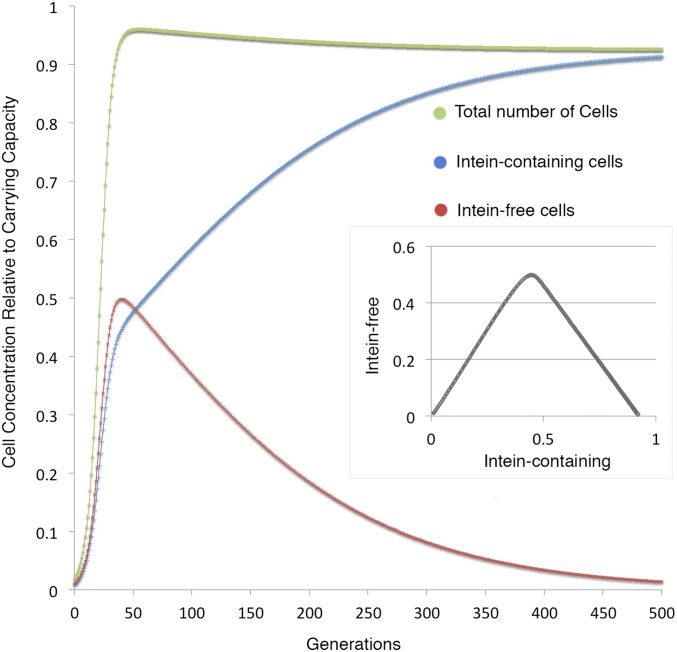
Fig. S2.
Intein invasion dynamics in a population with carrying capacity. Number of cells relative to the carrying capacity is plotted for the total number of cells (green line), the intein-containing cells (blue line), and the intein-free cells (red line) over 500 generations. Inset shows the concentration of intein-free alleles plotted against the concentration of intein-containing alleles. The simulation assumes that the fitness cost lowers the growth rate as well as the carrying capacity of the intein-containing cells. Parameters are as follows: homing efficiency = 0.01, fitness cost = 0.075, carrying capacity = 1, growth rate = 0.2 per generation, and starting concentration = 0.01 each for the intein-containing and intein-free cells.
Mating Between Intein-Positive and Intein-Negative Cells Results in Increased Recombination Frequencies.
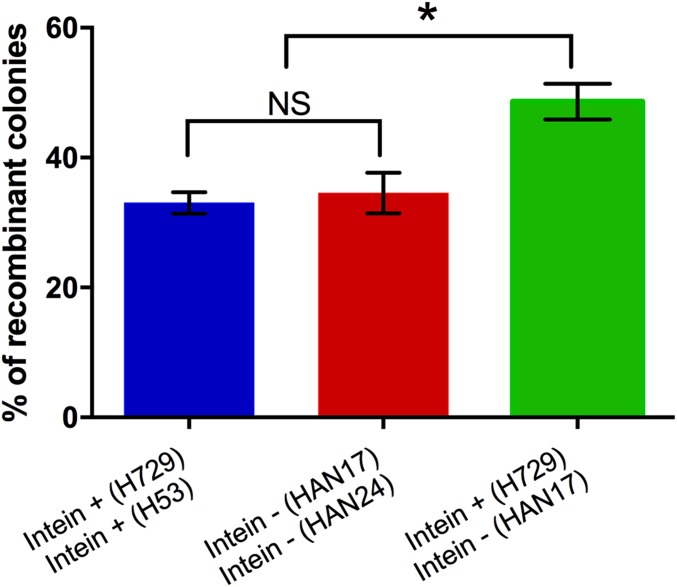
Because the HEN generates double strand breaks, which can be repaired by homologous recombination, we tested the link between recombination frequency and the presence of the intein/HEN. Recombination frequency can increase when intein/HEN-positive and intein/HEN-negative cells come together due to homing but potentially also due to off-target cleavage by the HEN at nonspecific sites. We tested the overall recombination frequency following natural cell fusion events, among the strains described above. Following mating, the cells first become heterodiploid (heterozygous), containing chromosomes that originated from two different cells. However, a substantial fraction of the population undergoes recombination and segregation so that the resulting cells contain a single chromosome type. We calculated the frequency of recombination, using PCR (as in ref. 19; Materials and Methods), by measuring the fraction of recombinant colonies from the general mated population (which contains a majority of heterodiploid cells). The fraction of recombinant colonies that have a single chromosome type but can grow on a medium lacking thymidine, tryptophan, and uracil is shown in Fig. 5. To estimate the effect of intein on recombination, we mated intein-positive cells with intein-negative cells (H729 and HAN17); in this scenario, one cell contains an intein with an active HEN, whereas the other chromosome contains an empty polB allele, which is the recognition site for this HEN. As control experiments we also performed such mating experiments between pairs of strains that were both either intein-positive (H729 and H53) or intein-negative (HAN17 and HAN24). Notably, the recombination frequency was similar for intein-positive and intein-negative pairs of strains, between 33% and 35% recombinants out of the entire mated population. This near identical rate of recombinants implies that off-target DNA cleavage by the PolB HEN is not sufficient to increase recombination rates and is therefore probably very low. However, when mating an intein-positive strain with an intein-negative partner, the recombination efficiency was markedly higher, with 48.6% of the mated cells being recombinants (Fig. 5). This suggests that the specific cleavage of the empty site in the polB gene (homing) by the HEN caused an increase in recombination frequency, probably because of the presence of double-strand breaks due to homing.
Fig. 5.
Recombination frequency in different mating experiments. Intein-free and intein-containing cells were used in mating experiments, and the number of recombinants was determined as described in ref. 19. Recombination rate is higher when intein-containing cells mate with intein-free cells. Statistical significance by Student t test: *P = 0.016; NS indicates not significant (P = 0.86).
Recombination Tracts in H. volcanii Following HEN Activity Can Extend over 50 kb.
These experiments raised the question whether the homologous recombination tract initiated at the intein insertion site can extend well beyond the hundreds of bases within the extein to thousands of bases away, as was previously observed in archaea (25, 26) and bacteria (27). To investigate this, we established a simpler intein invasion assay where the intein donor only has to transfer a small plasmid to the recipient strain and all donors cannot replicate because they cannot synthesize uracil or thymidine. This was achieved by mating a donor intein-positive strain carrying a mevinoline resistance plasmid that is hdrB−/pyrE−/trpA+ with a recipient strain that is mevinoline-sensitive and hdrB+/pyrE+/trpA−. By growing the mated cells on a medium that contains mevinoline and tryptophan and does not contain uracil or thymidine, donor cells were selected against. This effectively leaves only hdrB+/pyrE+ cells that received the mevinoline resistance plasmid because the chances of two recombination events at very distant loci (over 1 Mb) are much smaller than the chances of successful transfer of a small plasmid during mating (see also Fig. S3). Following selection, colonies were screened by PCR for intein presence or absence as described above, excluding heterozygous colonies (those that contained more than one genotype, less than 25% of the colonies). We observed that using this strain combination and plasmid transfer-based selection, only about 30% of the colonies were invaded by the intein (Table 2). However, when intein-invaded colonies were streaked on a medium that lacked tryptophan, 23 ± 6% of the intein-positive were trpA+ vs. 10 ± 3% of the intein-negative colonies. This demonstrates that the HEN target site, polB, and trpA gene are linked in recombination despite the fact that these loci are over 60 kb apart. When we performed an identical mating experiment selecting on a medium that lacks tryptophan (requiring recombination at the trpA locus because only trpA+ progeny survive), we observed that 85% of the colonies were intein-positive, further supporting the linkage.
Fig. S3.
The experimental system used to investigate the genetic linkage between the trpA and polB/intein loci.
Table 2.
Percent genotypes of colonies obtained in the plasmid-based mating assay for intein invasion
| Genotype | trpA+ Int+ | trpA+ Int− | trpA− Int+ | trpA− Int− |
| Mean (three biological repeats) | 23 (18) | 10 (8) | 7 (5.7) | 60 (47.3) |
| SD | 6 | 3 | 3 | 5 |
Number of colonies are in parentheses.
These findings are in agreement with previous results showing recombination tracts of hundreds of kb between H. volcanii and H. mediterranei (19).
Inteins in Natural Populations.
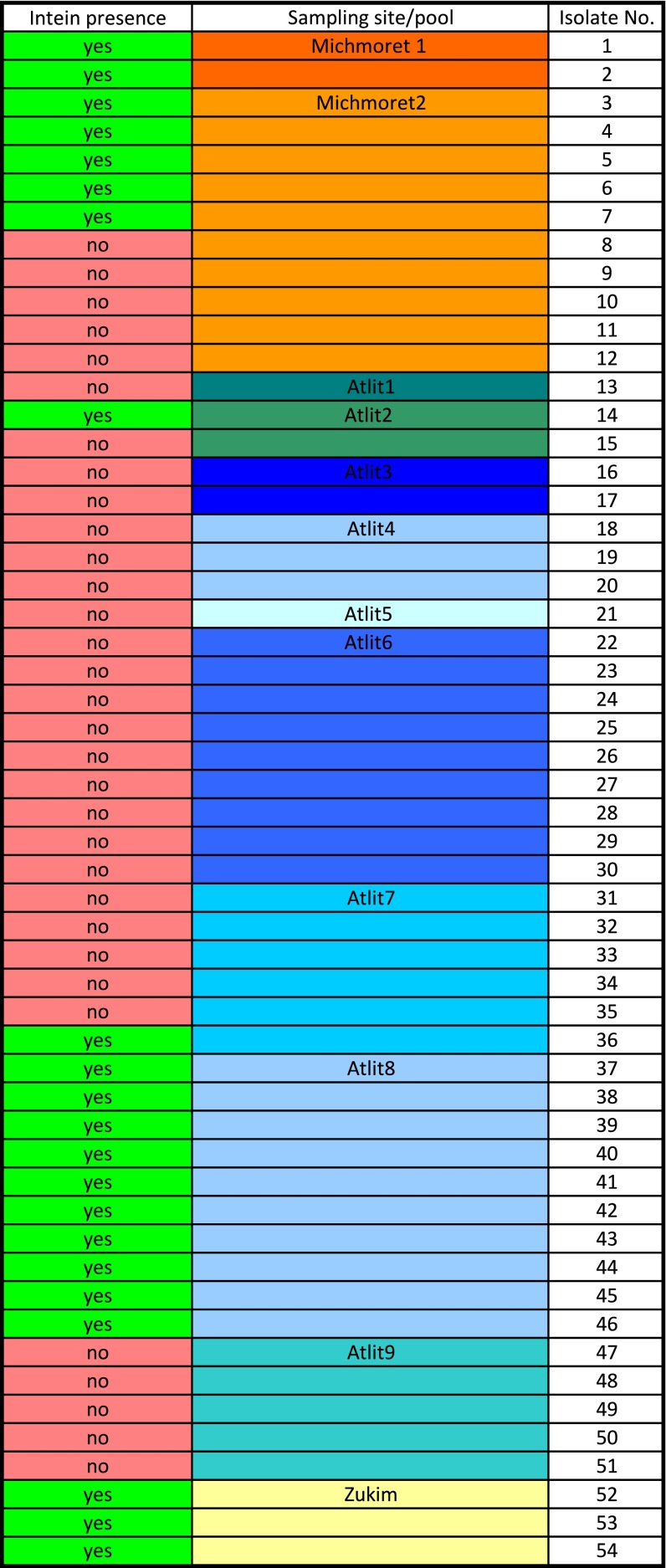
If indeed not every mating event leads to successful invasion and given that under natural conditions, only a fraction of cells will fuse with one another, we predicted that not all natural isolates will present the same polB-c intein genotype. We therefore isolated from tidal pools along Israel’s coastline Haloferax strains that can be regarded as belonging to H. volcanii or closely related to it (defined as having identical 16S rRNA gene sequences to that of a Haloferax sp.).
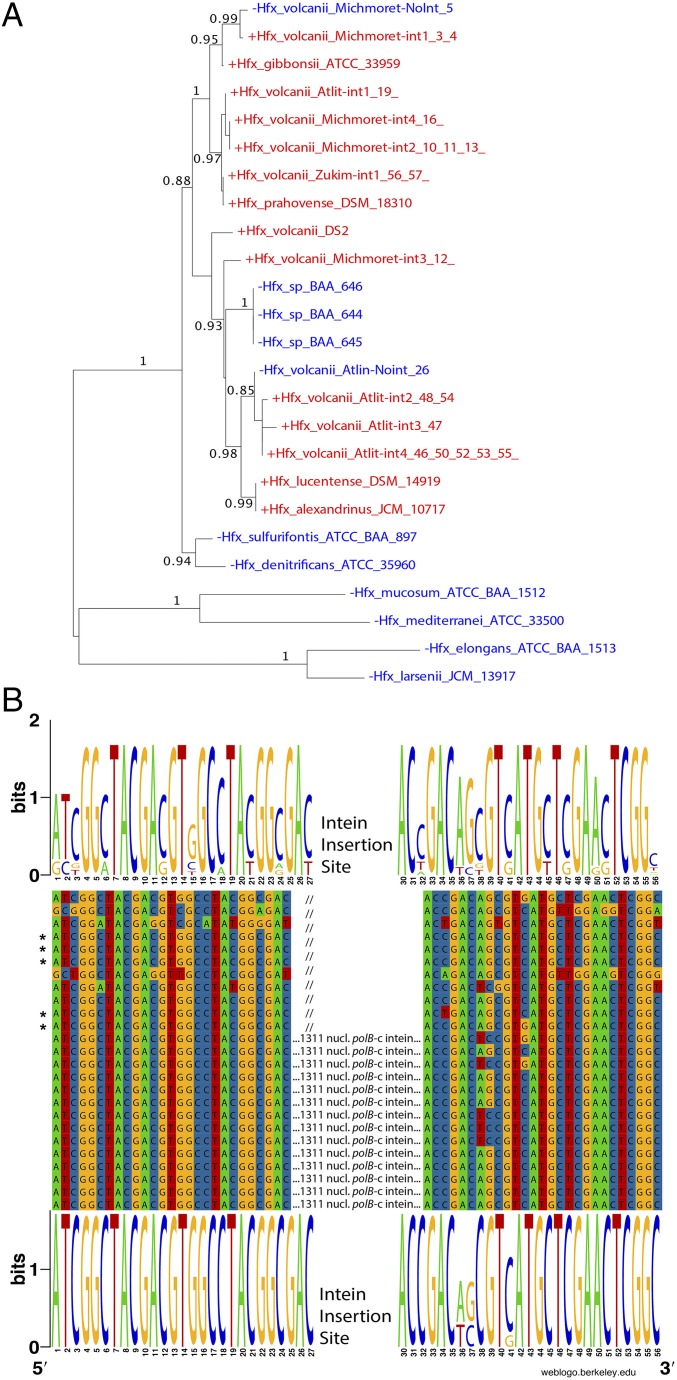
At the geographic level, two out of the three coastal sampling locations had both intein-containing and intein-free isolates, based on PCR amplification of the polB gene fragment that may contain the H. volcanii intein. Of the 14 tidal pools we sampled in the three locations, 12 contained isolates with polB sequences of high similarity to H. volcanii polB. Of these sites, nine contained either intein-containing isolates or intein-free isolates but not both, whereas three pools contained a mixed population (Fig. S4). Fifty-four different isolates had their entire polB genes amplified and sequenced, and two different intein-free genotypes were identified, as well as seven distinct intein-containing genotypes. A phylogenetic reconstruction of extein sequences revealed that inteins have recently invaded inteinless sites in this population (Fig. 6). Further examination of intein sequence revealed that all nucleic acid substitutions within the intein and HEN motif blocks resulted in synonymous (silent) mutations with the exception of a single proline to serine substitution in a conserved region of the HEN, known as block B (21), which was present in two genotypes. Thus, most of the isolates that are intein/HEN-positive appear to have a genetically intact HEN, showing no sign of degeneration.
Fig. S4.
Breakdown of intein-positive and intein-negative Haloferax isolates at the three sampling sites, based on PCR amplification of the polB gene.
Fig. 6.
Maximum likelihood phylogeny for polB extein sequences (A) and conservation of polB-c intein insertion sites (B). Numbers give support values calculated using the approximate likelihood ratio test as implemented in phyml 3.0 (51). Although drawn as rooted, the tree should be considered unrooted. The finding that sequences without (blue) and with (red) intein do not always form distinct clans (53) reveals that invasion of the Haloferax genus with the polB-c intein is an ongoing process. B shows a polB nucleotide sequence alignment around the intein insertion site c. Web logos (52) give the site conservation for intein minus (Top) and intein plus (Bottom) sequences. The five intein minus sequences that group within the cluster of intein plus sequences are marked with asterisks. The intein minus sequences show greater nucleotide diversity surrounding the intein insertion site, mainly in synonymous positions—only two positions at the 5′ and close to the 3′ end of the alignment represent nonsynonymous changes. Homing endonuclease site specificity was shown to tolerate substitutions that result in nonsynonymous changes (54), suggesting that none of the depicted Haloferax sequences may be immune to intein invasion.
Discussion
None of the current models for the maintenance of inteins with HENs captures the full complexity of these selfish elements in nature. If one assumes efficient invasion, as in the original homing cycle, one would expect most intein-containing cells in nature to have degenerate HENs waiting to be rescued from this dead end state by a precise deletion event. Here we show that in fact even under ideal conditions—we look only at the cells in which intein-positive and intein-negative cells have to come into contact—a substantial fraction of cells are not invaded. We also show that a vacant site can be recreated, if rarely, simply by a random (i.e., not targeted) homologous recombination event that converts the intein-positive allele to an intein-negative, as a by-product of polyploidy. In agreement with our genetic experiments, natural populations of H. volcanii-related strains can contain both intein-positive and intein-negative cells, and intein-positive isolates show little to no signs of HEN degeneration.
Our simulations exploring the effect of ploidy on intein invasion suggest that polyploidy facilitates invasion by the HEN (Fig. 3). Higher levels of ploidy slow down the rate of complete invasion (more successful homing events need to occur before invasion of chromosomes is complete); however, assuming random assortment of chromosomes into the daughter cells, higher ploidy levels lead to a higher frequency of complete invasion in the long run. The finding that in our mating experiments (Fig. 2) the colonies obtained from heterozygous mated cells after five doubling periods no longer reveal any heterozygosity with regards to the intein suggests that the effective ploidy level of the fused cell may be lower than expected from the measurement of chromosomes (22). A high rate of forming homozygotes from heterozygous H. volcanii has been described for engineered heterozygotes in the absence of maintaining selection (28) and was attributed to a high rate of gene conversion. Alternatively or in addition, it is possible that chromosomes in the fusion cell do not mix efficiently and that the first cell division tends to segregate the chromosomes in a way that recreates the genotypes of the two parents. A low level of effective ploidy could also explain why inteins were fixed in only about 80% of the cells following mating (compare Figs. 2 and 3).
Alternatives to a homing cycle model where intein invasion goes to completion posit a fitness cost for the intein (15, 17) and a copersistence of intein-free and intein-containing alleles (with and without functioning HEN) in the population. Our simulations show that during exponential growth, intein-free alleles can outcompete intein-containing ones (Fig. 4). The fitness cost of the intein is balanced by conversion of intein-free alleles through homing. We experimentally determined a surprisingly high fitness cost of having an intein-positive allele, at least under laboratory conditions and averaged over the growth cycle. Growth retardation was previously shown during expression of group I introns in the 23S rRNA gene of the bacterium Coxiella burnetii (29). In accordance with an equilibrium resulting from the fitness cost of the intein, balanced by the spread of the intein, and in accordance with the simulations performed in refs. 15 and 17, we find that many natural isolates have survived retaining intein-positive alleles.
Inteins with HEN can be described as parasitic genetic elements (1). Parasites often affect the phenotype of their host to increase the chance of their own propagation (30), and an effect on host behavior and development has also been described for the Saccharomyces intein (vma1-a) (31). The observed increase in recombination when only one of the genotypes of the mating cells is intein-positive (Fig. 5) may be an illustration of this principle. Clearly, the propagation of the intein is enhanced through increased recombination, and therefore, the increased recombination rate is in the best interest of the intein. However, the recombination tract that is generated by the homing activity is by no means limited to the immediate vicinity of the polB gene because the locus we used to test recombination is located over 60 kb away (Fig. S1) from the homing site. Thus, despite the high specificity of the homing site, homing events can exert indirect effects on gene exchange frequency across large regions of the chromosome, and assessing the distance dependence of these effects requires further study. Nevertheless, although recombination will be primarily induced close to the homing site, it could also be more generally increased because as long as DNA breaks are generated, this enhances transcription of homologous recombination genes, such as radA, the archaeal homolog of the bacterial recA recombinase. Indeed, recA transcription is induced in bacteria following DNA breaks (32), and radA was the most strongly up-regulated gene in the halophilic archaeon Halobacterium salinarum following UV-B exposure (33). Such induction of radA is thus expected to increase recombination rates globally and not just around the homing site. A relationship between intein presence and increased rate of recombination is also suggested by the mating type switching HO endonuclease in yeast, which also facilitates mating and homologous recombination between cells through cuts made at a single locus (34). This HO endonuclease is a domesticated intein with endonuclease activity, which is most closely related to the vma1-a intein (see above). In addition to the endonuclease domain, the HO endonuclease still contains a self-splicing domain, albeit not shown to be active (12).
The finding of intein-containing alleles with active HENs that coexist in the same population with empty target sites (Fig. 6) (35) suggests that the long-term survival of inteins does not include the homing cycle going to completion; instead, fitness differences between organisms with and without inteins appear to play a role in the long-term coexistence of the HEN with alleles containing empty target sites. In our simplifying simulations, the HEN was found to completely invade a homogeneous population close to its carrying capacity; however, natural populations of H. volcanii are neither homogeneous, nor do they exist in a constant environment. In the rock–paper–scissors models describing long term coexistence of HENs with empty target sites (15, 17), inteins without HEN activity provide the crucial link in the intransitive fitness relationships leading to long-term coexistence. Our findings show that under conditions that include occasional growth of the population, the high fitness cost of the intein guarantees the survival of alleles with the empty target site, even in the absence of inteins without HEN activity. Although here we showed a fitness cost for the intein, several reports have indicated a potential positive role for intein presence, claiming a regulatory function under stress conditions (36–40). However, our finding that natural H. volcanii populations in the same sampling location tend to be mixed, having both intein-positive and intein-negative strains sharing the same niche, does not support a strong selective pressure for intein retention in this case.
It is tempting to speculate that the increased recombination rate might provide a benefit to the group and that possibly the intein is maintained in the population through group selection. This reasoning is reminiscent of a cost of sex scenario (e.g., refs. 41 and 42), where under some conditions, such as stress or environmental change, benefits at the population level, resulting from the increased genetic variation, outweigh the costs (for example, the cost of males in parthenogenetic insect species). However, given that the increased recombination rate provides a direct benefit to the molecular parasite, invoking group selection, i.e., the competition between a group that harbors inteins and groups that do not, seems an unnecessary complication; instead, this appears as another case (43, 44) where the gene’s selfishness drives the survival of the trait at a cost to the individual. Although there is a benefit to the larger group, it is likely the benefit to the molecular parasite, not the benefit to the group, that guarantees the survival of the parasite.
Materials and Methods
Culture Conditions.
H. volcanii cells were routinely grown in rich (YPC) medium or in CAS medium (see ref. 21). Nucleic acids and amino acids were added to a final concentration of 50 µg/mL (Sigma).
Competition Assays.
H26 and H12 (Table 1) cultures were grown overnight, and when they reached an OD600 ∼1, the cells were diluted to OD600 ∼0.2 and allowed growth until OD600 ∼0.4, where they were all diluted to OD600 ∼0.1 and mixed 1:1. The cells were plated to obtain a measurement of ratios at time 0. The cultures were diluted 1:50 every 24 h for 6 d. The experiment was performed with four biological repeats, and each repeat was done in duplicates. To distinguish between intein-containing cells and intein-deleted cells, we performed PCR analysis using primers AP409 and AP410, which amplify the polB region surrounding the intein, and that enabled the assessment of the number of colonies on each plate that originated from intein-positive or intein-negative cells. From the 0 and 3 d time points, about 95 colonies were screened for each experiment, and for the 6 d time point, 48 colonies were screened. We defined fitness cost of genotype x vs. genotype y as the relative change in average growth rate calculated as in ref. 23 using the cell numbers at days 0, 3, and 6 with the appropriate dilution factors (see spreadsheet in Dataset S1).
Mating Protocol.
Each culture was grown to an OD600nm of 1.1–1.3, and 2-mL samples were taken from both strains and applied to a 0.2-μm filter connected to a vacuum to eliminate excess medium. The filter was then placed on a Petri dish containing a rich medium (YPC medium + thymidine, see below) for 48 h at 42 °C. The cells were washed and then resuspended in casamino (CA) broth, washed twice more in the same medium, and plated on selective media.
Examination of Intein Spread.
To estimate the efficiency with which this intein/HEN spreads, we mated strains H729 and HAN17 (Table 1 and Fig. S1). Following mating, the cells were plated using selection for thymidine and uracil (using CAS medium supplemented with tryptophan), and thus, only mated cells could form colonies. From these colonies, which must contain both selection markers, eight single colonies were picked per mating experiment (four independent mating experiments were performed) and were grown in liquid medium in the absence of selection for four to six generations, allowing all cell types to grow and heterodiploid cells to segregate. Subsequently, cultures were replated, and from each of these 32 mated cultures, 11–50 single colonies were analyzed to establish their genotype and intein status. To establish whether each colony originated from the Δintein (HAN17) strain or from the wild-type polB strain (H729; Table 1), we examined the trpA marker, which is 62 kb from polB and therefore generally unlinked to it. Each colony was streaked on media with and without tryptophan, and the polB allele was tested using PCR (primers AP409 and AP410); thus, trpA+ cells are ones that originated from H729 (with intein), and trpA− cells originate from HAN17.
Determination of Recombination Frequencies.
To estimate recombination frequencies, we performed three mating experiments, all under the same conditions, repeated three to five times. The cells after mating were plated using selection for thymidine and uracil, and the medium used contained tryptophan. The mated cells were analyzed as previously described (19), by testing the trpA marker by PCR (using primers AP214 and AP215), where colonies containing two copies of the gene, both trpA+ and trpA−, are heterodiploid colonies, and ones containing either trpA+ or trpA− are recombinants. We have previously shown that cells that are either trpA+ or trpA− show a single type of chromosome in other locations along the chromosomes and are true recombinants (19).
Mating Experiments to Test the Linkage Between the Intein/polB and trpA Loci.
To further quantify the success of intein invasion using selection for growth on a selective medium lacking tryptophan, strain HAN17 (Table 1) was transformed with a suicide plasmid containing 500 bp each of the upstream and downstream flanking sequences of the gene Hvo0894 cloned into pTA131 (45), thus generating strain UG417 that is pyrE2+ (Fig. S3). Strain UG417 was then mated with strain H729 (trpA+int+; Table 1 and Fig. S3), and mated cells were selected on CAS medium lacking thymidine, uracil, and tryptophan. This medium only allows growth of heterozygous/heterodipoloids that have both of the parental genotypes or recombinant cells that have wt pyrE2, hdrB, and trpA. Because we selected for three genomic markers, two of which are on the UG417 genome, we expected that all recombinant cells would be UG417 that have replaced their trpA locus with the trpA+ from H729 via homologous recombination. Ninety-six colonies from three biological repeats were screened with hdrB primers AP121 (5′ CCCGCCTCGCCGACGTGCAGT 3′) and AP122 (5′ GGAGTTGGTCTGCGAGTGTCG 3′) and were all found to be homozygous and hence recombinant. The colonies were further screened with intein primers AP409 and Ap410.
To examine intein invasion rate independently of the trpA locus, strain H729 was transformed with pWL102 (46), a shuttle vector that contains a mevinolin resistance marker. H729–pWL102 was then mated with strain UG417. Mated cells were selected on CAS medium supplemented with tryptophan and mevinolin lacking thymidine and uracil, hence selecting for hdrB and pyrE2, two genomic markers on UG417 (Fig. S3). Using this selection, only heterozygous/heterodipoloids or cells with the UG417 that obtained pWL102 during mating could grow. This mating assay was repeated three times, and from each biological repeat, 96 colonies were selected and streaked on CAS medium lacking tryptophan as well as CAS medium supplemented with tryptophan to assess recombination at the trpA locus. All colonies were also subjected to PCR analysis using intein primers AP409 and Ap410 to assess intein invasion.
Sample Collection and 16S rRNA and polB Gene Sequencing.
Stagnant seawater or dry salt samples were collected from 14 tidal/spray pools from three rocky shores along Israel’s Mediterranean coastline. Five milliliters of seawater from each pool was spread onto a YPC plate containing ampicillin. Dry salt samples were dissolved in sterile seawater before spreading on plates. Colonies with typical Haloferax coloring and shape were further examined by 16S rRNA gene PCR using Halobacteriales-specific 16S rRNA gene primers 287F and 958R (47). The ∼600-bp amplicon was sequenced by ABI 3730XL sequencers at MCLAB DNA Sequencing Services (San Francisco). Colonies that proved to belong to the Haloferax genus (100% identity to a Haloferax sp. in NCBI nucleotide BLASTN) were subjected to PCR analysis and sequencing of the polB gene. The polB gene was amplified using primers AP8 and AP11. Intein presence or absence was determined according to the length of the amplified polB product. The polB sequence of the isolates was determined using primers AP8, AP11, AP439, AP409, and AP410. For sequence verification, each isolate was sequenced twice using all five primers, and only areas covered by at least two overlapping sequences were considered error free and used for genotype determination. Distinct polB genotypes showed sequence difference of at least two nucleotides in at least two locations (namely, two adjacent nucleotides did not count as two different nucleotides but only as one).
Phylogenetic Reconstruction and Sequence Logos.
polB sequences were aligned in MUSCLE (48) using the default parameters as implemented in Seaview4.3 (49) (Dataset S3). The phylogeny of extein sequences was calculated with Phyml (50) in Seaview4.3 (49) under the GTR + Gamma + I model. Numbers give support values calculated using the approximate likelihood ratio test as implemented in Phyml 3.0 (51). Sequences surrounding the insertion site c were extracted, and weblogos (52) were calculated from the aligned extein sequences.
Simulations to Explore the Effects of Ploidy.
Intein invasion during mating and colony growth was simulated to determine the effects of ploidy, homing endonuclease efficiency, and fitness cost of the intein on the rate of intein invasion. We do not know how the chromosomes segregate following fusion; therefore, we modeled the events following cell fusion in two different ways. In the first, two cells, one intein-positive and one intein-negative cell, form a fused single cell, and each chromosome segregates independently into the daughter cells. In the second simulation, chromosomes from each parental type are randomly assorted into daughter cells, and one of the daughter cells is chosen at random and followed for a given number of generations. For both simulations during each generation, the probability that an intein-negative chromosome is invaded is calculated as follows: Each uninvaded chromosome may be cut with probability
where E is the efficiency of the homing endonuclease, Xn is the number of homing endonucleases in the nth generation, and N is the total number of chromosomes in the fused cell. This normalizes the total activity so that the HEN activity in the fully invaded cell is the same for different ploidy levels (i.e., the amount of cytoplasm per chromosome is constant).
If a cut occurs on a given chromosome, the repair of the cut with an invaded sequence is dependent on the number of invaded sequences in the cell. The repair of the cut site with an invaded sequence occurs with probability
The minus 1 in the denominator excludes the cut chromosome from the calculation (i.e., it cannot act as template for its own repair).
Cells with a newly invaded chromosome divide with probability 1 − f, where f is the fitness cost per intein.
Parameter Estimation.
The estimated number of generations of growth after mating is 34. This is based on ∼2 × 108 cells per colony, or ∼28 generations of growth. The cells were grown for another 6 generations after the colonies were picked, giving a total of 34 generations.
The fitness cost (f) is the decrease in organismal fitness per intein acquired. Based on the coculture competition experiments (Fig. 2), a completely invaded 20-ploid cell has a fitness decrease of 7.2% compared with an identical uninvaded cell. We use a fitness cost of 0.075% per cell carrying the intein in the population.
The efficiency of the homing endonuclease (E) was calculated using deterministic simulations in Excel. The number of invaded chromosomes in a given generation was calculated according to the formula
where N is the number of chromosomes per cell, Xn is the number of chromosomes with intein for these mating simulations, n is the generation number, N − Xn is the number of alleles that can be invaded, and E is the efficiency of the homing endonuclease. The factor Xn/N reflects the amount of HE per genome. The factor Xn/(N − 1) gives the fraction of templates that contain the intein and would lead to insertion of the intein if the template were used to repair the double strand cut. f is the fitness cost per intein. Notice that this formula is equivalent to .
In these deterministic calculations, a homing endonuclease efficiency (E) of 0.06 leads to invasion of 81% in the case of a 20-ploid cell (spreadsheet in Dataset S4). This is consistent with results that 68.5% of uninvaded cells become invaded and 92.5% of invaded cells remain invaded after mating, for a total of 80.5% of cells invaded after mating.
Simulating Intein Invasion in a Population with a Limited Carrying Capacity.
We simulated intein invasion in populations that had a specific carrying capacity using a modified discrete logistic equation. We use the following formulas:
where p denotes the proportion of the population that contains the intein and q is the proportion of the population that does not contain the intein, hm represents the compound probability of an intein-positive and intein-negative cell exchanging DNA and the efficiency of the HEN domain making a double strand break and the intein invading, f represents the fitness cost to individual cells for carrying the intein, r is the rate of population growth, and k is the carrying capacity (we set k = 1 to express populations as fractions of the carrying capacity). The growth rate of the intein-free and intein-containing cells is r and (1 − f)r, respectively. Following the logistic equation, the growth rate is multiplied by k minus the current population size. For the simulations depicted in Fig. 4 we considered the contribution of intein-containing and intein-free cells to the carrying capacity as equal. To incorporate an effect of the intein on both the growth rate and the carrying capacity, we modified the equation to weigh the contributions of p and q to the carrying capacity (Fig. S2):
Supplementary Material
Acknowledgments
We thank Ella Shtifman Segal for help with strain construction. The University of Connecticut Bioinformatics Facility provided computing resources for the analyses reported in this manuscript. This work was supported through a grant from the Bi-national Science Foundation (BSF 2013061). Work in the J.P.G. laboratory was supported in part through grants from the National Science Foundation (DEB 0830024) and NASA Astrobiology: Exobiology and Evolutionary Biology (NNX13AI03G). Work in the U.G. laboratory was supported by the Israel Science Foundation Grant 201/12. A.N. was supported by the Dan David Prize scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606416113/-/DCSupplemental.
References
- 1.Gogarten JP, Hilario E. Inteins, introns, and homing endonucleases: Recent revelations about the life cycle of parasitic genetic elements. BMC Evol Biol. 2006;6(1):94. doi: 10.1186/1471-2148-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burt A, Koufopanou V. Homing endonuclease genes: The rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14(6):609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Bonocora RP, Shub DA. A likely pathway for formation of mobile group I introns. Curr Biol. 2009;19(3):223–228. doi: 10.1016/j.cub.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hausner G, Hafez M, Edgell DR. Bacterial group I introns: Mobile RNA catalysts. Mob DNA. 2014;5(1):8. doi: 10.1186/1759-8753-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietrokovski S. Intein spread and extinction in evolution. Trends Genet. 2001;17(8):465–472. doi: 10.1016/s0168-9525(01)02365-4. [DOI] [PubMed] [Google Scholar]
- 6.Novikova O, Topilina N, Belfort M. Enigmatic distribution, evolution, and function of inteins. J Biol Chem. 2014;289(21):14490–14497. doi: 10.1074/jbc.R114.548255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy SR, Gold L. The phage T4 nrdB intron: A deletion mutant of a version found in the wild. Genes Dev. 1991;5(6):1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- 8.Foley S, Bruttin A, Brüssow H. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J Virol. 2000;74(2):611–618. doi: 10.1128/jvi.74.2.611-618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wikmark O-G, Einvik C, De Jonckheere JF, Johansen SD. Short-term sequence evolution and vertical inheritance of the Naegleria twin-ribozyme group I intron. BMC Evol Biol. 2006;6(1):39. doi: 10.1186/1471-2148-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nomura N, et al. Heterogeneous yet similar introns reside in identical positions of the rRNA genes in natural isolates of the archaeon Aeropyrum pernix. Gene. 2002;295(1):43–50. doi: 10.1016/s0378-1119(02)00802-8. [DOI] [PubMed] [Google Scholar]
- 11.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA. 1999;96(24):13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. Inteins: Structure, function, and evolution. Annu Rev Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- 13.Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22(1):16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Derr LK, Strathern JN. A role for reverse transcripts in gene conversion. Nature. 1993;361(6408):170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- 15.Barzel A, Obolski U, Gogarten JP, Kupiec M, Hadany L. Home and away—The evolutionary dynamics of homing endonucleases. BMC Evol Biol. 2011;11:324. doi: 10.1186/1471-2148-11-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swithers KS, Senejani AG, Fournier GP, Gogarten JP. Conservation of intron and intein insertion sites: Implications for life histories of parasitic genetic elements. BMC Evol Biol. 2009;9:303. doi: 10.1186/1471-2148-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yahara K, Fukuyo M, Sasaki A, Kobayashi I. Evolutionary maintenance of selfish homing endonuclease genes in the absence of horizontal transfer. Proc Natl Acad Sci USA. 2009;106(44):18861–18866. doi: 10.1073/pnas.0908404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naor A, Gophna U. Cell fusion and hybrids in Archaea: Prospects for genome shuffling and accelerated strain development for biotechnology. Bioengineered. 2013;4(3):126–129. doi: 10.4161/bioe.22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22(15):1444–1448. doi: 10.1016/j.cub.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 20.Rosenshine I, Tchelet R, Mevarech M. The mechanism of DNA transfer in the mating system of an archaebacterium. Science. 1989;245(4924):1387–1389. doi: 10.1126/science.2818746. [DOI] [PubMed] [Google Scholar]
- 21.Naor A, Lazary R, Barzel A, Papke RT, Gophna U. In vivo characterization of the homing endonuclease within the polB gene in the halophilic archaeon Haloferax volcanii. PLoS One. 2011;6(1):e15833. doi: 10.1371/journal.pone.0015833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breuert S, Allers T, Spohn G, Soppa J. Regulated polyploidy in halophilic archaea. PLoS One. 2006;1:e92. doi: 10.1371/journal.pone.0000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138(6):1315–1341. [Google Scholar]
- 24.DeMaere MZ, et al. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc Natl Acad Sci USA. 2013;110(42):16939–16944. doi: 10.1073/pnas.1307090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grogan DW, Rockwood J. Discontinuity and limited linkage in the homologous recombination system of a hyperthermophilic archaeon. J Bacteriol. 2010;192(18):4660–4668. doi: 10.1128/JB.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams D, Gogarten JP, Papke RT. Quantifying homologous replacement of loci between haloarchaeal species. Genome Biol Evol. 2012;4(12):1223–1244. doi: 10.1093/gbe/evs098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MCJ. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol Biol Evol. 2005;22(3):562–569. doi: 10.1093/molbev/msi041. [DOI] [PubMed] [Google Scholar]
- 28.Lange C, Zerulla K, Breuert S, Soppa J. Gene conversion results in the equalization of genome copies in the polyploid haloarchaeon Haloferax volcanii. Mol Microbiol. 2011;80(3):666–677. doi: 10.1111/j.1365-2958.2011.07600.x. [DOI] [PubMed] [Google Scholar]
- 29.Raghavan R, Hicks LD, Minnick MF. Toxic introns and parasitic intein in Coxiella burnetii: Legacies of a promiscuous past. J Bacteriol. 2008;190(17):5934–5943. doi: 10.1128/JB.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes DP, Brodeur J, Thomas F. Host Manipulation by Parasites. Oxford Univ Press; Oxford: 2012. [Google Scholar]
- 31.Giraldo-Perez P, Goddard MR. A parasitic selfish gene that affects host promiscuity. Proc Biol Sci. 2013;280(1770):20131875. doi: 10.1098/rspb.2013.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel B. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 2005;3(7):e255. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boubriak I, et al. Transcriptional responses to biologically relevant doses of UV-B radiation in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 2008;4:13. doi: 10.1186/1746-1448-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strathern JN, et al. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 35.Fullmer MS, et al. Population and genomic analysis of the genus Halorubrum. Front Microbiol. 2014;5:140. doi: 10.3389/fmicb.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyake T, Hiraishi H, Sammoto H, Ono B. Involvement of the VDE homing endonuclease and rapamycin in regulation of the Saccharomyces cerevisiae GSH11 gene encoding the high affinity glutathione transporter. J Biol Chem. 2003;278(41):39632–39636. doi: 10.1074/jbc.M302084200. [DOI] [PubMed] [Google Scholar]
- 37.Callahan BP, Topilina NI, Stanger MJ, Van Roey P, Belfort M. Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat Struct Mol Biol. 2011;18(5):630–633. doi: 10.1038/nsmb.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topilina NI, et al. SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses. Proc Natl Acad Sci USA. 2015;112(33):10348–10353. doi: 10.1073/pnas.1512777112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topilina NI, Novikova O, Stanger M, Banavali NK, Belfort M. Post-translational environmental switch of RadA activity by extein-intein interactions in protein splicing. Nucleic Acids Res. 2015;43(13):6631–6648. doi: 10.1093/nar/gkv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novikova O, et al. Intein clustering suggests functional importance in different domains of life. Mol Biol Evol. 2016;33(3):783–799. doi: 10.1093/molbev/msv271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Vienne DM, Giraud T, Gouyon P-H. Lineage selection and the maintenance of sex. PLoS One. 2013;8(6):e66906. doi: 10.1371/journal.pone.0066906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nunney L. The maintenance of sex by group selection. Evolution. 1989;43(2):245–257. doi: 10.1111/j.1558-5646.1989.tb04225.x. [DOI] [PubMed] [Google Scholar]
- 43.Olendzenski L, Gogarten JP. Gene transfer: Who benefits? Methods Mol Biol. 2009;532:3–9. doi: 10.1007/978-1-60327-853-9_1. [DOI] [PubMed] [Google Scholar]
- 44.Olendzenski L, Gogarten JP. Evolution of genes and organisms: The tree/web of life in light of horizontal gene transfer. Ann N Y Acad Sci. 2009;1178:137–145. doi: 10.1111/j.1749-6632.2009.04998.x. [DOI] [PubMed] [Google Scholar]
- 45.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70(2):943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blaseio U, Pfeifer F. Transformation of Halobacterium halobium: Development of vectors and investigation of gas vesicle synthesis. Proc Natl Acad Sci USA. 1990;87(17):6772–6776. doi: 10.1073/pnas.87.17.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Youssef NH, Ashlock-Savage KN, Elshahed MS. Phylogenetic diversities and community structure of members of the extremely halophilic Archaea (order Halobacteriales) in multiple saline sediment habitats. Appl Environ Microbiol. 2012;78(5):1332–1344. doi: 10.1128/AEM.07420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 51.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 52.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilkinson M, McInerney JO, Hirt RP, Foster PG, Embley TM. Of clades and clans: Terms for phylogenetic relationships in unrooted trees. Trends Ecol Evol. 2007;22(3):114–115. doi: 10.1016/j.tree.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J Mol Biol. 2007;372(5):1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.