Significance
In many human B-cell cancers, a complex signaling cascade called NF-κB is abnormally activated by genetic mutations. The uncontrolled activity of NF-κB because of genetic mutations promotes the formation of B-cell tumors. The NF-κB cascade is comprised of two distinct pathways. We here define the role of one of these routes, called the alternative NF-κB pathway, in the normal cells from which these B-cell tumors are derived, namely germinal center (GC) B cells or plasma cells (PCs). We found that the inactivation of the alternative NF-κB pathway led to the loss of GC B cells and impaired PC development. Understanding the role of this pathway in normal cells may provide important insights into how aberrant activation promotes B-cell tumors.
Keywords: germinal center B cell, plasma cell, NF-κB transcription factors
Abstract
The NF-κB signaling cascade relays external signals essential for B-cell growth and survival. This cascade is frequently hijacked by cancers that arise from the malignant transformation of germinal center (GC) B cells, underscoring the importance of deciphering the function of NF-κB in these cells. The NF-κB signaling cascade is comprised of two branches, the canonical and alternative NF-κB pathways, mediated by distinct transcription factors. The expression and function of the transcription factors of the alternative pathway, RELB and NF-κB2, in late B-cell development is incompletely understood. Using conditional deletion of relb and nfkb2 in GC B cells, we here report that ablation of both RELB and NF-κB2, but not of the single transcription factors, resulted in the collapse of established GCs. RELB/NF-κB2 deficiency in GC B cells was associated with impaired cell-cycle entry and reduced expression of the cell-surface receptor inducible T-cell costimulator ligand that promotes optimal interactions between B and T cells. Analysis of human tonsillar tissue revealed that plasma cells and their precursors in the GC expressed high levels of NF-κB2 relative to surrounding lymphocytes. Accordingly, deletion of nfkb2 in murine GC B cells resulted in a dramatic reduction of antigen-specific antibody-secreting cells, whereas deletion of relb had no effect. These results demonstrate that the transcription factors of the alternative NF-κB pathway control distinct stages of late B-cell development, which may have implications for B-cell malignancies that aberrantly activate this pathway.
During T-cell–dependent immune responses, B cells diversify their antigen receptors by somatic hypermutation (SHM) of the Ig variable region (IgV) genes (1). SHM and selection of B cells with increased antigen affinity occurs within germinal centers (GCs). The efficiency of the GC reaction is enhanced by topological and temporal segregation of proliferation and SHM within the dark zone (DZ) and antigen selection within the light zone (LZ) (2–4). Recirculation of GC B cells between these zones results in the generation of high-affinity, often isotype-switched memory B cells and plasma cells (PCs) (2–5). The GC reaction is critical for immunity; however, errors during SHM and class-switch recombination can lead to genetic aberrations that promote lymphomagenesis (6, 7). Recently, genetic mutations resulting in constitutive activation of the NF-κB signaling cascade were identified in a large fraction of GC-derived B-cell lymphomas and multiple myeloma (MM) (8–16).
Activation of NF-κB signaling results in the transcription of NF-κB target genes that regulate many cellular processes, including cell survival and proliferation (17, 18). The NF-κB signaling cascade comprises two branches, the canonical and alternative (or noncanonical) NF-κB pathways, which activate specific NF-κB transcription factor subunits that occur mainly as heterodimers. Canonical NF-κB pathway activation leads to the nuclear translocation of v-rel avian reticuloendotheliosis viral oncogene homolog c-REL, RELA, and p50, whereas alternative pathway activation causes nuclear translocation of RELB and p52. In normal cells, NF-κB activation is transient and tightly controlled. Conversely, constitutive NF-κB activation due to genetic alterations in NF-κB pathway components is pathogenic (8, 9). Mutations affecting multiple different NF-κB signaling components have been identified in several GC-derived B-cell malignancies, which can lead to the constitutive activation of the canonical and/or alternative NF-κB pathways (8–16). The selection of these mutations implies that NF-κB signaling may have an important biological role during normal GC B-cell development that is “hijacked” in tumors (7, 8).
Distinguishing the functions of the canonical and alternative NF-κB pathways by studying upstream regulators may be complicated by the possibility of pathway cross-talk. Therefore, focusing on the downstream transcription factor subunits may help to clarify the specific roles of the separate NF-κB pathways. Toward this aim, early work on human lymphoid tissue revealed that nuclear translocation of canonical NF-κB subunits within GCs occurred only within a subset of cells in the LZ (19). By ablating the canonical NF-κB transcription factors c-REL or RELA specifically in GC B cells, we recently showed that c-REL was essential for GC maintenance, whereas RELA was required for PC development (20). The expression, activation status, and function of the alternative NF-κB transcription factors RELB and p52 in GC B cells remain largely unknown. Due to the diverse functions of the alternative NF-κB pathway in a range of cell types, mice with constitutional knockout of either relb or nfkb2 (the gene encoding the p100/p52 precursor, referred to as NF-κB2, from which p52 is generated upon activation) have severe defects in lymphoid organization (21–23), thus hampering the analysis of GC B-cell development in these mice. We here determined the expression pattern of the alternative NF-κB subunits in human lymphoid tissue and investigated their roles during GC B-cell development in vivo by crossing conditional relb and/or nfkb2 alleles to mice that express Cre-recombinase in GC B cells. We found that RELB and NF-κB2 were jointly required to maintain the GC B-cell reaction, whereas the development of antigen-specific PCs was impaired upon deletion of only nfkb2 in GC B cells.
Results
Expression and Activation of Alternative NF-κB Subunits in Human GC B Cells.
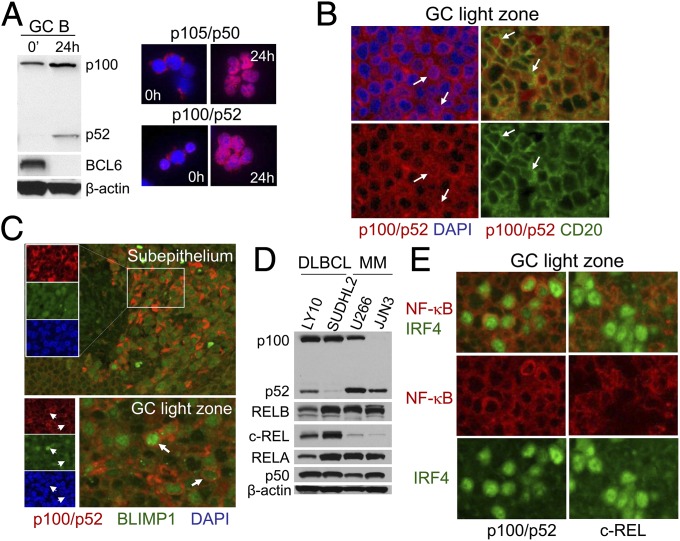
The expression and activation of the alternative NF-κB subunits in GC B cells has not been investigated. Because CD40 stimulation strongly activates both NF-κB pathways (24, 25), the CD40–CD40 ligand (CD40L) interaction occurring between LZ B cells and T-follicular helper (Tfh) cells is expected to activate alternative NF-κB signaling. Indeed, Western blot analysis of human tonsillar GC B cells cultured on CD40L-expressing fibroblasts demonstrated p100→p52 processing (Fig. 1A, Left) and thus alternative pathway activation. This was accompanied by the down-regulation of the GC master regulator BCL6, an event believed to occur during LZ selection (7), and resulted in nuclear translocation of p52 along with the canonical subunit p50 (Fig. 1A, Right). In accordance with the in vitro findings, nuclear translocation of p52 could be observed in vivo in tonsillar GCs within LZ B cells by immunofluorescence (IF) analysis (Fig. 1B). Thus, nuclear translocation of p52 was detected within a small subset of LZ B cells and therefore suggests that the alternative NF-κB pathway may have a functional role in LZ B cells.
Fig. 1.
Expression and activation of alternative NF-κB subunits in normal and transformed human GC B cells and PCs. (A) Human tonsillar GC B cells ex vivo or following 24 h of coculture on CD40L-expressing feeders were subjected to Western blot analysis for p100/p52 and BCL6 (Left) and IF analysis for p105/p50 and p100/p52 (Right, red) and DAPI (blue). (B) IF analysis of tonsil sections for p100/p52 and DAPI or CD20 in the GC LZ. (C) IF analysis of tonsil sections for p100/p52, BLIMP1, and DAPI in the subepithelium and GC LZ. (D) Western blot analysis of DLBCL and MM cell lines for p100/p52, RELB, c-REL, RELA, and p105/p50. (E) IF analysis of tonsil sections for IRF4 and NF-κB subunits (either p100/p52 or c-REL) in the GC LZ. (Magnification: A–C and E, 400×.)
Interestingly, we observed strong staining of p100/p52 in PCs localizing in the tonsillar subepithelium that were identified by staining for the major PC regulator BLIMP1 (26, 27), relative to lymphocytes at the border of the subepithelium (Fig. 1C, Upper). The same staining pattern was observed in BLIMP1+ PC precursors in the LZ of tonsillar GCs (Fig. 1C, Lower). These observations may point toward a potential role of the alternative NF-κB pathway in the development of normal PCs and their precursors in the GC. The large amount of NF-κB2 in the cytoplasm may predispose BLIMP1+ PC precursors and PCs to undergo strong signaling via the alternative NF-κB pathway upon stimulation. Alternative pathway activation was observed via strong p100→p52 processing in two MM cell lines (Fig. 1D), and to a lesser extent in a cell line corresponding to diffuse large B-cell lymphoma (DLBCL), where mutations leading to activation of the alternative pathway have been observed in a subset of cell lines and primary cases (13, 15). Of note, Western blot analysis revealed that the canonical NF-κB subunit c-REL was expressed at dramatically lower levels in the MM lines compared with the DLBCL lines (Fig. 1D). A low expression of c-REL relative to surrounding lymphocytes appears to also be a feature of normal PC precursors in the LZ (Fig. 1E, Right), as identified by strong staining for IRF4, which at high expression levels promotes PC differentiation along with BLIMP1 (27, 28). In contrast, cytoplasmic p100/p52 expression is increased in several IRF4+ cells (Fig. 1E, Left), similar to the corresponding BLIMP1 staining (Fig. 1C, Lower). Collectively, these data suggest that relative to mature B cells, PCs and their precursors in the GC are characterized by a distinct expression pattern of NF-κB subunits; high expression of NF-κB2 and low expression of c-REL.
Combined GC B-Cell–Specific Deletion of relb and nfkb2 Impairs the GC Reaction.
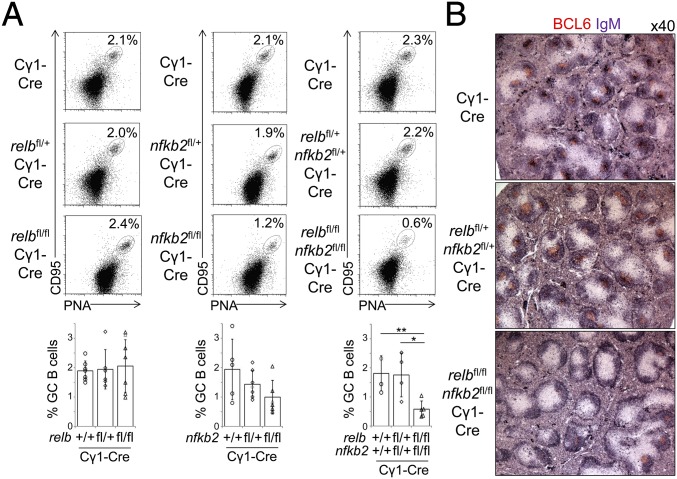
To determine the in vivo role of RELB and NF-κB2 during GC B-cell development, we crossed conditional relb and nfkb2 alleles (29) to Cγ1-Cre mice (30), either alone or in combination, to delete the genes in GC B cells. Expression of Cγ1-Cre is induced upon T-cell–dependent immunization, resulting in the Cre-mediated deletion of loxP-flanked genes in the majority of GC B cells (30). relbfl/flCγ1-Cre, nfkb2fl/flCγ1-Cre, or relbfl/flnfkb2fl/flCγ1-Cre mice and the corresponding heterozygous and Cγ1-Cre control mice were immunized with sheep red blood cells (SRBCs) to induce a robust GC response. Fourteen days following immunization, the fractions of splenic CD95hiPNAhi GC B cells in relbfl/flCγ1-Cre and nfkb2fl/flCγ1-Cre were not significantly different from those in the controls (Fig. 2A, Left and Center). In contrast, the fraction of splenic GC B cells in relbfl/flnfkb2fl/flCγ1-Cre mice was markedly reduced in comparison with relbfl/+nfkb2fl/+Cγ1-Cre and Cγ1-Cre mice 14 d postimmunization (Fig. 2A, Right). Accordingly, immunohistochemistry (IHC) revealed reduced BCL6+ GCs within B-cell follicles in relbfl/flnfkb2fl/flCγ1-Cre mice compared with controls at day 14 postimmunization (Fig. 2B). Together, these findings demonstrate that single ablation of either RELB or NF-κB2 in GC B cells had no significant impact on the GC reaction. Instead, combined ablation of RELB and NF-κB2 in GC B cells strongly impaired the GC reaction, demonstrating that both subunits of the alternative NF-κB pathway are required for GC maintenance.
Fig. 2.
Combined GC B-cell–specific deletion of relb and nfkb2 impairs the GC reaction. (A) relbfl/flCγ1-Cre, nfkb2fl/flCγ1-Cre, or relbfl/flnfkb2fl/flCγ1-Cre mice and the corresponding heterozygous and Cγ1-Cre control mice were analyzed by flow cytometry 14 d following immunization with SRBCs for CD95hiPNAhi splenic GC B cells. Summary of the frequencies of GC B cells (Lower). Each symbol represents a mouse. Statistical significance was determined by Student’s t test (*P < 0.05; **P < 0.01). Data are shown as mean ± SD. (B) Spleen sections from the indicated genotypes were analyzed for the expression of BCL6 and IgM via IHC.
To define the temporal kinetics of the impaired GC reaction observed in relbfl/flnfkb2fl/flCγ1-Cre mice, we determined the fractions of splenic GC B cells at various time points following immunization with SRBCs. Seven days postimmunization, the fraction of GC B cells in relbfl/flnfkb2fl/flCγ1-Cre mice was comparable to that observed in the controls (SI Appendix, Fig. S1A). Because the conditional relb and nfkb2 alleles were constructed such that Cre-mediated recombination of loxP sites is accompanied by expression of an enhanced-GFP (eGFP) (29), it was possible to trace the gene-deleted GC B cells by flow cytometry for eGFP expression. Analysis for eGFP expression in GC B cells from relbfl/flnfkb2fl/flCγ1-Cre and relbfl/+nfkb2fl/+Cγ1-Cre mice revealed single peaks of expression (SI Appendix, Fig. S1A, Bottom Right), indicating that the vast majority of GC B cells have deleted the relb and nfkb2 alleles at day 7. GC B-cell development therefore occurred normally in relbfl/flnfkb2fl/flCγ1-Cre mice up to day 7 of the GC reaction, after which GC B cells were progressively lost.
To investigate the possibility of a selective loss of a particular GC B-cell subpopulation, we determined the fractions of CXCR4hiCD86lo DZ and CXCR4loCD86hi LZ B-cell fractions (31) over time (SI Appendix, Fig. S1B). Statistically significant differences in the DZ and LZ B-cell fractions between relbfl/flnfkb2fl/flCγ1-Cre and Cγ1-Cre control mice were observed; however, these differences were minor and do not point toward a preferential loss of a specific GC B-cell subpopulation. Thus, these data suggest that RELB and NF-κB2 are required for the maintenance of both DZ and LZ subpopulations past day 7 of the GC reaction.
Identification of Genes Controlled by the Alternative NF-κB Subunits RELB and NF-κB2 in GC B Cells.
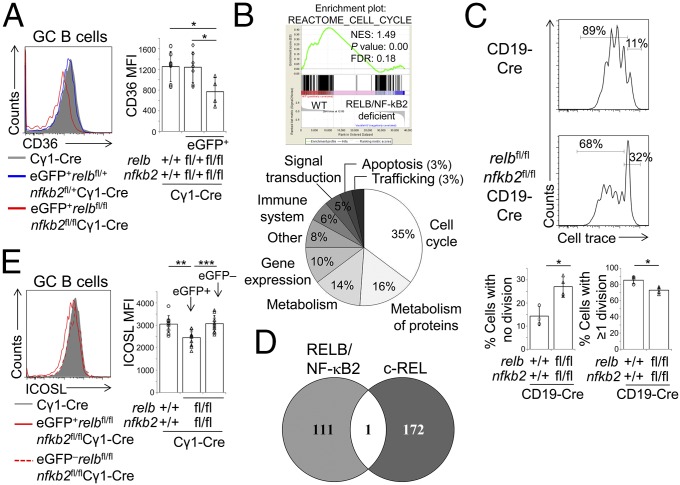
To identify the biological programs controlled by RELB and NF-κB2 that are required for GC maintenance, we isolated GC B cells from relbfl/flnfkb2fl/flCγ1-Cre and Cγ1-Cre control mice 7 d postimmunization and conducted an RNA-sequencing (RNA-seq) analysis. We reasoned that gene expression changes that ultimately contribute to the loss of relb/nfkb2-deleted GC B cells at later time points would already be detectable in these cells at day 7. Splenic eGFP+ GC B cells were flow cytometrically sorted from two relbfl/flnfkb2fl/flCγ1-Cre mice, and GC B cells were isolated from three Cγ1-Cre mice 7 d postimmunization with SRBCs and subjected to RNA-seq analysis. Reduced transcript counts of the relb and nfkb2 genes were identified in GC B cells from relbfl/flnfkb2fl/flCγ1-Cre compared with Cγ1-Cre mice (SI Appendix, Fig. S2A), and the relbfl/flnfkb2fl/flCγ1-Cre and Cγ1-Cre samples clustered into two separate groups in an unsupervised analysis (SI Appendix, Fig. S2B). Because a monoclonal antibody was available for the surface molecule CD36, a putative fatty acid translocase (32), we could confirm reduced protein expression of CD36 on eGFP+ GC B cells from relbfl/flnfkb2fl/flCγ1-Cre mice in comparison with GC B cells from Cγ1-Cre mice and eGFP+ GC B cells from relbfl/+nfkb2fl/+Cγ1-Cre mice (Fig. 3A). Together, these observations validate the robustness of the RNA-seq dataset.
Fig. 3.
Identification of genes controlled by the alternative NF-κB subunits RELB and NF-κB2 in GC B cells. (A) relbfl/flnfkb2fl/flCγ1-Cre mice and the corresponding heterozygous and Cγ1-Cre control mice were analyzed via flow cytometry 8–12 d following immunization with SRBCs for the expression of CD36 on CD95hiCD38lo GC B cells. Summary of the corresponding median fluorescence intensities (MFI) (Right). (B) GSEA was used to identify gene signatures that were enriched in GC B cells from Cγ1-Cre vs. relbfl/flnfkb2fl/flCγ1-Cre mice. (Upper) Representative example of a cell-cycle regulation signature. (Lower) Gene sets showing significant enrichment were grouped into functional categories. For the identity of the gene sets, see SI Appendix, Dataset S3. (C) CellTrace Violet dilution in CD40 + IL-4–stimulated B cells of the indicated genotypes (day 3). (Upper) Representative examples. Gates on the Right identify nondividing cells and gates on the Left, cells that have undergone divisions. (Lower) Summary of the results. (D) Venn diagram showing the overlap of genes with reduced expression in RELB/NF-κB2 or c-REL–deficient GC B cells vs. controls. (E) relbfl/flnfkb2fl/flCγ1-Cre mice and Cγ1-Cre control mice were analyzed via flow cytometry 10 d following immunization with SRBCs for the expression of ICOSL on CD95hiCD38lo GC B cells. Summary of the corresponding MFI in GC B cells from Cγ1-Cre mice and eGFP+ and eGFP– GC B cells from relbfl/flnfkb2fl/flCγ1-Cre mice (Right). (A, C, and E) Each symbol represents a mouse. Statistical significance was determined by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). Data are shown as mean ± SD.
Differentially expressed sequence analysis (DESeq) of RELB/NF-κB2–proficient vs. RELB/NF-κB2–deficient GC B cells identified 59 transcripts with greater than 2.5-fold reduced expression and 84 transcripts with greater than 2.5-fold increased expression in the relb/nfkb2-deleted B cells at a significance threshold of P < 0.01 (for the identity of the corresponding genes, fold-change and P values, see SI Appendix, Datasets S1 and S2). Transcripts with reduced expression could be assigned to functional categories, with genes involved in the immune response and metabolism representing the largest categories (SI Appendix, Fig. S2C; for the identity of the genes, see SI Appendix, Dataset S1). The metabolism category was largely comprised of two groups of genes with presumptive roles in either protein or lipid metabolism.
We next used gene set enrichment analysis (GSEA) (33) to further investigate the gene expression changes among the genotypes. The largest group of signatures enriched in the control cells vs. RELB/NF-κB2–deficient cells was associated with cell-cycle regulation (Fig. 3B and SI Appendix, Dataset S3), suggesting that RELB/NF-κB2 may control genes involved in proliferation. To test this possibility, we cultured RELB/NF-κB2–deficient B cells purified from relbfl/fl nfbk2fl/flCD19-Cre mice and control B cells isolated from CD19-Cre mice with CD40 and IL-4, a combination that provides a strong proliferative signal and activates the alternative pathway via CD40. By analyzing the proliferation profile of these cells at day 3 of stimulation, we found that a significantly smaller fraction of eGFP+RELB/NF-κB2–deficient B cells entered the cell cycle in comparison with control B cells (Fig. 3C). Of note, the relb/nfkb2-deleted B cells that were able to enter the cell cycle appeared to proliferate slightly more than controls. Together, the data suggest that RELB/NF-κB2–deficient B cells have a reduced ability to enter the cell cycle, which may contribute to the loss of RELB/NF-κB2–deficient GC B cells over time that we observed in vivo.
To investigate whether RELB/NF-κB2–deficient GC B cells proliferate less than WT GC B cells in vivo, we assessed BrdU incorporation at day 10. At day 10, a reduction in relb/nfkb2-deleted vs. WT GC B cells is already evident (SI Appendix, Fig. S3A). We chose this time point for analysis because it is in between day 7 (when relb/nfkb2-deleted GC B cells are present at normal frequencies) (SI Appendix, Fig. S1A) and day 14 (when relb/nfkb2-deleted GC B cells are greatly reduced) (Fig. 2). We observed a trend toward decreased BrdU incorporation in relb/nfkb2-deleted vs. WT GC B cells identified as CD19+GL7hi cells (SI Appendix, Fig. S3C) (for gating strategy, see SI Appendix, Fig. S3B). We believe these differences are minor because the percentage of GFP+ cells is variable between different mice (SI Appendix, Fig. S3D), which could reflect counterselection against relb/nfkb2-deleted GC B cells. Because the BrdU protocol involves fixation, we were unable to specifically measure BrdU incorporation in GFP+ GC B cells, which could dilute the actual difference in the fraction of cells that have incorporated BrdU.
The second largest group of signatures identified in the GSEA analysis was associated with the metabolism of proteins (Fig. 3B). In addition, when we compared our RNA-seq dataset to a library of normal and pathological lymphoid gene expression signatures (34), five signatures were found to be enriched in RELB/NF-κB2–proficient vs. -deficient GC B cells (SI Appendix, Fig. S4), including a ribosomal protein signature and two X box-binding protein 1 (XBP1)-associated gene expression signatures. XBP1 is required for the unfolded protein response and is essential for the development of PCs capable of secreting large amounts of antibodies (26, 27). This suggests that in GC B cells, the alternative NF-κB subunits may be required to set up a program that allows for the efficient production of proteins and facilitates antibody secretion, presumably in GC B cells destined to become plasmablasts (see below).
Finally, we have previously shown that deletion of the gene encoding the canonical NF-κB subunit c-REL (rel) in GC B cells leads to the involution of GCs (20) similar to what we observed upon relb/nfkb2 deletion, suggesting that c-REL and the alternative NF-κB subunits exert nonredundant functions during the GC reaction. In support of this notion, genes with reduced expression in relb/nfkb2 or rel-deleted GC B cells vs. controls were found to be largely mutually exclusive (Fig. 3D), indicating that the different canonical and alternative NF-κB subunits control distinct transcriptional programs within the same GC context.
RELB/NF-κB2-Deficient GC B Cells Have Reduced Cell-Surface Expression of Inducible T-Cell Costimulator Ligand.
The interaction between inducible T-cell costimulator (ICOS), expressed on Tfh cells, and ICOS ligand (ICOSL), expressed on GC B cells, promotes the selection of high-affinity B cells (35). The expression of ICOSL is regulated by the alternative NF-κB subunits in response to B-cell activating factor receptor (BAFF-R) stimulation (36) and also CD40 stimulation (29) in murine B cells. To determine the extent to which the deletion of relb and nfkb2 in GC B cells affects ICOSL expression on GC B cells in vivo, we stained splenic mononuclear cells from relbfl/flnfkb2fl/flCγ1-Cre and Cγ1-Cre control mice for ICOSL and GC markers 10 d following SRBC immunization. eGFP+RELB/NF-κB2–deficient GC B cells from relbfl/flnfkb2fl/flCγ1-Cre mice showed a slight but significant reduction in the surface expression of ICOSL compared with WT GC B cells and eGFP–RELB/NF-κB2–proficient GC B cells from the same mice (Fig. 3E and SI Appendix, Fig. S2D). Reduced cell-surface expression of ICOSL on RELB/NF-κB2–deficient GC B cells may impair optimal GC B cell–Tfh cell interactions within the GC, which may contribute to their gradual disappearance after day 7 of the GC reaction.
Deletion of nfkb2 in GC B Cells Impairs the Development of Antigen-Specific PCs.
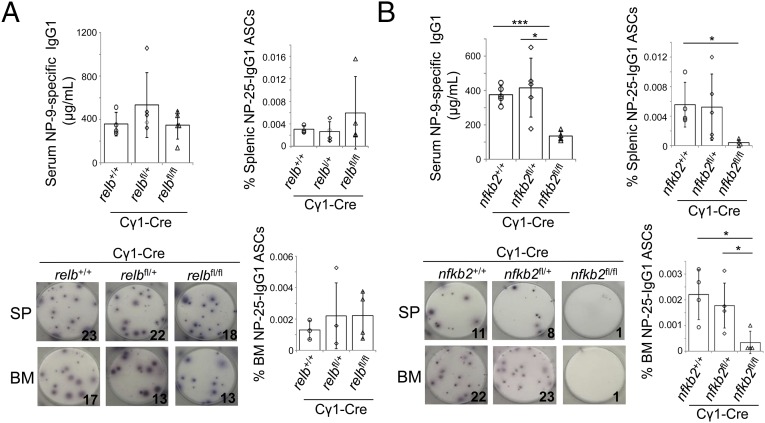
The combined deletion of relb and nfkb2 resulted in the involution of established GCs and, as expected, PCs in relbfl/flnfkb2fl/flCγ1-Cre mice were also reduced compared with relbfl/+nfkb2fl/+Cγ1-Cre and Cγ1-Cre mice (SI Appendix, Fig. S5). The deletion of relb or nfkb2 alone in GC B cells, however, did not affect GC B-cell maintenance upon SRBC immunization (Fig. 2A) or immunization with 4-hydroxy-3-nitrophenyl-acetyl coupled to keyhole limpet hemocyanin (NP-KLH) (SI Appendix, Fig. S6). Therefore, to determine whether RELB or NF-κB2 are required for the generation of antigen-specific PCs in the GC reaction in vivo, we immunized relbfl/flCγ1-Cre or nfkb2fl/flCγ1-Cre and the corresponding control mice with NP-KLH and performed ELISA and ELISPOT analyses. At 28 d postimmunization, we found that, whereas deletion of relb in GC B cells did not significantly reduce NP-specific IgG1 serum levels or the number of NP-specific IgG1 antibody-secreting cells (ASCs) in the spleen and bone marrow (Fig. 4A), GC-specific deletion of nfkb2 led to an ∼3-fold reduction in NP-specific IgG1 serum levels and an 8- to 10-fold reduction in ASCs compared with the control mice (Fig. 4B). This defect does not appear to be due to the loss of PCs, as we were able to detect eGFP+ and therefore nfkb2-deleted, CD138+ PCs (SI Appendix, Figs. S7 and S8). Although the basis for this observation remains to be determined, these results provide functional evidence for a biological role of NF-κB2, which is highly expressed in PCs and their precursors, in the development of PCs that cannot be complemented by other NF-κB subunits.
Fig. 4.
Deletion of nfkb2 in GC B cells impairs the development of antigen-specific PCs. (A) relbfl/flCγ1-Cre and (B) nfkb2fl/flCγ1-Cre and the corresponding heterozygous and Cγ1-Cre control mice were analyzed for NP9-specific IgG1 levels via ELISA (Upper Left) and NP25-specific IgG1 ASCs via ELISPOT (Lower) 28 d following immunization with NP-KLH. Summary of the frequencies of NP25-specific IgG1 ASCs (Right). Each symbol represents a mouse. Statistical significance was determined by Student’s t test (*P < 0.05; ***P < 0.001). Data are shown as mean ± SD.
Discussion
In agreement with previous work using bone-marrow chimeras (21, 23), we found that GC B-cell development proceeds normally in mice with GC B-cell–specific ablation of either RELB or NF-κB2 alone. In contrast, combined ablation of RELB and NF-κB2 resulted in the progressive loss of GC B cells. Therefore, RELB and NF-κB2 are jointly required for the maintenance of the GC reaction.
Among the alternative NF-κB subunits, only RELB is a transcriptional activator. It was therefore perhaps surprising to observe that ablation of RELB alone did not impair the GC reaction, revealing redundancy between RELB and NF-κB2 in GC B cells. This redundancy in the absence of either subunit may be explained by dimerization of the remaining transcription factor with subunits of the canonical NF-κB pathway (37). It is clear, however, that redundancy does not exist between the canonical and alternative NF-κB pathways, because the GC maintenance defect observed in the combined absence of RELB and NF-κB2 was not compensated for by canonical NF-κB subunits.
Evidence suggests that CD40 stimulation by Tfh cells leads to activation of both the canonical and alternative NF-κB pathways in LZ B cells. An additional signal that may activate the alternative pathway in LZ B cells is stimulation by BAFF (38). Recent work provides evidence that Tfh cells secrete BAFF locally to adjacent LZ B cells (39). Of note, whereas abolishing BAFF secretion by Tfh cells impaired the selection of high-affinity GC B cells, it had no impact on the maintenance of the GC reaction. This finding suggests that the inability of LZ B cells to transmit signals through the BAFF-R is unlikely to contribute to the loss of GCs observed upon GC B-cell–specific deletion of relb and nfkb2. Because follicular dendritic cells may contribute to BAFF production in the GC (40), the conclusive determination of the function of BAFF signaling during the GC reaction would therefore require conditional deletion of the BAFF-R in GC B cells.
GCs are believed to reach maturity at approximately day 7 of the GC reaction, the time point at which DZ/LZ polarization has been established and when selection of high-affinity GC B-cell mutants, followed by cyclic reentry, is initiated (2, 3). It is clear that continuous or periodic signals are required for the maintenance of mature GCs, as the involution of established GCs has been observed upon inhibition of CD40 signaling (41). Via specific conditional gene deletion within GC B cells, it has been shown that c-MYC, c-REL, and NF-κB–induced kinase, an upstream regulator of the alternative NF-κB pathway that can also activate the canonical pathway (42, 43), are all required for the maintenance of established GCs (20, 44–47). We here demonstrated that RELB and NF-κB2 have a similarly critical role in this process.
It is becoming increasingly clear that individual NF-κB subunits have divergent roles in GC and post-GC development. In the case of the canonical subunits, RELA is dispensable for the GC reaction but promotes PC development, whereas c-REL is required for GC maintenance (20) similar to what we demonstrated here for the alternative NF-κB subunits RELB and NF-κB2. Interestingly, however, gene expression profiling analysis revealed that the genes controlled by c-REL and RELB/NF-κB2 are largely distinct. This finding suggests that the respective transcription factors regulate complementary biological programs that are independently required for the GC reaction to persist over time. Impaired cell proliferation and reduced cell-surface expression of ICOSL on LZ B cells may contribute to the progressive loss of RELB/NF-κB2–deficient GC B cells. LZ B cells undergoing selection receive signals from Tfh cells that promote their survival and license cyclic reentry and division in the DZ. Our results suggest that LZ B cells lacking the alternative subunits may respond improperly to these signals, resulting in fewer cells reentering the cell cycle and seeding the GC over time. In addition, reduced cell-surface expression of ICOSL could lead to suboptimal interactions with Tfh cells, further depriving these cells of critical signals necessary for GC maintenance.
Several observations suggest a biological role for the alternative NF-κB pathway in PCs. Our finding of strong protein expression of the NF-κB2 subunit in tonsillar PCs is in accordance with a gene expression profile analysis that reported an up-regulation of mRNA encoding NF-κB2 and RELB in human tonsillar and bone-marrow PCs relative to other B-cell subsets (48). Moreover, murine plasmacytoma lines were characterized by the nuclear translocation of RELB/p52 (49). These observations are supported by the in vivo data reported here, demonstrating a requirement for NF-κB2 in PC development. The results of our GSEA analysis on RELB/NF-κB2–deficient GC B cells raise the intriguing possibility that the alternative NF-κB pathway may have a role in establishing a genetic program that facilitates the production of high amounts of antibodies in GC B cells destined to develop into PCs.
Materials and Methods
Mice.
The conditional relb and nfkb2 alleles, Cγ1-Cre, and CD19-cre mice have been described (29, 30, 50). Mice were housed and treated in compliance with the guidelines of Columbia University. The animal protocol was approved by Columbia University’s Institutional Animal Care and Use Committee. Mice were immunized with SRBCs or NP-KLH in complete Freund’s adjuvant as described (20).
Cell Culture.
Discarded leftovers from routine tonsillectomies performed on children at Columbia-Presbyterian Medical Center were obtained. Institutional Review Board approval was obtained for all procedures. Consent was not required because all patient identifiers were deidentified and specimens anonymized before use. Human GC B cells were isolated as described (51). Human GC B cells and the CD40L-expressing mouse feeder cell lines (52) were cultured in RPMI/10% (vol/vol) FBS. P3HR1, SUDHL2, and JJN3 lines were cultured in Iscove’s modified Dulbecco’s medium (IMDM)/10% FBS. U266 was cultured in IMDM/20% FBS and LY10 in IMDM with 15% human serum (New York Blood Center). Murine B cells were purified and cultured with CD40 and/or IL-4, as described previously (20).
Immunoblot Analysis.
Cell lines or human GC B cells were subjected to immunoblot analysis as described (20). For antibodies used, see SI Appendix, Table S1.
Flow Cytometry.
Spleen cell suspensions were stained and analyzed as described (20). For antibodies used, see SI Appendix, Table S1. The CellTrace Violet Proliferation Kit (Thermo Fisher Scientific) was used for cell trace experiments. For the analysis of BrdU incorporation in GC B cells in vivo, mice were injected with 2 mg of BrdU and killed 6 h later. Staining for BrdU was conducted using the APC–BrdU kit (Becton Dickinson).
Histology and Immunohistochemistry.
Spleen sections (4 μm) were H&E stained for morphological evaluation. IHC staining analysis was performed as described (20). For antibodies used, see SI Appendix, Table S1.
Immunofluorescence.
For single-cell staining, cells were spun onto slides using a cytocentrifuge and fixed in 10% formalin for 20 min followed by 20 min of methanol fixation. Nuclear permeabilization was achieved via incubation with 0.2% Triton/PBS. Cytospin slides were incubated with primary antibodies overnight followed by incubation with a Cy3-conjugated antibody. Slides were counterstained with DAPI (Molecular Probes). Images were acquired with an Eclipse E400 microscope (Nikon). Tissue sections were prepared and stained as described (51). For antibodies used, see SI Appendix, Table S1.
Gene Expression Analysis.
B cells were isolated from spleens of relbfl/flnfkb2fl/flCγ1-Cre and Cγ1-Cre mice as described (20). eGFP+CD95hiPNAhi GC B cells were flow-cytometrically sorted from splenic B cells of relbfl/flnfkb2fl/flCγ1-Cre mice and GC B cells were sorted from splenic B cells of Cγ1-Cre mice. Total RNA was isolated using the Nucleospin RNA XS Isolation Kit (Macherey-Nagel). New York Genome Center amplified RNA using the NuGEN Ovation RNA-Seq System V2 before RNA-seq. A total of 35–40 million 2 × 50-bp paired-end reads were sequenced per sample on an HiSeq2500 (Illumina). DESeq analysis identified differentially expressed genes. Genes identified via RNA-seq analysis with reduced expression in RELB/NF-κB2–deficient GC B cells (SI Appendix, Dataset S1) and genes identified via DNA microarray analysis with reduced expression in c-REL–deficient GC B cells (20) were compared after filtering out genes identified via the RNA-seq analysis that were not represented on the microarray. The overlap between the datasets was determined using Venny 2.1.0 available at bioinfogp.cnb.csic.es/tools/venny/index.html. GSEA (33) was used to identify signatures enriched in control vs. relb/nfkb2-deleted GC B cells. We screened the collection of signatures under the category CP:REACTOME, CP:KEGG, CP:BIOCARTA, BP:GO, MF:GO, CC:GO and signatures from a library of normal and pathological lymphoid gene expression signatures (34) to determine significant enrichment (false discovery rate < 25%, P ≤ 0.05).
ELISA and ELISPOT.
ELISA and ELISPOT analyses for NP-specific IgG1 or NP-specific IgG1 ASCs, respectively, was conducted as described previously (20).
Supplementary Material
Acknowledgments
We thank L. Pasqualucci and members of the U.K. laboratory for insights and discussion; J. Zhang, M. Cato, and R. Rickert for helpful experimental suggestions; A. Holmes for bioinformatics support; Q. Shen and H. Tang for advice on staining procedures; and Y.-J. Liu for providing the CD40L feeder cell line. This work was supported by National Cancer Institute (NCI)/NIH Grant R01-CA157660 (to U.K.); a grant from the Alexander and Margaret Stewart Trust (Washington, DC); the Herbert Irving Comprehensive Cancer Center; a Cancer Biology Training Program Fellowship (NCI/NIH 19 Grant 5T32-CA009503-26) (to N.S.D.S.); and a fellowship from the German Research Council (to N.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE77374).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602728113/-/DCSupplemental.
References
- 1.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381(6585):751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 3.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15(3):137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27(2):190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247(1):52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 6.Küppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5(4):251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 7.Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol. 2015;15(3):172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- 8.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasqualucci L, Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin Hematol. 2015;52(2):67–76. doi: 10.1053/j.seminhematol.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12(2):115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12(2):131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demchenko YN, et al. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood. 2010;115(17):3541–3552. doi: 10.1182/blood-2009-09-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bushell KR, et al. Genetic inactivation of TRAF3 in canine and human B-cell lymphoma. Blood. 2015;125(6):999–1005. doi: 10.1182/blood-2014-10-602714. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, et al. An oncogenic role for alternative NF-κB signaling in DLBCL revealed upon deregulated BCL6 expression. Cell Reports. 2015;11(5):715–726. doi: 10.1016/j.celrep.2015.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein U, Heise N. Unexpected functions of nuclear factor-κB during germinal center B-cell development: Implications for lymphomagenesis. Curr Opin Hematol. 2015;22(4):379–387. doi: 10.1097/MOH.0000000000000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaileh M, Sen R. NF-κB function in B lymphocytes. Immunol Rev. 2012;246(1):254–271. doi: 10.1111/j.1600-065X.2012.01106.x. [DOI] [PubMed] [Google Scholar]
- 18.Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol. 2010;2(5):a000182. doi: 10.1101/cshperspect.a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basso K, et al. Tracking CD40 signaling during germinal center development. Blood. 2004;104(13):4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- 20.Heise N, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-κB transcription factor subunits. J Exp Med. 2014;211(10):2103–2118. doi: 10.1084/jem.20132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franzoso G, et al. Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187(2):147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caamaño JH, et al. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J Exp Med. 1998;187(2):185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weih DS, Yilmaz ZB, Weih F. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J Immunol. 2001;167(4):1909–1919. doi: 10.4049/jimmunol.167.4.1909. [DOI] [PubMed] [Google Scholar]
- 24.Coope HJ, et al. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 2002;21(20):5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qing G, Qu Z, Xiao G. Stabilization of basally translated NF-kappaB-inducing kinase (NIK) protein functions as a molecular switch of processing of NF-kappaB2 p100. J Biol Chem. 2005;280(49):40578–40582. doi: 10.1074/jbc.M508776200. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5(3):230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 27.Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15(3):160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer AL, Emre NC, Romesser PB, Staudt LM. IRF4: Immunity. Malignancy! Therapy? Clin Cancer Res. 2009;15(9):2954–2961. doi: 10.1158/1078-0432.CCR-08-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Silva NS, Silva K, Anderson MM, Bhagat G, Klein U. Impairment of mature B Cell maintenance upon combined deletion of the alternative NF-κB transcription factors RELB and NF-κB2 in B cells. J Immunol. 2016;196(6):2591–2601. doi: 10.4049/jimmunol.1501120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casola S, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci USA. 2006;103(19):7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2(72):re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaffer AL, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 35.Liu D, et al. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 2015;517(7533):214–218. doi: 10.1038/nature13803. [DOI] [PubMed] [Google Scholar]
- 36.Hu H, et al. Noncanonical NF-kappaB regulates inducible costimulator (ICOS) ligand expression and T follicular helper cell development. Proc Natl Acad Sci USA. 2011;108(31):12827–12832. doi: 10.1073/pnas.1105774108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih VF, Tsui R, Caldwell A, Hoffmann A. A single NFκB system for both canonical and non-canonical signaling. Cell Res. 2011;21(1):86–102. doi: 10.1038/cr.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatto D, Brink R. B cell localization: Regulation by EBI2 and its oxysterol ligand. Trends Immunol. 2013;34(7):336–341. doi: 10.1016/j.it.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Goenka R, et al. Local BLyS production by T follicular cells mediates retention of high affinity B cells during affinity maturation. J Exp Med. 2014;211(1):45–56. doi: 10.1084/jem.20130505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hase H, et al. BAFF/BLyS can potentiate B-cell selection with the B-cell coreceptor complex. Blood. 2004;103(6):2257–2265. doi: 10.1182/blood-2003-08-2694. [DOI] [PubMed] [Google Scholar]
- 41.Han S, et al. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J Immunol. 1995;155(2):556–567. [PubMed] [Google Scholar]
- 42.Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity. 2004;21(4):477–489. doi: 10.1016/j.immuni.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Zarnegar B, Yamazaki S, He JQ, Cheng G. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci USA. 2008;105(9):3503–3508. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominguez-Sola D, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13(11):1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calado DP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13(11):1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hahn M, Macht A, Waisman A, Hövelmeyer N. NF-κB-inducing kinase is essential for B-cell maintenance in mice. Eur J Immunol. 2016;46(3):732–741. doi: 10.1002/eji.201546081. [DOI] [PubMed] [Google Scholar]
- 47.Brightbill HD, et al. Conditional deletion of NF-κB-inducing kinase (NIK) in adult mice disrupts mature B cell survival and activation. J Immunol. 2015;195(3):953–964. doi: 10.4049/jimmunol.1401514. [DOI] [PubMed] [Google Scholar]
- 48.Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J., Jr Gene expression profiling of plasma cells and plasmablasts: Toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102(2):592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- 49.Liou HC, Sha WC, Scott ML, Baltimore D. Sequential induction of NF-kappa B/Rel family proteins during B-cell terminal differentiation. Mol Cell Biol. 1994;14(8):5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito M, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12(3):280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Liu YJ, et al. Mechanism of antigen-driven selection in germinal centres. Nature. 1989;342(6252):929–931. doi: 10.1038/342929a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.