Significance
Most pathogenic enveloped viruses crucially depend on the quality control (QC) machinery in the endoplasmic reticulum (ER) of the host cell. ERQC inhibitors therefore have the double potential benefit of targeting a wide variety of viruses (“broad-spectrum antivirals”) without the risk of losing efficacy due to escape mutations in the viral genome. Our recent work has proven that inhibition of the central enzyme of ERQC, α-glucosidase II (α-GluII), is sufficient for antiviral activity against dengue fever in vitro and in vivo. Here, we show how antiviral inhibitors bind to portions of α-GluII that are unique to this enzyme, and we open the way to the development of potent and selective antivirals against existing and emerging infectious disease.
Keywords: broad-spectrum antiviral, ER α-glucosidase II, eukaryotic secretion, glycoprotein folding, iminosugar
Abstract
The biosynthesis of enveloped viruses depends heavily on the host cell endoplasmic reticulum (ER) glycoprotein quality control (QC) machinery. This dependency exceeds the dependency of host glycoproteins, offering a window for the targeting of ERQC for the development of broad-spectrum antivirals. We determined small-angle X-ray scattering (SAXS) and crystal structures of the main ERQC enzyme, ER α-glucosidase II (α-GluII; from mouse), alone and in complex with key ligands of its catalytic cycle and antiviral iminosugars, including two that are in clinical trials for the treatment of dengue fever. The SAXS data capture the enzyme’s quaternary structure and suggest a conformational rearrangement is needed for the simultaneous binding of a monoglucosylated glycan to both subunits. The X-ray structures with key catalytic cycle intermediates highlight that an insertion between the +1 and +2 subsites contributes to the enzyme’s activity and substrate specificity, and reveal that the presence of d-mannose at the +1 subsite renders the acid catalyst less efficient during the cleavage of the monoglucosylated substrate. The complexes with iminosugar antivirals suggest that inhibitors targeting a conserved ring of aromatic residues between the α-GluII +1 and +2 subsites would have increased potency and selectivity, thus providing a template for further rational drug design.
Most antiviral drugs target viral proteins critical to the viral life cycle. In recent years, increasing levels of resistance to direct-acting antivirals have become a major public health concern, highlighting the urgent need for the development of alternative treatments (1). One appealing strategy to avoid antiviral drug resistance is the targeting of virus–host interactions (2). Drugs that act by blocking specific host functions required by many different viruses have the additional potential to treat a wide range of viral infections and coinfections (broad-spectrum antivirals) (3). Inhibitors of the endoplasmic reticulum (ER) glycoprotein quality control (QC) machinery, used by most enveloped viruses for the correct folding of their surface glycoproteins (4), represent one class of promising host-targeting, broad-spectrum antivirals. ER α-glucosidase II (α-GluII) is the main ERQC glycosyl hydrolase, admitting folding client glycoproteins into ERQC and releasing them from it (4–9). Inhibition of the ERQC machinery, including α-GluII inhibition, causes viral glycoproteins to misfold and reduces virion secretion and/or infectivity (2). The centrality of α-GluII to viral glycoprotein folding and secretion makes it an appealing target for host-targeting antivirals, with clinical relevance for broad-spectrum antiviral therapy. ER α-GluII is a heterodimer composed of a catalytic α-subunit (104–116 kDa) and an accessory β-subunit (58–80 kDa) (5, 8, 10, 11). The enzyme first removes a glucose residue from the conserved Glc2Man9GlcNAc2 N-linked glycan attached to a nascent glycoprotein (“first cleavage”), and the resulting Glc1Man9GlcNAc2 glycan enables glycoprotein binding to the ER lectins calnexin and calreticulin and associated chaperones and refolding isomerases (5, 12). After another α-GluII–mediated glycosidic bond hydrolysis (“second cleavage”), which removes the innermost Glc residue, the glycoprotein is left with a Man9GlcNAc2 glycan and loses binding affinity for the ER lectins and refolding machinery. At this stage, if folded correctly, the α-GluII substrate glycoprotein is free to proceed toward the Golgi apparatus and secretion. Iminosugars, glycomimetics originally isolated from plants, represent a promising class of ERQC inhibitors that are well tolerated in mammals (13). Altering glycoprotein processing via inhibition of the ER α-GluI and α-GluII by iminosugars has demonstrated in vitro and in vivo efficacy against multiple virus families (14, 15), including Herpes-, Hepadna-, Toga-, Rhabdo-, Orthomyxo-, Paramyxo- and Retroviridae, as well as those virus families that cause hemorrhagic fever (16, 17), including Arena-, Flavi- and Filoviridae. The deoxynojirimycin iminosugar derivative N-9′-methoxynonyl 1-deoxynojirimycin (MON-DNJ) has antiviral activity in vivo, against both dengue virus (18) and influenza A (H1N1) (19) and influenza B (17), with the latter being the first reported incidence of antiviral activity of iminosugars against these strains of influenza. Two iminosugars with known inhibitory activity against α-GluII are in clinical trials against dengue fever (https://clinicaltrials.gov/ct2/show/record/NCT02569827 and https://clinicaltrials.gov/ct2/show/NCT02696291). The α-GluII α-subunit belongs to the GH31 family of glycosyl hydrolases, which also comprises intestinal maltase-glucoamylase (MGAM) and sucrase-isomaltase (SI). Both glycosidic bonds specifically cleaved by α-GluII have α(1,3) linkages, unlike the α(1,4)-maltose–like or α(1,6) bonds hydrolyzed by intestinal GH31 α-glucosidases. However, iminosugars, which are glucose mimetics and inhibit α-GluII, also target intestinal α-glucosidases and the ceramide-specific glucosyltransferase involved in glycosphingolipid biosynthesis (20). Toward an understanding of the molecular basis of iminosugar binding to their targets, and the development of better inhibitors of α-GluII, we have successfully established and describe here the first, to our knowledge, expression system capable of producing milligram quantities of an intact recombinant mammalian α-GluII heterodimer. This expression system has enabled us to carry out extensive biochemical and biophysical characterization of the enzyme. Eight crystal structures with a variety of ligands, including the two iminosugars in clinical trials against dengue fever, indicate that molecules targeting the +1 and +2 subsites of the enzyme will improve both potency and selectivity.
Results and Discussion
The Full-Length Murine α-GluII α/β Holoenzyme Has an Elongated Shape.
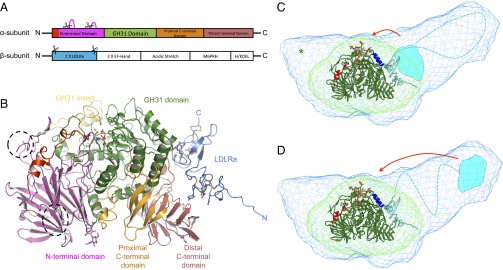
Both α- and β-subunit α-GluII sequences are highly conserved across eukaryotes. Initial tests screened expression of soluble α-GluII constructs from Saccharomyces cerevisiae, Caenorhabditis elegans, Mus musculus, and Homo sapiens. Wild-type (wt) recombinant mouse α-GluII (Mmα-GluII) gave the best preliminary results and was expressed and purified on a larger scale, together with its stable trypsinolytic fragment Mmα-GluIITryps (21) (SI Appendix, Fig. S1A). The murine and human α-subunit sequences share 92% identity, and the β-subunit sequences share 87% identity. Size exclusion chromatography multiangle laser light scattering (SEC-MALLS) and electrospray ionization mass spectrometry (ESI-MS) confirmed folded α/β heterodimers of 166 kDa and 109 kDa, respectively, for Mmα-GluII and Mmα-GluIITryps (SI Appendix, Fig. S1B). The Mmα-GluIITryps small-angle X-ray scattering (SAXS) data (SI Appendix, Fig. S1C and Table S1) reveal a globular molecule, with approximate dimensions of 11 × 8 × 7 nm (green mesh in Fig. 1 C and D and SI Appendix, Fig. S1D). The SAXS data for the wt Mmα-GluII full-length heterodimer yield an elongated shape that extends off one end of the globular structure of the trypsinized fragment to an overall length of 22 nm (light blue mesh in Fig. 1 C and D and SI Appendix, Fig. S1D). This shape and volume are in agreement with the sedimentation velocity ultracentrifugation measurements on the intact heterodimeric rat liver enzyme purified from tissue (21).
Fig. 1.
SAXS and crystal structures of Mmα-GluII. (A) A 1-Dimensional representation of the α-GluII subunits. The trypsinolysis sites are symbolized by scissors, and portions of the α-subunit removed by trypsin are represented by magenta loops. (B) Crystal structure of Mmα-GluIITryps in cartoon representation. The unique α-subunit N-terminal extension (residues 33–54) is shown in red, and the N-terminal domain (residues 55–392) is shown in purple. Dotted lines circle the α-subunit portions removed by trypsinolysis. The α-subunit GH31 catalytic domain (residues 393–748) is shown in green, with its insertion subdomain shown in yellow (residues 492–531). The proximal and distal α-subunit C-terminal domains (residues 749–828 and 829–966, respectively) are shown in orange and pink, respectively. After trypsinization, the α/β Mmα-GluII dimer retains the N-terminal domain of the β-subunit, adopting two tandem LDLRa folds (blue), with the trypsinized β-subunit N and C termini marked by a letter. Two calcium ions in the β-subunit LDLRa subdomains are shown as green spheres. The α-subunit active site residues, the N-linked glycan at N97, and the calcium-coordinating residues in the β-subunit are depicted in stick representation. (C and D) Crystal structure of Mmα-GluIITryps (in cartoon representation) overlaid on the SAXS-hydrated envelopes for Mmα-GluIITryps (green mesh) and Mmα-GluII (light blue mesh). The green cartoon illustrates the α-subunit and the cyan cartoon illustrates the β-subunit (N-terminal LDLRa domains), the dashed line illustrates the central part of the protein (unannotated), and the cyan shape illustrates the C-terminal M6PRH domain. The α-GluII–specific α-subunit N-terminal extension is painted red. The green asterisk illustrates the volume taken up by the residues α 186–240, removed from Mmα-GluIITryps by trypsinolysis. Helices α 427–441 and α 470–482, which the β-subunit protects from HDX, are shown in dark blue. A stereoregular, extended conformation of the Glc2Man9GlcNAc2 glycan, with the terminal Glc residues inserted in the active site, is represented in sticks (yellow, carbon atoms; red, oxygen atoms). Two possible locations of the β-subunit M6PRH domain (cyan shape) are shown, either proximal to the α-subunit active site (C) or distal to the α-subunit active site (D), at the far end of the light blue holoenzyme SAXS envelope. The red arrows indicate hypothetical conformational changes in the β-subunit that would bring its M6PRH domain closer to the enzyme’s α-subunit active site if simultaneous binding of both subunits to a single monoglucosylated glycan was necessary for cleavage.
The α-GluII β-Subunit Associates with the Catalytic α-Subunit Via Its N-Terminal Domains.
Full-length Mmα-GluII did not yield crystals; however, Mmα-GluIITryps crystals enabled structure solution by molecular replacement at a resolution of 1.74 Å (Fig. 1B and SI Appendix, Table S2). The Mmα-GluII catalytic α-subunit overall fold (green in Fig. 1 C–D) and catalytic pocket (Fig. 2A) confirm a GH31 family fold. Known α-subunit activity-impairing mutations corresponding to Mmα-GluII E568Q (22), S569F, and S651F (23) localize to regions proximal to the active site or destabilize the fold of the α-subunit (SI Appendix, Fig. S2A). Unique to ER α-GluII α-subunit is its N terminus (residues 33–54), which reaches the brim of the catalytic pocket (red in Fig. 1 B–D) and supports a loop (residues 305–317 in Mmα-GluII) contributing to the +1 and +2 enzyme subsites (Fig. 3 A and B). In addition to the α-subunit, our Mmα-GluIITryps crystals contain two tandem low-density lipoprotein receptor class A (LDLRa) subdomains, the N-terminal and only portion of the β-subunit still associated with the α-subunit after trypsinolysis (light blue in Fig. 1 A–D and SI Appendix, Fig. S4A), in keeping with the predictions from previous biochemical studies (24–27). Each β-subunit LDLRa module folds around an octahedrally coordinated calcium ion (SI Appendix, Figs. S2B and S3 A and B). The interface between the α-subunit and the β-subunit LDLRa modules spans 700 Å2 of solvent-accessible area, and is organized around salt bridges between conserved aspartic acid residues in the β-subunit and positively charged residues in the α-subunit, clustered around residues α R837 and α R840 (SI Appendix, Figs. S4A and S5 A and B). Docking of the Mmα-GluIITryps crystal structure onto the wt Mmα-GluII heterodimer SAXS envelope reveals that, contiguous to the β-subunit N-terminal LDLRa subdomains of Mmα-GluIITryps, the Mmα-GluII full-length heterodimer has a protuberance corresponding to the trypsin-sensitive portion of the β-subunit that is missing in the crystals (Fig. 1 C and D). Hydrogen deuterium exchange (HDX) MS conducted on the full-length Mmα-GluII heterodimer supports the observed α/β crystal interface (SI Appendix, Fig. S6). Additionally, helices α 427–441 and α 470–482, which contact each other on the surface of the α-subunit between the active site and the β-subunit, are protected from HDX (SI Appendix, Fig. S6 A and C). In the context of the full-length Mmα-GluII heterodimer, these α-subunit helices (dark blue in Fig. 1 C and D) are buried inside the SAXS envelope, indicating that they also form part of the α/β interface. The interaction between subunits at this spot must be weak/transient, because the portion of β-subunit involved in it does not survive trypsinolysis.
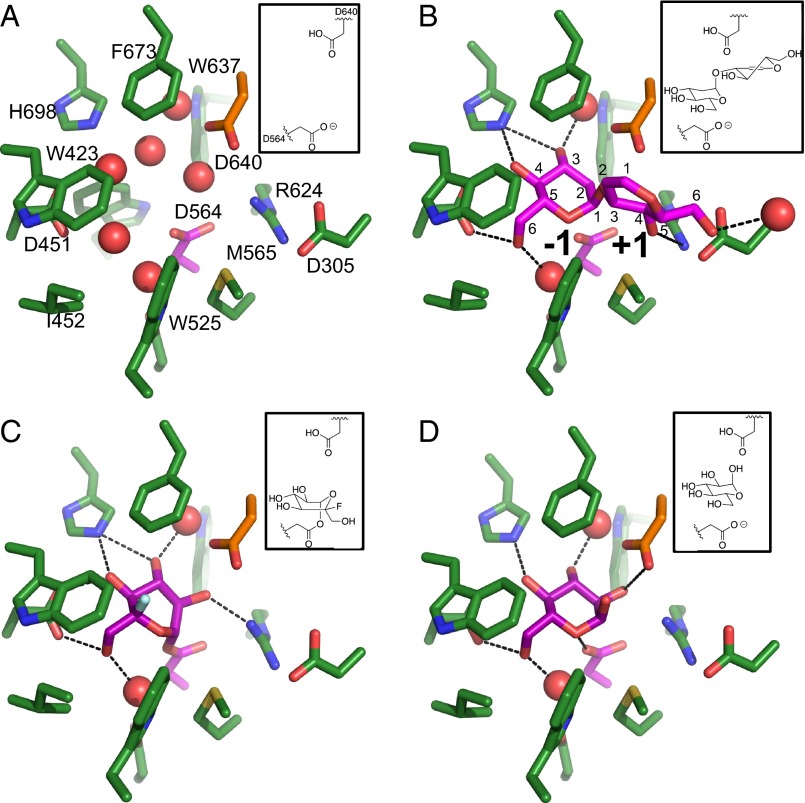
Fig. 2.
Details of the Mmα-GluII α-subunit active site at key stages during its catalytic cycle (with corresponding chemical schemes shown in the upper right corners). The −1 and +1 subsites are marked in B. (A) Apo enzyme; the α-subunit pocket is filled with ordered water molecules (red spheres). (B) Complex with the d-glucal transglucosylation product, an analog of an α(1,3)-disaccharide substrate. (C) Covalent complex after nucleophilic attack from D564 to 5F-Glc fluoride (SI Appendix, Fig. S9; NMR spectra of 1), forming a reaction intermediate analog. (D) Complex with d-glucose, one of the products of the enzyme’s reactions. Protein stick representation: green, carbon; red, oxygen; blue, nitrogen. Hydrogen atoms are omitted. The carbon atoms of the catalytic nucleophile D564 and the ligands are colored purple. The carbon atoms of the catalytic acid/base D640 are colored orange. The fluorine atom in C is colored light green.
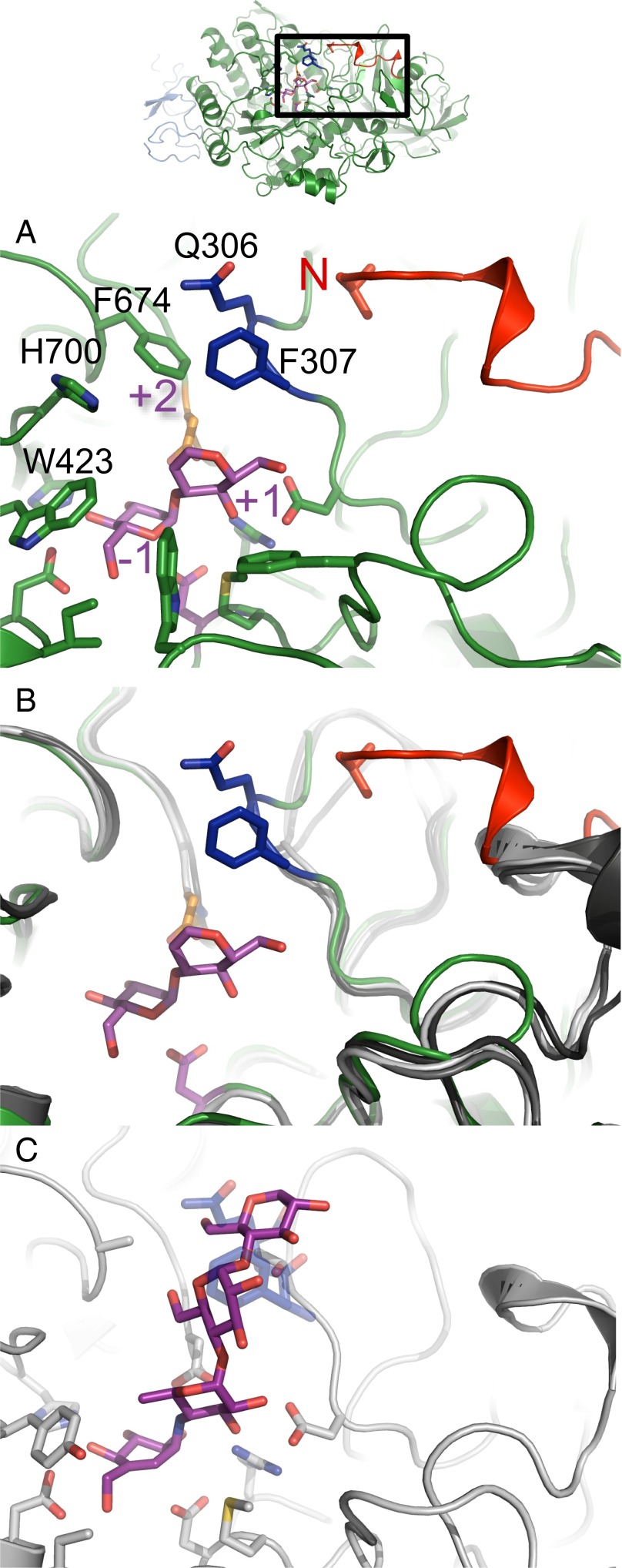
Fig. 3.
α-Subunit N terminus extension and exclusion loop insertion are previously unidentified α-GluII-specific determinants of enzyme activity. (A) Mmα-GluIITryps α-subunit (green) with its bound disaccharide substrate analog (purple). The enzyme’s subsites −1, +1, and +2 are indicated in purple lettering. The position of the α-subunit N terminus is highlighted in red, next to the exclusion loop insertion F307 and Q308 (dark blue). The exclusion loop insertion F307, Q308 is only present in α-GluII. The N-terminal V33 residue is shown in stick representation. (B) Structural alignment of Mmα-GluIITryps α-subunit (green) with its bound disaccharide substrate analog (purple) against the structures of H. sapiens intestinal α-glucosidases: N-terminal MGAM (light gray, PDB ID code 2QMJ), C-terminal MGAM (dark gray, PDB ID code 3TOP), and N-terminal SI (black, PDB ID code 3LPP). (C) Details of acarbose [purple (C atoms) and red (O atoms)] binding to human MGAM in PDB entry (gray, ID code 2QMJ). This mode of binding of acarbose is not possible in α-GluII due to steric hindrance from the exclusion loop residues F307 and Q308 (transparent dark blue).
Arginine Residues on the α-GluII α-Subunit Mediate Its ER Retention.
The α-GluII β-subunit carries a C-terminal ER-retention signal that ensures ER retention of the GluII heterodimer (28). The Mmα-GluII α-subunit single mutant R840E, designed on the basis of the crystal structure to disrupt α/β association, loses binding capacity for the β-subunit in vitro. The latter no longer copurifies with the 6× His-tagged α-subunit R840E mutant as assayed by immobilized metal affinity chromatography (SI Appendix, Fig. S4G). Similar results were obtained in vivo for the equivalent Arabidopsis thaliana (At) α-GluII α-subunit single mutant R787E, transfected into tobacco leaves as a GFP fusion (SI Appendix, Fig. S4). The mutant shows weaker association with the β-subunit, as judged by fluorescence microscopy and (co)immunoprecipitation experiments (SI Appendix, Fig. S4 C, E, and F). The Atα-GluII α-subunit R784E/R787E double mutant (corresponding to the Mmα-GluII α-subunit double-mutant R837E/R840E) completely abrogates the α/β association in vivo, again as shown by fluorescence microscopy and (co)immunoprecipitation experiments (SI Appendix, Fig. S4 D–F). The murine α-GluII α-subunit R840E mutant that abrogates the α/β interaction in vitro and corresponds to the plant R787E mutant showing reduced ER localization in planta, shows only 10% of the wt enzyme activity in an in vitro assay (SI Appendix, Table S3), suggesting that ligands targeting the α-GluII α/β interface would constitute selective α-GluII inhibitors.
The α-GluII β-Subunit LDLRa Subdomains Stabilize the α-Subunit in a Catalytically Competent Conformation.
The differences in the activity of the enzyme against glycoproteins carrying a variable number of N-linked glycans have led to the suggestion that more than one glycan is needed for α-GluII to allow entry of a glycoprotein into the calnexin/calreticulin cycle (29). In this case, the mannose 6-phosphate receptor homology (M6PRH) C-terminal domain of the β-subunit would bind a terminal α(1,2)-linked mannose residue on one N-linked glycan (30, 31), helping the recruitment of the other N-linked glycan to the active site (29, 32). The shape and dimensions of our Mmα-GluII SAXS envelope and Mmα-GluIITryps crystal structure are compatible with a role for the M6PRH C-terminal domain of the β-subunit assisting glucose hydrolysis by binding a second glycan on glycoproteins carrying more than one glycan (29, 32). In the absence of the β-subunit, α-GluII activity is reduced in vivo (27) and in vitro (33) to about 5–10% of wt levels and the recombinant α-subunit is unstable unless associated with the β-subunit (4, 28). In our hands, Mmα-GluIITryps is still able to hydrolyze Glc1Man7GlcNAc2 and Glc2Man7GlcNAc2, has identical binding affinity for the 4-methylumbelliferyl α-d-glucopyranoside (4-MUG) synthetic substrate, and retains 70% of the wt catalytic efficiency against 4-MUG (SI Appendix, Table S3). Because the first cleavage seems to proceed even when the β-subunit is disrupted (26, 27, 29), it appears the C-terminal M6PHR domain of the β-subunit may simply increase the avidity of the α-subunit for the substrate glycoprotein-linked Glc2ManxGlcNAc2 (32). Conversely, the N-terminal portion of the β-subunit is necessary for α-GluII catalysis by the α-subunit in vitro and in vivo, and acts by stabilizing the α-subunit in a catalytically competent conformation.
A Conformational Change is Likely to Be Needed for Simultaneous Binding of a Glc1ManxGlcNAc2 Glycan to Both α-GluII Subunits.
In light of the NMR structure of the core N-glycan (34), it has been proposed that the monoglucosylated glycan must undergo a conformational change for its hydrolysis to take place (26, 27, 29). The NMR structure of the core N-glycan showed that the Glc-α(1,3)-Glc moiety (substrate for “cleavage 1”) is exposed on the external face of the 3′ N-linked glycan branch, whereas the Glc α(1,3)-Man moiety (substrate for “cleavage 2”) is on its internal face. It is therefore likely that the two substrates are inserted in the active site with the N-linked glycan bound to α-GluII in different orientations (34). These observations, in turn, have led to models in which a single Glc1ManxGlcNAc2 glycan is simultaneously bound to the α-subunit active site and the M6PRH C-terminal domain of the β-subunit, which would assist the reorientation of the glycan needed for the second cleavage (29, 32). Our docking of the Mmα-GluIITryps crystal structure into the Mmα-GluII SAXS envelope shows that, in the absence of a major conformational change, the distance between the active site of the α-subunit and the closest portion of the β-subunit is comparable to the length of a single N-glycan in extended conformation. The span of an N-linked glycan engaged in the α-subunit active site can be appreciated by looking at manually positioned Glc2Man9GlcNAc2 glycan emerging from the active site in stick representation in Fig. 1 C and D. In view of our data, it is therefore feasible that the same N-linked glycan is shared between the catalytic site and the M6PRH domain when the enzyme is engaged to cleave the inner glucose; however, for this sharing of a single glycan between the two subunits to be possible, the β-subunit would need to undergo a conformational rearrangement. The extent of this hypothetical movement could be minimal, for example, if the β-subunit were bent onto itself in a U-shape (Fig. 1C). A conformational change has been reported by circular dichroism for rat liver α-GluII in the presence of the molecular crowding agent PEG 20,000; indeed, crowding did not affect the first cleavage, but it greatly enhanced the rate of the second cleavage (35). An arrangement in which the N- and C-terminal regions of the β-subunit were proximal in space has already been suggested on the grounds that the region β 273–400 has been reported to interact with the α-subunit (25). Notably, a subset of that region, residues β 357–390, seems to be protected from HDX exchange in our HDX experiment (SI Appendix, Fig. S6), although further experiments are necessary to determine if they contact the α-subunit or a different portion of the β-subunit. Alternatively, if the β-subunit domains are arranged in a linear fashion, only a major conformational change (also yet to be observed) could bring the C terminus of the β-subunit closer to the α-subunit (Fig. 1D).
Key Snapshots from the α-GluII Catalytic Cycle.
For the first time to our knowledge, all key stages of the catalytic cycle of a single member of the GH31 family have been trapped in crystal structures by soaking of ligands in the Mmα-GluIITryps crystals (SI Appendix, Table S2). The interactions of the enzyme with the ligands are illustrated in detail in Fig. 2 and SI Appendix, Fig. S7. All GH31 family members fold as retaining α-glycosidases following the classical Koshland double-displacement mechanism (36) and can catalyze transglycosylation reactions at high substrate concentrations, forming disaccharides. Instead of hydrolysis of a disaccharide moiety, the enzyme in this case catalyzes the formation of a glycosidic bond between two monosaccharides at subsites −1 and +1 (37, 38). Transglycosylation enables the structural study of the complex of a native glycosyl hydrolase with a substrate analog, without the need for an inactive mutant trapping a substrate. Indeed, by soaking the Mmα-GluIITryps crystals with the 1,2-unsaturated glucose analog d-glucal, we obtained a complex with a bound α(1,3)-linked d-glucal pseudodisaccharide (Fig. 2B). This substrate analog is sandwiched between D564 and D640, which sequence analysis predicted to be the catalytic nucleophile and general acid/base catalyst, respectively. Selection of α- over β-anomers is mediated by the side chains of conserved residues R617 and M565, which would clash against the ring at the reducing end of a β(1,3)-bound disaccharide. In the hexose ring at the −1 subsite, the three equatorial hydroxyl moieties of at C-2, C-3, and C-4 make H-bonds to the conserved R624, H698, and D451, respectively (Fig. 2 B–D and SI Appendix, Fig. S7 A–C), which explains the selection of d-glucose over d-mannose and d-galactose at this subsite. To confirm that D564 is the catalytic residue, and to obtain the crystal structure of an analog of the first covalent reaction intermediate, we have synthesized and soaked into the Mmα-GluIITryps crystals a difluorinated d-glucose analog, 5-fluoro-α-d-glucopyranosyl (5F-Glc) fluoride. Compounds of this class are known to undergo nucleophilic attack at C-1 in a manner analogous to natural substrates, with the F atom at C-1 as the leaving group; the remaining F atom at C-5 stabilizes the covalent intermediate, trapping it in the −1 subsite (39). The complex obtained by soaking the Mmα-GluIITryps crystals with 5F-Glc fluoride (Fig. 2C and SI Appendix, Fig. S7B) shows the reaction intermediate in a skew boat 1S3 conformation (40), which is also observed in the structures of other GH31 enzymes with trapped covalent fluoroglycosyl/enzyme intermediates. The rmsd for the overlay of the non-H atoms of 5F-Glc adduct in our structure onto the equivalent ones in the ligands of Protein Data Bank (PDB) ID codes 2XVK, 1XSK, and 4BA0 are 0.07 Å, 0.12 Å, and 0.09 Å, respectively. The 5F-Glc is covalently bound to D564 after the first nucleophilic attack displaces the fluorine atom from C-1, proving that Mmα-GluII D564 is the catalytic nucleophile. Mmα-GluII mutants D564N (and D640N) are both inactive against 4-MUG and the two physiological substrates tested (SI Appendix, Table S3). Finally, cleavage of the natural substrates liberates d-glucose, which is present in a 4C1 chair conformation bound to the −1 pocket of the enzyme after glycosidic bond hydrolysis in our structure (Fig. 2D and SI Appendix, Fig. S7C). The α-anomer configuration of the hydroxyl at C-1 in this structure confirms that the catalytic site of this retaining glucosidase stabilizes the same C-1 configuration in substrate and product.
Glc-α(1,3)-Man Stabilizes the Protonated Form of the D640 Acid/Base Catalyst.
The two successive cleavages effected by ER α-GluII have different catalytic rates, pH dependency, and inhibitor sensitivities (41), but the enzyme has a single catalytic site that can accommodate two distinct terminal sugar moieties, Glc-α(1,3)-Glc and Glc-α(1,3)-Man, during the first cleavage and second cleavage, respectively. Because d-mannose and d-glucose differ in the stereochemical configuration of the C-2 atom, at the +1 subsite the hexose ring of the natural substrates presents an equatorial or an axial hydroxyl group at C-2. The ring of the d-glucal pseudodisaccharide in the +1 subsite forms H-bonds to the conserved R624 and D305 with the hydroxyl groups at C-4 and C-6, respectively (Fig. 2B and SI Appendix, Fig. S7A), whereas no contact with the enzyme is seen at the unsaturated C-2/C-3 side of the same ring. Absence of close contacts on this side of the substrate analog at the +1 subsite supports tolerance of either stereochemistry at C-2 of the hexose ring at the +1 subsite. Modeling Glc-α(1,3)-Glc and Glc-α(1,3)-Man based on the d-glucal pseudodisaccharide in the active site suggests that the equatorial hydroxyl group at C-2 of a d-glucose ring in the +1 subsite would also make no significant contacts with the protein. Conversely, the axial hydroxyl group on the C-2 of a d-mannose ring in the +1 subsite could accept an H-bond from the acid/base catalytic residue D640. This observation raises the possibility that the catalytic acid/base could be a less efficient acid in assisting hydrolysis of the Glc-α(1,3)-Man vs. the Glc-α(1,3)-Glc bond in the glycan, consistent with the observed differences in the rates of hydrolysis of the two natural substrates (41). The pKa of the general acid/base carboxyl group of a glycosidase cycles during catalysis, because its dual role places specific demands upon its ionization states, which, in turn, depend on the structural changes in the active site during catalysis (42). If the pH is raised beyond a glycosidase’s pH optimum, the alkaline conditions will eventually strip the acid/base catalytic residue of its proton, preventing it from acting as an acid and abrogating activity (43). Different substrates in the active site may influence the pH dependence of the activity by altering the acid/base residue’s environment in different ways, and leading to changes in the apparent pKa value of this residue (42). To investigate the effects of interactions between the α-GluII acid/base catalytic residue and the substrates, and to probe their contribution to the observed differential rates of α-GluII’s first and second cleavages (41), we have studied the pH dependence of the Mmα-GluII activity and the specificity constant kcat/Km against Glc-α(1,3)-Glc and Glc-α(1,3)-Man disaccharide substrates. In the pH interval of 6.9–7.9, the drop in Mmα-GluII substrate turnover is more pronounced for the cleavage of Glc-α(1,3)-Glc than Glc-α(1,3)-Man (SI Appendix, Fig. S10B). Similarly, the specificity constant kcat/Km of the enzyme against the Glc-α(1,3)-Glc substrate in the pH interval of 6.9–7.9 drops more steeply than the kcat/Km for the Glc-α(1,3)-Man substrate (SI Appendix, Fig. S10C). In keeping with the known higher rate for the first cleavage, the enzyme is more efficient in cleaving the Glc-α(1,3)-Glc than the Glc-α(1,3)-Man disaccharide at physiological pH, but the differences in Km for the two substrates become negligible at higher pH (SI Appendix, Fig. S10D). Taken together, these observations are consistent with Glc-α(1,3)-Man [but not Glc-α(1,3)-Glc] stabilizing the protonated form of D640 (SI Appendix, Fig. S10 E and F). While this paper was in preparation, crystal structures of an inactive mutant of the Chaetomium thermophilum α-GluII α-subunit in complex with Glc-α(1,3)-Glc and Glc-α(1,3)-Man2 have been described (44). The structures agree with the mode of binding of the disaccharide substrate analog in our Mmα-GluIITryps crystal structure and provide further data in support of the model put forward here, highlighting the contribution of D640–substrate interactions to the different specificity constants of the enzyme’s first and second cleavages.
α-GluII Selectivity Determinants Reside Beyond the −1 Subsite Catalytic Pocket.
In our hands, both full-length Mmα-GluII and its trypsinized fragment are active against natural glycoprotein substrates (GlcMan7GlcNAc2 and Glc2Man7GlcNAc2) and against the synthetic fluorescent substrate, 4-MUG (SI Appendix, Table S3). Kinetic measurements on a range of differently linked diglucosides reveal that the specificity of Mmα-GluII is not restricted to α(1,3) bonds. The enzyme can cleave α(1,4) and, to a lesser extent, α(1,2) glycosidic bonds (SI Appendix, Fig. S10A and Table S4). At higher substrate concentrations (2–10 mM), the best diglucoside substrate, the α(1,3)-linked nigerose, shows kinetics indicative of substrate inhibition (41), compatible with two overlapping substrate-binding sites of different affinity (45, 46). Our crystal structures reveal a high degree of structural similarity between the Mmα-GluII catalytic pocket −1 subsite and the ones of intestinal α-glucosidases, and rationalize the observed reactivity against maltose (45–47) and the cross-reactivity of inhibitors within this family of enzymes (48). Most residues lining the Mmα-GluII α-subunit −1 and +1 subsites are identical to the ones in the human intestinal MGAM and SI enzymes (Fig. 3B and SI Appendix, Fig. S5C). It is likely that the absence of α(1,4) glycosidic bonds in the ER-localized glycan pool has lifted from α-GluII any evolutionary pressure to lose reactivity against maltose-like substrates. However, toward the brim of the active site, the Mmα-GluII α-subunit +2 subsite is smaller than in MGAM and SI, due to the conserved residue H700 (which is S, T, or G in the intestinal glucosidases) and a conserved two-residue insertion, which is absent in other GH31 family members, jutting into the +2 subsite (Fig. 3A and SI Appendix, Fig. S5C). This insertion (F307 and Q308 in Mmα-GluII; blue in Fig. 3) is located in a loop of the N-terminal domain (residues 305–317 in Mmα-GluII) that docks against the Mmα-GluIITryps N terminus and flanks the catalytic pocket. The insertion is critical for selectivity against α(1,4)-linked substrates. We overlaid our structure of the Mmα-GluII α-subunit onto the structure of the N-terminal subunit of human MGAM in complex with acarbose, a noncleavable α(1,4)-tetrasaccharide mimic and well-known inhibitor of intestinal α-glucosidases (PDB ID code 2QMJ). Acarbose cannot fit into the substrate-binding pocket of the Mmα-GluII α-subunit because of steric hindrance due to the insertion loop residues (Fig. 3C). Because it is this loop that prevents acarbose from binding and inhibiting Mmα-GluII, we name the Mmα-GluII α-subunit loop with the F307, Q308 insertion the “exclusion loop.” In our hands, the Mmα-GluII mutant F307G-Δ308 retains only 17% of wt activity against 4-MUG and the Mmα-GluII deletion mutant Δ307–308 is completely inactive against the same substrate (SI Appendix, Table S3), probably because the mutations interfere with the access of ligands to the +1 subsite.
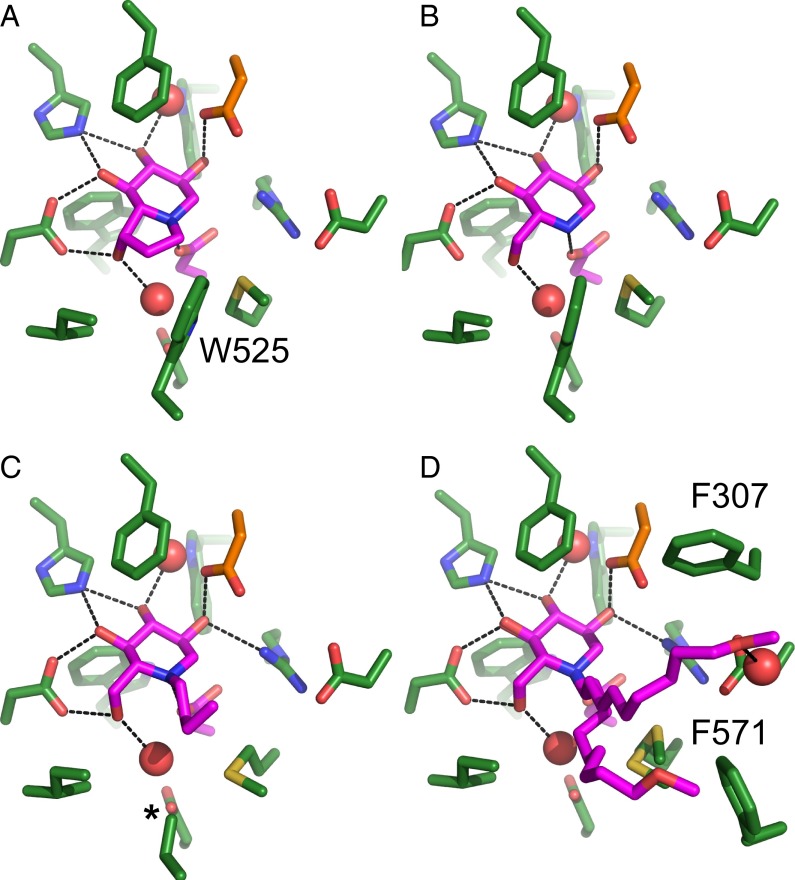
Alkylated Iminosugar Inhibitors Block Access to the +1 Subsite.
In our recent work, we have identified α-GluII in particular as a suitable target for the development of broad-spectrum antiviral agents. Inhibition of α-GluII is sufficient to inhibit, for example, dengue virus in vitro and in dengue disease mouse models (18). To improve our understanding of α-GluII inhibition, we have determined the crystal structures of Mmα-GluIITryps in complex with four iminosugars that possess antiviral activity: castanospermine; DNJ; and the alkylated derivatives of DNJ, N-butyl-1-deoxynojirimycin (NB-DNJ, also known as miglustat) and MON-DNJ (also known as UV-4) (14) (SI Appendix, Table S5). MON-DNJ is the best-known inhibitor of α-GluII in vivo (18), is active against dengue and influenza (17, 19), and is currently in clinical trials for dengue fever. The iminosugars all occupy the −1 subsite, and their hydroxyl moieties interact with the enzyme similar to glucose, 5F-glucose, and the d-glucal disaccharide (Fig. 4 A–D and SI Appendix, Fig. S8 A–D). Their endocyclic nitrogen atom is in close proximity to the catalytic D564 (in a similar orientation to the orientation observed for the iminosugar miglitol in the active site of the intestinal MGAM and SI enzymes), compatible with the idea that the molecules act as transition state mimics of the first step of glycosidic bond hydrolysis (49). The castanospermine hydrophobic five-membered ring fits in a pocket formed by the conserved W423, I448, and W525 (Fig. 4A and SI Appendix, Fig. S8A). The alkyl tail of NB-DNJ moves toward the +1 subsite and displaces the side chain of W525, disordering it (Fig. 4C and SI Appendix, Fig. S8C). In the crystal structure of the N-terminal domain of human MGAM in complex with the iminosugar miglitol (PDB ID code 3L4W) (50), a similar movement of the side chain of W406 (equivalent to Mmα-GluII W525) was caused by the N-hydroxyethyl group of the ligand. The alkyl chain of NB-DNJ is longer than the one in miglitol, and it stretches toward the side chains of the conserved residues F307 and F571, suggesting that N-alkylated iminosugar derivatives act by blocking access to the +1 subsite as well as occupying the −1 subsite with the iminosugar ring. This mode of alkylated iminosugar binding is confirmed by the structure with MON-DNJ (Fig. 4D and SI Appendix, Fig. S8D), which has a longer alkyl chain, and is the most potent of these DNJ derivatives [MON-DNJ has an IC50 against isolated Mmα-GluII of about 1.8 ± 0.3 μM (51) vs. 5.2 ± 1.0 μM for NB-DNJ and 11.4 ± 4.3 μM for DNJ (48)]. MON-DNJ displaces the whole loop, α 523–528, which changes conformation and whose terminal hairpin, α 525–527, is disordered in the crystal after MON-DNJ soaking. The alkyl chain of the iminosugar is in two main conformations of refined occupancies 0.61 and 0.39, with the major conformer docking against the exclusion loop F307 and the minor one docking against the hydrophobic side chain of F571, therefore reaching toward the +2 subsite (Fig. 4D). It appears that these N-alkylated iminosugars profit from favorable entropic contributions to the free energy of binding, both because of the disorder induced in the α 525–527 loop and because their alkyl chain adopts multiple conformations. Based on these observations, the ring of aromatic residues between the α-GluII +1 and +2 subsites (residues F307, W423, F674, and H700, conserved across α-GluII of several eukaryotes; Fig. 3A and yellow triangle in SI Appendix, Fig. S5A) would be a good target for inhibitors with increased potency and selectivity for α-GluII. In particular, the exclusion loop F307 is unique to ER α-GluII enzymes and is absent from intestinal glucosidases (SI Appendix, Fig. S5C). Together, the crystal structures of Mmα-GluIITryps in complex with NB-DNJ and MON-DNJ broaden the pharmacological search for selective α-GluII inhibitors, extending it to the enzyme’s +1 and +2 subsites. These subsites are shaped by the exclusion loop and the α-subunit N terminus (i.e., portions of α-GluII that this work has uncovered as specific to α-GluII) and constitute valid druggable targets, increasing the size and scope of the α-GluII drug epitope space. Our Mmα-GluIITryps crystal structures will assist in drug candidate discovery against existing and emerging infectious disease.
Fig. 4.
Crystal structures of Mmα-GluIITryps in complex with iminosugar inhibitors. Structures of complexes of Mmα-GluIITryps with castanospermine (A), DNJ (B), NB-DNJ (C), and MON-DNJ (D). Upon alkylation of the endocyclic nitrogen, the W525 side chain in the NB-DNJ complex is disordered (asterisk in C), whereas the longer chain of MON-DNJ causes complete disordering of the α 525–527 hairpin loop (D). The side chain of MON-DNJ adopts two main conformations, which interact with F307 and F571.
Materials and Methods
Cloning of Mmα-GluII.
Amplification of the M. musculus ganab (α-GluII α-subunit, isoform 2) and prkcsh (α-GluII β-subunit) genes (UniProt accession nos. Q8BHN3-2 and O08795) was achieved by PCR using a standard Phusion Flash (ThermoFisher Scientific) protocol. A DpnI digestion was performed to prevent template DNA from contaminating the newly assembled product. Purification of the PCR products was achieved by AMPure XP magnetic beads (Beckman Coulter) following the manufacturer’s protocol. Assembly of the constructs was carried out by mixing 1 μL of the linearized vector (pOPINGS and pOPING for ganab and prkcsh, respectively) with 5 μL of the purified PCR product to a total volume of 10 μL, which was added to lyophilized In-Fusion HD EcoDry enzyme mix (Clontech). pOPINGS bears C-terminal Strep-II and hexahistidine tags, and pOPING bears a C-terminal hexahistidine tag.
Expression of Mmα-GluII.
Cotransfection into the FreeStyle 293 Expression System (Life Technologies) was carried out according to the manufacturer’s protocol. The transfection reagent was used at 0.125% (vol/vol) of the culture volume. The plasmids were used in equimolar amounts at a total of 0.1% (wt/vol) of the culture volume. Cells were maintained at 37 °C, 5% CO2, and shaking at 135 rpm.
Purification of Mmα-GluII.
After 4 d, the cells were harvested by centrifugation at 3,000 × g for 15 min. The supernatant was adjusted to 1× PBS and 5 mM imidazole and a final pH above 7.5. The supernatant was flowed through a 5-mL HisTrap excel (GE LifeSciences) column, washed, and finally eluted with 10 column volume of 400 mM imidazole in PBS supplemented with 5% (wt/vol) glycerol. The imidazole was removed from the eluate by dialysis into Strep Wash Buffer [100 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA]. The Mmα-GluII was bound to 10 mL of StrepTactin Superflow High Capacity resin (IBA). The resin was washed and eluted following the manufacturer’s protocol. The concentrated enzyme was applied to a Superdex 200 16/600 column (GE Lifesciences) in 20 mM Hepes (pH 7.5) and 150 mM NaCl. Yield of the full-length Mmα-GluII is 8 mg/L culture.
Trypsinolysis of Mmα-GluII.
Mmα-GluII was treated with sequencing grade modified trypsin (Promega) at a 1:100 trypsin/Mmα-GluII mass ratio, supplemented with 2 mM CaCl2 for 4 h at room temperature. The trypsinized material was purified on a Superdex 200 column as above.
X-Ray Crystal Structures Determination.
Crystal growth.
All crystallization solutions were purchased from Molecular Dimensions. Filtered Mmα-GluIITryps at 5.6 mg/mL was crystallized by vapor diffusion with 21% (vol/vol) ethylene glycol, 11% (wt/vol) PEG 8000 (from Morpheus precipitant mix 2), 50 mM Morpheus carboxylic acids mix, and 100 mM Morpheus buffer system 1 (pH 6.25) (all solutions from Molecular Dimensions) in a 3:1 protein/precipitant ratio. Two hundred-micrometer-long rods formed after about 1 wk at 18 °C. Crystals were transferred into a solution of 16% (wt/vol) PEG 8000, 50 mM Morpheus carboxylic acids mix, and 100 mM Morpheus buffer system 2 (pH 7.2) and 20% (vol/vol) PEG 400, with or without ligands, before cooling in liquid nitrogen.
X-ray diffraction.
Diffraction from Mmα-GluIITryps crystals was measured at the Diamond Light Source, except for the apo and NB-DNJ structures, data for which were collected on beamlines ID30-1 and BM14, respectively, at the European Synchrotron Radiation Facility (ESRF). All experiments were carried out at a temperature of 90 K in a stream of cryogenic N2 gas.
Data Processing, Structure Determination, and Refinement.
X-ray diffraction images were processed using the autoPROC (52) or the xia2 (53) suite of programs, both of which index and integrate with XDS (54), and were scaled and merged using the CCP4 (55) suite of programs: Pointless, Aimless, and Truncate. Molecular replacement leading to structure determination of the Apo form was performed with Phaser (56), which is also part of the CCP4 suite, run with the automated MR pipeline MrBUMP (57), using chain A of PDB ID code 3L4Z as a search model. Model building was performed with Coot and Buccaneer (58, 59), and refinement was performed with autoBUSTER, using local structure similarity restraints (LSSR) (60, 61). Model validation was carried out with internal modules of Coot and through the MolProbity server (62). Initial sets of phases for the 5F-glucosyl fluoride, glucal, glucose, and iminosugar soaks were obtained by molecular replacement from the apo structure. Idealized coordinates and stereochemical dictionaries for ligands not present in the autoBUSTER libraries and nonstandard ligands were generated using the GRADE server, starting from SMILES strings (grade.globalphasing.org/). Each ligand was docked in the unbiased Fo-Fc difference electron density map calculated from the phases at the end of the iterative protein-only model building and refinement. A final round of refinement of the protein and docked ligand used ligand stereochemical restraints from the GRADE-generated dictionary. All figures were produced in PyMOL.
SAXS of Mmα-GluII and Mmα-GluIITryps.
SAXS data for Mmα-GluII and Mmα-GluIITryps were collected at the BM29 beamline at the ESRF. The wavelength was set at 0.992 Å, and transmission was at 100%, with images recorded on a Pilatus 1M detector set to a distance of 2.886 m. Calibration was conducted with measurements on albumin or glucose oxidase to derive molecular weights from intensity at zero scattering angle (I0) values. All measurements were carried out in 150 mM NaCl and 20 mM Hepes at pH 7.4. A twofold dilution series with six concentrations between 4.12 mg/mL and 0.13 mg/mL was measured for Mmα-GluIITryps, and a twofold dilution series with five concentrations between 2.97 mg/mL and 0.17 mg/mL was measured for Mm α-GluII. Thirty microliters of each sample was flowed through a quartz capillary taking 10 × 1-s images. Automated image processing followed by buffer subtraction, as part of the processing pipeline at the beamline, allowed scattering curves to be used for further data processing. SAXS data were processed using the ATSAS (63) software suite. Using PRIMUS (64), for both Mmα-GluII and Mmα-GluIITryps samples, the low-angle regions of the low-concentration scattering curves were merged with the high-angle regions of the high-concentration profile. This merging procedure was done to compensate for interparticle effects at high concentration. The radius of gyration was determined using PRIMUS (64), and the maximum particle size Dmax was calculated from the pair distribution function calculated by GNOM (65). Ten bead models were created for each structure by DAMMIN (66), and then aligned and averaged using DAMAVER (67). DAMMIN was then used to compare the averaged model against raw data using reduced χ2 values. All models possess 0.9 < χ2 < 1.1 against raw data. The Mmα-GluIITryps crystal structure was initially fitted to the Mmα-GluIITryps SAXS envelope using SUPCOMB (68). Chimera (69) was used to convert the SAXS envelopes to maps (command MOLMAP, using a 2.5-nm filter) and to superpose the Mmα-GluII SAXS map to the Mmα-GluIITryps SAXS map and model.
The full, detailed methods for cloning, expression, purification, enzymology, HDX-MS, in planta confocal microscopy and (co)immunoprecipitations, X-ray crystal structure determination, and SAXS used in this study can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Raymond Dwek, Mark Wormald, Max Crispin, David Harris, and Kathryn Scott for helpful discussions and comments on the manuscript, and the members of the Zitzmann laboratory for assistance with molecular biology and protein chemistry. Louise Bird, Heather Rada, and Ray Owens (Oxford Protein Production Facility at the Research Complex) helped with cloning and initial expression trials. We also thank Justin Benesch and Shane Chandler (Department of Chemistry, University of Oxford) for HDX support and access to the mass spectrometer. Snezana Vasiljević assisted with mammalian cell expression. David Staunton took the size exclusion chromatography multiangle laser light scattering measurements. The staff at beamlines I02, I03, I04, and I04-1 (Diamond Light Source) and at beamlines BM14, ID30-1, and BM29 (ESRF) helped with X-ray data collection. Jesse Gayk helped with glycan 2AA labeling. A.T.C. was funded by a Wellcome Trust 4-Year Studentship (097300/Z/11/Z). J.L.K. is a Lerner–Fink Fellow in Medicinal Chemistry. N.Z. is a Fellow of Merton College, Oxford.
Footnotes
Conflict of interest statement: N.Z. is a named inventor on the following patents, which describe iminosugars and methods of treating viral diseases: WO/2001/010429(2001) and EP1210082(2002); WO/1999/029321(1999), EP1037636(2000) and KR1020010033028(2001); WO/2010/096764(2010) and EP2398321(2011); WO/2011/028775(2011) and EP2473482(2012); WO/2011/028779(2011) and EP2473046(2012); WO/2011/028781(2011) and EP2473493(2012); WO/2010/144759(2010) and EP2440205(2012); and WO/2010/099064(2010) and EP2400843(2012).
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5F0E, 5HJO, 5HJR, 5H9O, 5IED, 5IEE, 5IEF, and 5IEG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604463113/-/DCSupplemental.
References
- 1.Wisskirchen K, Lucifora J, Michler T, Protzer U. New pharmacological strategies to fight enveloped viruses. Trends Pharmacol Sci. 2014;35(9):470–478. doi: 10.1016/j.tips.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sayce AC, Miller JL, Zitzmann N. Targeting a host process as an antiviral approach against dengue virus. Trends Microbiol. 2010;18(7):323–330. doi: 10.1016/j.tim.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Martinez JP, Sasse F, Brönstrup M, Diez J, Meyerhans A. Antiviral drug discovery: Broad-spectrum drugs from nature. Nat Prod Rep. 2015;32(1):29–48. doi: 10.1039/c4np00085d. [DOI] [PubMed] [Google Scholar]
- 4.Pieren M, Galli C, Denzel A, Molinari M. The use of calnexin and calreticulin by cellular and viral glycoproteins. J Biol Chem. 2005;280(31):28265–28271. doi: 10.1074/jbc.M501020200. [DOI] [PubMed] [Google Scholar]
- 5.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91(3):913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci Rep. 2011;1:90. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 8.D’Alessio C, Caramelo JJ, Parodi AJ. UDP-GlC:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin Cell Dev Biol. 2010;21(5):491–499. doi: 10.1016/j.semcdb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauc G, Pezer M, Rudan I, Campbell H. Mechanisms of disease: The human N-glycome. Biochim Biophys Acta. 2016;1860(8):1574–1582. doi: 10.1016/j.bbagen.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Trombetta ES, Simons JF, Helenius A. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J Biol Chem. 1996;271(44):27509–27516. doi: 10.1074/jbc.271.44.27509. [DOI] [PubMed] [Google Scholar]
- 11.Roth J, Ziak M, Zuber C. The role of glucosidase II and endomannosidase in glucose trimming of asparagine-linked oligosaccharides. Biochimie. 2003;85(3-4):287–294. doi: 10.1016/s0300-9084(03)00049-x. [DOI] [PubMed] [Google Scholar]
- 12.Lucocq JM, Brada D, Roth J. Immunolocalization of the oligosaccharide trimming enzyme glucosidase II. J Cell Biol. 1986;102(6):2137–2146. doi: 10.1083/jcb.102.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeyakumar M, Dwek RA, Butters TD, Platt FM. Storage solutions: Treating lysosomal disorders of the brain. Nat Rev Neurosci. 2005;6(9):713–725. doi: 10.1038/nrn1725. [DOI] [PubMed] [Google Scholar]
- 14.Chang J, Block TM, Guo J-T. Antiviral therapies targeting host ER alpha-glucosidases: Current status and future directions. Antiviral Res. 2013;99(3):251–260. doi: 10.1016/j.antiviral.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343(6166):1235681. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- 16.Chang J, et al. Small molecule inhibitors of ER α-glucosidases are active against multiple hemorrhagic fever viruses. Antiviral Res. 2013;98(3):432–440. doi: 10.1016/j.antiviral.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warfield KL, et al. A novel iminosugar UV-12 with activity against the diverse viruses influenza and dengue (novel iminosugar antiviral for influenza and dengue) Viruses. 2015;7(5):2404–2427. doi: 10.3390/v7052404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry ST, et al. An iminosugar with potent inhibition of dengue virus infection in vivo. Antiviral Res. 2013;98(1):35–43. doi: 10.1016/j.antiviral.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Stavale EJ, Vu H, Sampath A, Ramstedt U, Warfield KL. In vivo therapeutic protection against influenza A (H1N1) oseltamivir-sensitive and resistant viruses by the iminosugar UV-4. PLoS One. 2015;10(3):e0121662. doi: 10.1371/journal.pone.0121662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butters TD, Van den Broek L, Fleet G, Krulle TM. Molecular requirements of imino sugars for the selective control of N-linked glycosylation and glycosphingolipid biosynthesis. Tetrahedron. 2000;11(1):113–124. [Google Scholar]
- 21.Trombetta ES, Fleming KG, Helenius A. Quaternary and domain structure of glycoprotein processing glucosidase II. Biochemistry. 2001;40(35):10717–10722. doi: 10.1021/bi010629u. [DOI] [PubMed] [Google Scholar]
- 22.Feng J, Romaniouk AV, Samal SK, Vijay IK. Processing enzyme glucosidase II: Proposed catalytic residues and developmental regulation during the ontogeny of the mouse mammary gland. Glycobiology. 2004;14(10):909–921. doi: 10.1093/glycob/cwh110. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, et al. Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Natl Acad Sci USA. 2009;106(52):22522–22527. doi: 10.1073/pnas.0907711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelletier MF, et al. The heterodimeric structure of glucosidase II is required for its activity, solubility, and localization in vivo. Glycobiology. 2000;10(8):815–827. doi: 10.1093/glycob/10.8.815. [DOI] [PubMed] [Google Scholar]
- 25.Arendt CW, Ostergaard HL. Two distinct domains of the beta-subunit of glucosidase II interact with the catalytic alpha-subunit. Glycobiology. 2000;10(5):487–492. doi: 10.1093/glycob/10.5.487. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson BM, Purswani J, Stirling CJ. Yeast GTB1 encodes a subunit of glucosidase II required for glycoprotein processing in the endoplasmic reticulum. J Biol Chem. 2006;281(10):6325–6333. doi: 10.1074/jbc.M510455200. [DOI] [PubMed] [Google Scholar]
- 27.Stigliano ID, Alculumbre SG, Labriola CA, Parodi AJ, D’Alessio C. Glucosidase II and N-glycan mannose content regulate the half-lives of monoglucosylated species in vivo. Mol Biol Cell. 2011;22(11):1810–1823. doi: 10.1091/mbc.E11-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arendt CW, Ostergaard HL. Identification of the CD45-associated 116-kDa and 80-kDa proteins as the alpha- and beta-subunits of alpha-glucosidase II. J Biol Chem. 1997;272(20):13117–13125. doi: 10.1074/jbc.272.20.13117. [DOI] [PubMed] [Google Scholar]
- 29.Deprez P, Gautschi M, Helenius A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell. 2005;19(2):183–195. doi: 10.1016/j.molcel.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 30.Totani K, Ihara Y, Matsuo I, Ito Y. Substrate specificity analysis of endoplasmic reticulum glucosidase II using synthetic high mannose-type glycans. J Biol Chem. 2006;281(42):31502–31508. doi: 10.1074/jbc.M605457200. [DOI] [PubMed] [Google Scholar]
- 31.Ito Y, Takeda Y. Analysis of glycoprotein processing in the endoplasmic reticulum using synthetic oligosaccharides. Proc Jpn Acad Ser B Phys Bio Sci. 2012;88(2):31–40. doi: 10.2183/pjab.88.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson LJ, et al. Crystal structure and functional analyses of the lectin domain of glucosidase II: Insights into oligomannose recognition. Biochemistry. 2015;54(26):4097–4111. doi: 10.1021/acs.biochem.5b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson LJ, et al. Structure of the lectin mannose 6-phosphate receptor homology (MRH) domain of glucosidase II, an enzyme that regulates glycoprotein folding quality control in the endoplasmic reticulum. J Biol Chem. 2013;288(23):16460–16475. doi: 10.1074/jbc.M113.450239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrescu AJ, et al. The solution NMR structure of glucosylated N-glycans involved in the early stages of glycoprotein biosynthesis and folding. EMBO J. 1997;16(14):4302–4310. doi: 10.1093/emboj/16.14.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Totani K, Ihara Y, Matsuo I, Ito Y. Effects of macromolecular crowding on glycoprotein processing enzymes. J Am Chem Soc. 2008;130(6):2101–2107. doi: 10.1021/ja077570k. [DOI] [PubMed] [Google Scholar]
- 36.Koshland DE. Stereochemistry and the mechanism of enzymatic reactions. Biol Rev Camb Philos Soc. 1953;28(4):416–436. [Google Scholar]
- 37.Sinnott ML. Catalytic mechanism of enzymic glycosyl transfer. Chem Rev. 1990;90(7):1171–1202. [Google Scholar]
- 38.Buchowiecka A, Bielecki S. Determination of the regiochemistry of D-glucal glucosylation by endo-β-1,3-glucanase GA cellulomonas cellulans using CI MS. Biocatal Biotransformation. 2009;21(1):1–5. [Google Scholar]
- 39.McCarter JD, Withers SG. Unequivocal identification of Asp-214 as the catalytic nucleophile of Saccharomyces cerevisiae alpha-glucosidase using 5-fluoro glycosyl fluorides. J Biol Chem. 1996;271(12):6889–6894. doi: 10.1074/jbc.271.12.6889. [DOI] [PubMed] [Google Scholar]
- 40.Mayes HB, Broadbelt LJ, Beckham GT. How sugars pucker: Electronic structure calculations map the kinetic landscape of five biologically paramount monosaccharides and their implications for enzymatic catalysis. J Am Chem Soc. 2014;136(3):1008–1022. doi: 10.1021/ja410264d. [DOI] [PubMed] [Google Scholar]
- 41.Kaushal GP, Pastuszak I, Hatanaka K, Elbein AD. Purification to homogeneity and properties of glucosidase II from mung bean seedlings and suspension-cultured soybean cells. J Biol Chem. 1990;265(27):16271–16279. [PubMed] [Google Scholar]
- 42.McIntosh LP, et al. The pKa of the general acid/base carboxyl group of a glycosidase cycles during catalysis: A 13C-NMR study of bacillus circulans xylanase. Biochemistry. 1996;35(31):9958–9966. doi: 10.1021/bi9613234. [DOI] [PubMed] [Google Scholar]
- 43.Dixon M, Webb EC. Enzymes. Academic; New York: 1964. [Google Scholar]
- 44.Satoh T, Toshimori T, Yan G, Yamaguchi T, Kato K. Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control. Sci Rep. 2016;6:20575. doi: 10.1038/srep20575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alonso JM, Santa-Cecilia A, Calvo P. Glucosidase II from rat liver microsomes. Kinetic model for binding and hydrolysis. Biochem J. 1991;278(Pt 3):721–727. doi: 10.1042/bj2780721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonso JM, Santa-Cecilia A, Calvo P. Effect of bromoconduritol on glucosidase II from rat liver. A new kinetic model for the binding and hydrolysis of the substrate. Eur J Biochem. 1993;215(1):37–42. doi: 10.1111/j.1432-1033.1993.tb18004.x. [DOI] [PubMed] [Google Scholar]
- 47.Brada D, Dubach UC. Isolation of a homogeneous glucosidase II from pig kidney microsomes. Eur J Biochem. 1984;141(1):149–156. doi: 10.1111/j.1432-1033.1984.tb08169.x. [DOI] [PubMed] [Google Scholar]
- 48.Sayce AC, et al. Iminosugars inhibit dengue virus production via inhibition of ER alpha-glucosidases-not glycolipid processing enzymes. PLoS Negl Trop Dis. 2016;10(3):e0004524. doi: 10.1371/journal.pntd.0004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gloster TM, Vocadlo DJ. Developing inhibitors of glycan processing enzymes as tools for enabling glycobiology. Nat Chem Biol. 2012;8(8):683–694. doi: 10.1038/nchembio.1029. [DOI] [PubMed] [Google Scholar]
- 50.Sim L, et al. New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: Structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata. Biochemistry. 2010;49(3):443–451. doi: 10.1021/bi9016457. [DOI] [PubMed] [Google Scholar]
- 51.Warfield KL, et al. Inhibition of endoplasmic reticulum glucosidases is required for in vitro and in vivo dengue antiviral activity by the iminosugar UV-4. Antiviral Res. 2016;129:93–98. doi: 10.1016/j.antiviral.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winter G, Lobley CMC, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keegan RM, Winn MD. MrBUMP: An automated pipeline for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):119–124. doi: 10.1107/S0907444907037195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 60.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 61.Smart OS, et al. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen VB, Wedell JR, Wenger RK, Ulrich EL, Markley JL. MolProbity for the masses-of data. J Biomol NMR. 2015;63(1):77–83. doi: 10.1007/s10858-015-9969-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45(Pt 2):342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Konarev PV, et al. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36(5):1277–1282. [Google Scholar]
- 65.Svergun DI, Cr IU. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25(4):495–503. [Google Scholar]
- 66.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76(6):2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volkov VV, Svergun DI, Cr IU. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36(3):860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozin MB, Svergun DI, Cr IU. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34(1):33–41. [Google Scholar]
- 69.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.