Significance
The role of wild aquatic birds in perpetuating highly pathogenic avian influenza viruses (HPAIVs) is unresolved. We examined whether the subtype H5 clade 2.3.4.4 HPAIV that devastated the US poultry industry in 2015 is perpetuated in wild aquatic birds. Virologic surveillance in 2014/15 and over the previous 43 y failed to detect HPAIVs in wild aquatic birds before or after the poultry outbreak, supporting the premise that there are unresolved mechanisms preventing wild aquatic birds from perpetuating HPAIVs. The significance of these findings is that timely and efficient strategies used to successfully prevent and eradicate HPAIVs infecting poultry, without the use of vaccines, appear to complement natural biological mechanisms in disrupting the perpetuation and possible spread of HPAIVs by wild aquatic birds.
Keywords: highly pathogenic avian influenza, restricted perpetuation, migratory waterfowl, wild bird, surveillance
Abstract
One of the major unresolved questions in influenza A virus (IAV) ecology is exemplified by the apparent disappearance of highly pathogenic (HP) H5N1, H5N2, and H5N8 (H5Nx) viruses containing the Eurasian hemagglutinin 2.3.4.4 clade from wild bird populations in North America. The introduction of Eurasian lineage HP H5 clade 2.3.4.4 H5N8 IAV and subsequent reassortment with low-pathogenic H?N2 and H?N1 North American wild bird-origin IAVs in late 2014 resulted in widespread HP H5Nx IAV infections and outbreaks in poultry and wild birds across two-thirds of North America starting in November 2014 and continuing through June 2015. Although the stamping out strategies adopted by the poultry industry and animal health authorities in Canada and the United States—which included culling, quarantining, increased biosecurity, and abstention from vaccine use—were successful in eradicating the HP H5Nx viruses from poultry, these activities do not explain the apparent disappearance of these viruses from migratory waterfowl. Here we examine current and historical aquatic bird IAV surveillance and outbreaks of HP H5Nx in poultry in the United States and Canada, providing additional evidence of unresolved mechanisms that restrict the emergence and perpetuation of HP avian influenza viruses in these natural reservoirs.
The available evidence suggests that a Eurasian lineage highly pathogenic (HP) H5N8 influenza A virus (IAV) was introduced into the Americas by migratory waterfowl and subsequently reassorted with North American wild bird IAV lineages (1). The first detection of this Eurasian lineage HP H5Nx in North America occurred in November 2014, when an HP H5N2 virus was recovered from domestic turkeys in the Fraser Valley of Canada. This was followed by the isolation of HP H5N2 virus from migratory waterfowl near the British Columbia/Washington State border and HP H5N8 virus from a captive gyrfalcon in Washington State in mid-December 2014 (2, 3). All of these HP H5Nx viruses contained the HP clade 2.3.4.4 H5 hemagglutinin (HA) gene from the Eurasian H5N8 virus, and interestingly, the entire genomic constellation of the HP H5N8 virus detected in North America was of Eurasian lineage. In contrast, the HP H5N2 viruses contained the HA 2.3.4.4 genomic segment plus four gene segments encoding internal viral proteins—polymerase basic 2 (PB2), polymerase acidic (PA), matrix (M), and nonstructural (NS) from Eurasia—and three gene segments encoding viral proteins— polymerase basic 1 (PB1), neuraminidase (NA), and nucleoprotein (NP), of American wild aquatic bird origin (4). Another reassortant virus, HP H5N1, isolated from a hunter-shot green-winged teal on December 29, 2014, contained the HA 2.3.4.4 genomic segment plus three gene segments from the Eurasian HP H5N8 virus (PB2, NP, and M) and four North American gene segments (PB1, PA, NA, and NS) (5).
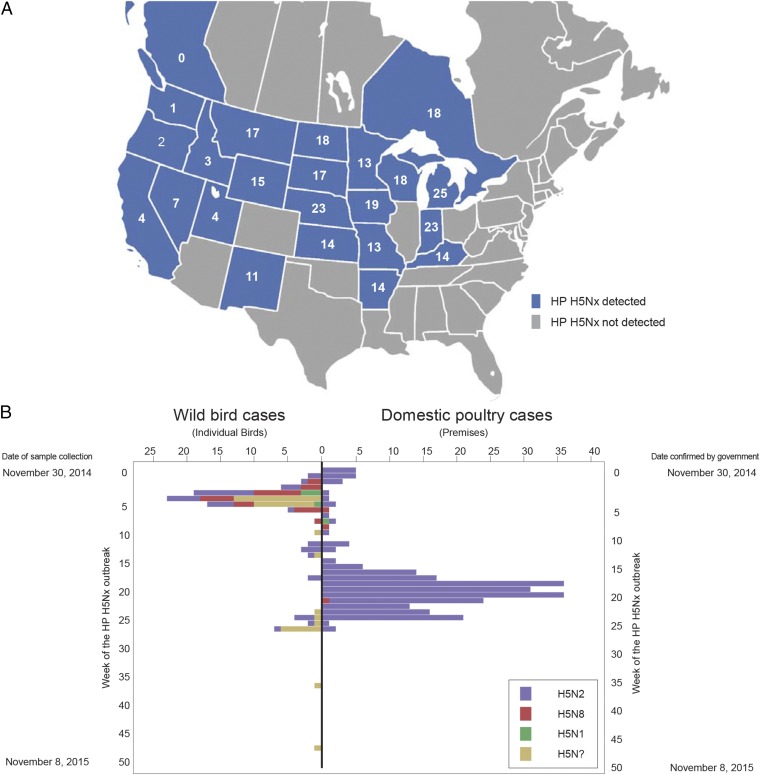
Of the originally introduced HP H5N8 IAV and the reassortants that emerged, the HP H5N2 reassortant was most frequently detected in wild birds (6, 7) and caused the vast majority of the outbreaks in domestic turkeys and chickens (8) (Fig. 1). Following the initial detection of HP H5N2 in British Columbia, IAV surveillance in wild bird populations led to the detection of HP H5Nx viruses in Washington, Oregon, Idaho, and California in December 2014 and in Utah, Nevada, and New Mexico by February 2015 (6, 7) (Fig. 1). The HP H5N2 virus was confirmed in samples collected from poultry on March 4, 2015, in Minnesota, and HP H5N2 IAV was recovered from wild bird samples collected on March 5, 2015, in the Mississippi flyway. It was suggested that the HP H5Nx viruses were carried north from the overwintering grounds during early spring migration of waterfowl (9). Overall, 102 H5Nx viruses were detected in wild birds in 15 states and one Canadian province (6, 7, 10), and 248 outbreaks occurred in commercial and backyard turkey and chicken farms (8, 11, 12), resulting in a cost of nearly $5 billion (13) to consumers, government agencies, and the poultry industry in the United States.
Fig. 1.
Detections of highly pathogenic H5Nx IAVs in wild birds and poultry in North America (6–8, 10–12). (A) The week of the outbreak, when the initial detection of HP H5Nx IAVs occurred in each state or province. The numbers indicate the week in which the H5Nx viruses spread to wild birds and poultry. Following the initial appearance of HP H5N2 in the upper midwest of the United States in week 13 (February 2015), the H5N2 virus quickly spread to numerous poultry premises within the region. (B) Cases of HP H5Nx IAV shown by week of detection, with individual wild birds displayed on the left and poultry cases on the right. Subtypes are represented by the color code. HP H5Nx IAVs were detected in migratory waterfowl as they moved south in the Pacific flyway of North America during the initial weeks of the outbreak, but the number and frequency of HP H5Nx detections quickly dwindled, even in the face of increased surveillance. The H5Nx reports from wild birds on weeks 37 and 48 were based only on RT-PCR detection; virus isolation and sequence analysis were unsuccessful, so the pathotype could not be determined.
Results
Wild bird testing was done in 2014 and 2015, before (8,962 samples), during (7,230 samples), and after (6,700 samples) the period December 2014 through June 2015, when HP H5Nx viruses were detected in poultry (Fig. 1 and Table 1). Surveillance following the HP H5Nx poultry outbreak in the upper midwestern United States was done before the southern migration during duck banding in Alberta and Minnesota in September 2015 and during the September 2015 to March 2016 hunting season along the Mississippi flyway. The surveillance sites, number of samples, and IAVs isolated are shown in Table 1 and Fig. 2. Multiple HA-NA combinations of low-pathogenic (LP) IAVs were recovered in the 2014–2015 surveillance, representing the HA subtypes H1–H14 and H16 as well as each of the nine NA subtypes. No highly pathogenic IAV isolates were detected (Table 2).
Table 1.
Avian influenza virus isolation from active surveillance in migratory aquatic birds in North America, 2014–2015
| Preoutbreak, January 1, 2014–November 30, 2014 | Outbreak, December 1, 2014–June 30, 2015 | Postoutbreak, July 1, 2015–December 31, 2015 | |||||
| Flyway | Species | Samples collected | IAV isolates | Samples collected | IAV isolates | Samples collected | IAV isolates |
| Atlantic | Aix sponsa (Wood duck) | 22 | 48 | 1 | 28 | 1 | |
| Anas acuta (Northern pintail) | 6 | ||||||
| Anas americana (American wigeon) | 38 | ||||||
| Anas clypeata (Northern shoveler) | 3 | 1 | 4 | ||||
| Anas discors (Blue-winged teal) | 1 | 4 | |||||
| Anas platyrhynchos (Mallard) | 666 | 41 | 246 | 2 | 358 | 21 | |
| Anas platyrhynchos × Anas rubripes | 5 | 1 | |||||
| Anas rubripes (American black duck) | 10 | 35 | 2 | ||||
| Anas strepera (Gadwall) | 1 | ||||||
| Arenaria interpres (Ruddy turnstone) | 325 | 13 | 681 | 107 | |||
| Aythya americana (Redhead) | 130 | 1 | |||||
| Aythya collaris (Ring-necked duck) | 2 | 18 | |||||
| Aythya marila (Greater scaup) | 3 | ||||||
| Aythya valisineria (Canvasback) | 1 | ||||||
| Chen caerulescens (Snow goose) | 65 | 55 | |||||
| Clangula hyemalis (Long-tailed duck) | 1 | 2 | |||||
| Environmental | 89 | 11 | 264 | ||||
| Gull feces | 162 | 23 | 318 | 8 | |||
| Larus argentatus (Herring gull) | 3 | ||||||
| Larus atricilla (Laughing gull) | 45 | 35 | |||||
| Melanitta deglandi (White-winged scoter) | 2 | ||||||
| Melanitta nigra (Common scoter) | 13 | 1 | 1 | ||||
| Melanitta perspicillata (Surf scoter) | 16 | 2 | 16 | ||||
| Phalacrocoracidae (Cormorant) | 30 | ||||||
| Shorebird feces | 949 | 95 | 1,599 | 120 | |||
| Mississippi | A. sponsa (Wood duck) | 234 | 36 | 116 | |||
| A. acuta (Northern pintail) | 54 | 4 | 9 | 78 | 4 | ||
| A. americana (American wigeon) | 35 | 4 | 54 | ||||
| Anas carolinensis (Green-winged teal) | 593 | 9 | 264 | 5 | 672 | 23 | |
| A. clypeata (Northern shoveler) | 149 | 7 | 207 | 6 | 139 | 5 | |
| A. discors (Blue-winged teal) | 2,574 | 140 | 1,886* | 62 | 1,929 | 129 | |
| Anas falcata (Falcated duck) | 1 | ||||||
| A. platyrhynchos (Mallard) | 1,282 | 242 | 375 | 2 | 1,873 | 158 | |
| A. platyrhynchos × A. rubripes | 1 | ||||||
| A. rubripes (American black duck) | 50 | 5 | 3 | 24 | 1 | ||
| A. strepera (Gadwall) | 167 | 62 | 1 | 264 | 1 | ||
| Anser albifrons (Greater white-fronted goose) | 2 | 2 | 3 | ||||
| Aythya affinis (Lesser scaup) | 13 | 1 | 9 | 35 | |||
| A. americana (Redhead) | 24 | 74 | 25 | ||||
| A. collaris (Ring-necked duck) | 33 | 1 | 30 | 26 | |||
| A. marila (Greater scaup) | 1 | ||||||
| A. valisineria (Canvasback) | 9 | 1 | 28 | 3 | |||
| Branta canadensis (Canada Goose) | 11 | 564* | 167 | ||||
| Bucephala albeola (Bufflehead) | 11 | 3 | 7 | ||||
| Bucephala clangula (Common goldeneye) | 4 | 1 | |||||
| C. caerulescens (Snow goose) | 441 | 1 | |||||
| Cygnus olor (Mute swan) | 1 | ||||||
| Fulica americana (American coot) | 10 | 1 | 55 | 1 | |||
| Lophodytes cucullatus (Hooded merganser) | 6 | 1 | 20 | ||||
| Mergus merganser (Common merganser) | 2 | ||||||
| Oxyura jamaicensis (Ruddy duck) | 2 | 1 | 3 | ||||
| Central | A. acuta (Northern pintail) | 14 | 1 | 12 | |||
| A. americana (American wigeon) | 2 | ||||||
| A. carolinensis (Green-winged teal) | 26 | ||||||
| A. discors (Blue-winged teal) | 4 | 9 | |||||
| A. platyrhynchos (Mallard) | 473 | 13 | 776 | 32 | |||
| A. affinis (Lesser scaup) | 16 | 3 | |||||
| A. americana (Redhead) | 6 | ||||||
| A. collaris (Ring-necked duck) | 6 | ||||||
| A. marila (Greater scaup) | 54 | 2 | |||||
| B. albeola (Bufflehead) | 2 | 1 | |||||
| C. caerulescens (Snow goose) | 162* | ||||||
| Environmental | 10 | ||||||
| O. jamaicensis (Ruddy duck) | 2 | ||||||
| Pacific | Cerorhinca monocerata (Rhinoceros auklet) | 42 | |||||
| Charadriiformes (unknown) | 2 | ||||||
| Fratercula cirrhata (Tufted puffin) | 3 | ||||||
| Rissa tridactyla (Black-legged kittiwake) | 168 | ||||||
| Uria aalge (Common murre) | 33 | ||||||
| Total samples collected | 8,962 | 7,230 | 6,700 | ||||
| Total IAV isolates | 614 | 315 | 376 | ||||
During the peak of the outbreak (April 1, 2015–June 30, 2015), swabs and species sampled were limited in the Mississippi and Central flyways. Only 484 of the 1,886 blue-winged teal were sampled between April 1 and June 30, 2015, and the majority of the remaining samples came from geese.
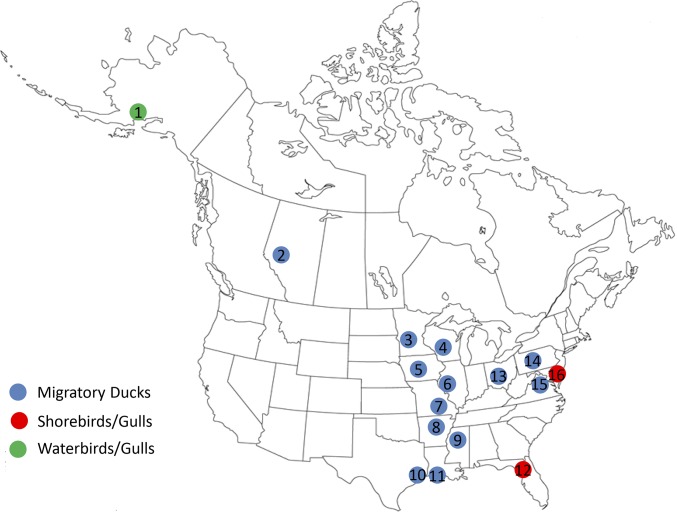
Fig. 2.
Surveillance sites for avian influenza viruses in wild aquatic birds in North America, 2014–2015. The surveillance focused primarily on the Mississippi flyway (3, Minnesota; 4, Wisconsin; 5, lowa; 6, lllinois; 7, Missouri; 8, Arkansas; 9, Mississippi; 10, Texas; 11, Louisiana; and 13, Ohio), but also included 1, Alaska; 2, Alberta, Canada; 16, Delaware Bay, New Jersey; 14, Pennsylvania; 15, Maryland; and 12, Florida. Samples collected were tracheal and/or cloacal swabs of trapped or hunter-harvested birds or fecal (environmental) swabs. The species of migratory ducks tested are specified in Materials and Methods.
Table 2.
Avian influenza virus HA-NA subtype combinations isolated from wild aquatic birds in North America, 2014–2015
| HA-NA | NA 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| HA 1 | H1N1 | H1N2 | H1N3 | H1N5 | H1N6 | H1N8 | H1N9 | ||
| 2 | H2N2 | H2N3 | H2N4 | H2N5 | H2N6 | H2N8 | H2N9 | ||
| 3 | H3N1 | H3N2 | H3N3 | H3N4 | H3N5 | H3N6 | H3N8 | H3N9 | |
| 4 | H4N1 | H4N2 | H4N3 | H4N4 | H4N5 | H4N6 | H4N8 | H4N9 | |
| LP 5* | H5N2 | H5N9 | |||||||
| 6 | H6N1 | H6N2 | H6N4 | H6N5 | H6N6 | H6N8 | H6N9 | ||
| LP 7* | H7N1 | H7N3 | H7N4 | H7N5 | H7N7 | H7N9 | |||
| 8 | H8N4 | ||||||||
| 9 | H9N1 | ||||||||
| 10 | H10N2 | H10N3 | H10N7 | H10N8 | H10N9 | ||||
| 11 | H11N1 | H11N2 | H11N3 | H11N6 | H11N8 | H11N9 | |||
| 12 | H12N1 | H12N4 | H12N5 | ||||||
| 13 | H13N3 | H13N6 | |||||||
| 14 | H14N7 | ||||||||
| 16 | H16N3 | H16N6 |
Viruses comprising HA subtypes H1–H14 and H16 and NA subtypes N1–N9 were identified. (Locations of surveillance sites and species sampled are described in Materials and Methods.) All subtype H5 viruses were determined to be low pathogenic using viral nucleotide sequencing performed by the National Veterinary Services Laboratories, US Department of Agriculture.
Low pathogenic.
Through increased biosecurity, quarantine, and culling, these HP H5Nx viruses were successfully eradicated from poultry in the United States and Canada and have not been detected in poultry since June 16, 2015; vaccines were not used. However, the documented widespread HP H5Nx infections in wild birds using the Pacific flyway in late 2014 and early 2015 and their sporadic detections in the Central and Mississippi flyways from late winter through early summer were the basis for concern that the HP H5Nx viruses would reemerge in poultry as a result of contact with infected wild birds returning south during the fall migration of 2015. Interestingly, this did not occur, and despite increased surveillance efforts, HP H5Nx viruses were not detected in wild bird populations in North America. The biological basis for this apparent disappearance of HP H5Nx viruses from North American wild birds is not understood, but the implication is that there are unresolved mechanisms that restrict the perpetuation of HPAI in these natural reservoirs.
LP H5 IAVs are routinely detected in wild waterfowl, but other than this recent event, no HP H5 or H7 IAVs have been detected in any wild birds in North America over the past 43 y. H5 IAV does not represent a predominant subtype in either historical (14) or recent surveillance (Table 2); however, when LP H5 viruses are detected, they most commonly occur paired with N2 (H5N2), and this combination appears to be consistent in wild birds of both North America and Eurasia (15). Thus, it might not be surprising that the HP Eurasian H5/North American N2 reassortant not only occurred early in this outbreak, but also was the most detected HA-NA combination isolated from both wild birds and poultry (Fig. 1).
Discussion
The premise that there are unresolved mechanisms in wild birds that restrict the emergence and perpetuation of HP IAVs in the natural IAV waterfowl reservoirs was developed based on the following criteria. First, we examined IAV surveillance data in wild aquatic birds in 2014 and 2015. Although all of the subtypes of IAV except H15, including H1–H14 and H16 and N1–N9, were detected, no HP IAVs were found (Table 2). Admittedly, we failed to detect HP IAVs in our surveillance of wild aquatic birds during the spring of 2015 (the height of the poultry outbreak), whereas other groups sporadically detected HP H5Nx in wild birds; however, this might be due to our limited number of samples collected in the Mississippi and Central flyways during that period (Table 1). Although we cannot conclude that the HP H5Nx viruses were eradicated from wild aquatic birds based on our 22,892 samples, we have 95% confidence (assuming a 90% test sensitivity) that we would have detected H5Nx at a prevalence <0.05% in each of the preoutbreak, outbreak, and postoutbreak periods.
Second, we examined our historical records of surveillance in wild aquatic birds in North America over 43 y, when we collectively tested more than 100,000 North American wild birds (14, 16–18). In these 43 y of wild aquatic bird surveillance, we isolated all of the avian-origin IAV subtypes (H1–H14, H16, and N1–N9) that we found in 2014–2015, but no HP IAVs. Third, and perhaps most strikingly, no HP H5Nx viruses have been detected in poultry or captive wild birds since June 11, 2015. Each of these criteria supports the premise stated above.
The epidemiologic patterns of LP IAVs in wild birds are consistent with the observed potential failure of HP H5Nx viruses to become established in the North American wild bird reservoir. Exchanges of IAV or genomic segments from Eurasia to North America are rare, and outside of Alaska (19, 20) there are no other documented cases of detection of a fully Eurasian lineage IAV in North American waterfowl. There is only one well-documented case of a successful genomic segment introduction, and this involved a Eurasian H6 IAV whose H6 gene replaced the North American H6 gene over a 20- to 30-y period (21, 22). Furthermore, recent evidence indicates that even the entire genomic constellations of North American lineage viruses rarely remain as a whole within the North American wild bird populations for any significant period or migratory distance (16).
Possible mechanisms explaining why HP IAVs do not establish in wild bird populations may include existing immunity to a wide range of LP HA and NA subtypes (Table 2), including H5 and H7 IAV, as well as to common antigenic determinants across all of the IAV subtypes distributed in the wild aquatic bird populations. Numerous IAV subtypes are maintained in North American waterfowl, but H3 and H4 subtypes are predominant, especially during fall migration (14, 15). Long-term surveillance of IAVs in migratory waterfowl has demonstrated higher infection rates in juvenile ducks compared with adult ducks (23) along with a cyclic dominance of subtypes, with one subtype dominating for 2 y, followed by replacement with another subtype. Both of these observations suggest a population immunity effect. In addition, recent field studies have demonstrated that reduced viral shedding can result from previous exposures to viruses representing heterospecific HAs (24). In addition, ducks are inherently more resistant to disease from HP IAV infections than gallinaceous poultry, because they have retained the IFN-inducing RIG-I family of genes lost from the jungle fowl that are the precursors of domestic chickens (25). Furthermore, population structure, host species diversity, and route of transmission likely make it difficult for HP IAVs to persist in wild aquatic birds (26). Additional mechanisms for “clearing out” HP H5 and H7 influenza viruses from migratory waterfowl populations likely exist and merit further study.
There is no doubt that wild birds, especially waterfowl, serve as the major reservoir of LP IAVs, and that they can be infected with and transport some HP IAVs, such as the Eurasian HP H5N1 viruses (27, 28); however, their potential role in the maintenance of HP IAVs in the influenza gene pool in wild aquatic birds remains controversial. The recent North American experience with HP H5Nx IAVs suggests that these viruses are not well fitted to persist in wild bird populations and likely will disappear without an endemic poultry source. It is likely that various poultry production systems and/or practices facilitate the perpetuation of HP IAVs, which can then spill over into wild birds and other host species (29). This observation is consistent with the apparent disappearance in Western Europe of HP H5N1 following detection in 2005, 2006 (30), and 2007 (31) and of H5N8 in 2014 (32). Although not completely resolved, it is very possible that these HP H5 clade 2.3.4.4 viruses were eradicated in North America through the use of quarantine and stamping out policies for poultry by US and Canadian animal health authorities without the use of vaccination. These eradication strategies, in combination with naturally occurring resistance, likely prevented HP IAVs from being incorporated into the North American wild bird IAV pool. Additional IAV surveillance and research is needed over multiple years to fully document this concept, for we know that the patterns of dominance in wild aquatic birds are cyclic (14). A more complete understanding of IAV fitness and diversity in the wild bird reservoir will have important implications for the prevention, control, and eradication of HP IAVs in poultry in other areas of the world.
Materials and Methods
Ethics Statement.
All procedures involving animals were approved by the Institutional Animal Care and Use Committees of the University of Georgia (A2013 05 0021), Ohio State University (2007A0148-R2), and St. Jude Children’s Research Hospital (546-100324).
Surveillance and Sampling of Wild Birds.
Longitudinal surveillance studies (1973–2015) for IAV and procedures for sample collection from wild aquatic birds (anseriforme and charadriiforme) in North America have been reported previously (14, 17, 33). In brief, the samples collected were tracheal and/or cloacal swabs of trapped or hunter-harvested birds or fecal (environmental) swabs. The samples were maintained on ice or in liquid nitrogen in the field and then transported to the laboratory. Samples were stored at −80 °C in the laboratory before testing. In 2014–2015, the sites for surveillance in migratory waterfowl, shorebirds, and gulls focused primarily on the Mississippi and Atlantic flyways. In addition, surveillance studies were conducted in Alaska and in Alberta, Canada. The predominant species of migratory ducks included American wigeon (Anas americana), Blue-winged teal (Anas discors), Canvasback duck (Aythya valisineria), American black duck (Anas rubripes), Gadwall duck (Anas strepera), Greater scaup (Aythya marila), Green-winged teal (Anas carolinensis), Mallard (Anas platyrhynchos), Northern pintail duck (Anas acuta), Northern shoveler (Anas clypeata), Redhead duck (Aythya americana), Ring-necked duck (Aythya collaris), and Wood duck (Aix sponsa). The species of gulls and terns included Black skimmer (Rynchops niger), Black-legged kittiwake (Rissa tridactyla), Herring gull (Larus argentatus), Laughing gull (Leucophaeus atricilla), Ring-billed gull (Larus delawarensis), and Royal tern (Thalasseus maximus). The species of shorebirds included Dunlin (Calidris alpina), Red knot (Calidris canutus), Ruddy turnstone (Arenaria interpres), Sanderling (Calidris alba), and Semipalmated sandpiper (Calidris pusilla). The species of pelagic water birds included Common murre (Uria aalge), Rhinocerous auklet (Cerorhinca monocerata), and Tufted puffin (Fratercula cirrhata).
Virus Isolation and Subtyping.
Molecular (PCR) and virologic testing (including virus isolation) was performed on tracheal, cloacal, and environmental swabs. Isolation of IAV was carried out following standard viral isolation procedures (34). In brief, samples were inoculated via the allantoic route into 9- or 10-d-old specific pathogen-free embryonating chicken eggs and then incubated at 37 °C for 72–120 h. Harvested allantoic fluid was tested for IAV using the hemagglutination test (34) and/or a FluDetect Kit (Synbiotics). All IAV subtypes were characterized antigenically (34) and/or molecularly (34) to define both the HA and NA subtypes. Pathotypes for H5 and H7 were determined based on the basic amino acid sequence at the connecting peptide of the HA gene (35).
Acknowledgments
We thank all of the wildlife and animal health scientists in Canada and the United States who provided data on influenza virus isolation and characterization. Without that material and information, this study would not have been possible. This work was funded by Contract HHSN272201400006C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, and the American Lebanese Syrian Associated Charities.
Footnotes
The authors declare no conflict of interest.
References
- 1.Lee DH, et al. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J Virol. 2015;89(12):6521–6524. doi: 10.1128/JVI.00728-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ip HS, et al. Novel Eurasian highly pathogenic avian influenza A H5 viruses in wild birds, Washington, USA, 2014. Emerg Infect Dis. 2015;21(5):886–890. doi: 10.3201/eid2105.142020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jhung MA, Nelson DI. Centers for Disease Control and Prevention (CDC) Outbreaks of avian influenza A (H5N2), (H5N8), and (H5N1) among birds—United States, December 2014-January 2015. MMWR Morb Mortal Wkly Rep. 2015;64(4):111. [PMC free article] [PubMed] [Google Scholar]
- 4.Pasick J, et al. Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to Eurasian H5N8 in British Columbia, Canada, 2014. Sci Rep. 2015;5:9484. doi: 10.1038/srep09484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torchetti MK, et al. Novel H5 clade 2.3.4.4 reassortant (H5N1) virus from a green-winged teal in Washington, USA. Genome Announc. 2015;3(2):e00195-15. doi: 10.1128/genomeA.00195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.US Department of Agriculture 2015 December 2014–June 2015 Wild Bird Highly Pathogenic Avian Influenza Cases in the United States. Available at www.aphis.usda.gov/wildlife_damage/downloads/DEC%202014%20-%20JUNE%202015%20WILD%20BIRD%20POSITIVE%20HIGHLY%20PATHOGENIC%20AVIAN%20INFLUENZA%20CASES%20IN%20THE%20UNITED%20STATES.pdf. Accessed February 5, 2016.
- 7.US Department of Agriculture 2016 July 2015–June 2016 Wild Bird Highly Pathogenic Avian Influenza Cases in the United States. Available at https://www.aphis.usda.gov/animal_health/downloads/animal_diseases/ai/july2015-june2016wildbirdpositivehpaicasesintheus.pdf. Accessed February 5, 2016.
- 8.US Department of Agriculture 2015 Highly Pathogenic Avian Influenza-Infected Premises, 2014–2015. Available at https://www.aphis.usda.gov/animal_health/animal_dis_spec/poultry/downloads/hpai-positive-premises-2014-2015.pdf. Accessed February 5, 2016.
- 9.Zimpfer NL, Rhodes WE, Silverman ED, Zimmerman GS, Richkus KD. 2015 Trends in Duck Breeding Populations, 1955–2015. US Fish and Wildlife Service Division of Migratory Bird Management Administrative Report, July 2, 2015. Available at flyways.us/sites/default/files/uploads/trends_in_duck_breeding_populations_2015.pdf. Accessed July 7, 2015.
- 10.Canadian Food Inspection Agency 2015 Avian Influenza H5N8 Confirmed in Wild Bird Carcass in British Columbia. Available at www.inspection.gc.ca/animals/terrestrial-animals/diseases/reportable/ai/2014-2015-ai-investigation-in-bc/statement-2015-05-01/eng/1430491222672/1430491223750. Accessed February 11, 2016.
- 11.Canadian Food Inspection Agency 2015 Avian Influenza—British Columbia Infected Premises. Available at www.inspection.gc.ca/animals/terrestrial-animals/diseases/reportable/ai/2014-2015-ai-investigation-in-bc/infected-premises/eng/1418340527324/1418340584180. Accessed February 11, 2016.
- 12.Canadian Food Inspection Agency 2015 Avian Influenza—Ontario Infected Premises. Available at www.inspection.gc.ca/animals/terrestrial-animals/diseases/reportable/ai/2015-investigation-ontario/infected-premises/eng/1428620608125/1428620733600. Accessed February 11, 2016.
- 13.Congressional Research Service 2015 Update on the Highly Pathogenic Avian Influenza Outbreak of 2014–2015. Available at fas.org/sgp/crs/misc/R44114.pdf. Accessed January 22, 2016.
- 14.Krauss S, et al. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 2004;4(3):177–189. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- 15.Wilcox BR, et al. Influenza-A viruses in ducks in northwestern Minnesota: Fine-scale spatial and temporal variation in prevalence and subtype diversity. PLoS One. 2011;6(9):e24010. doi: 10.1371/journal.pone.0024010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fries AC, et al. Spread and persistence of influenza A viruses in waterfowl hosts in the North American Mississippi migratory flyway. J Virol. 2015;89(10):5371–5381. doi: 10.1128/JVI.03249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slemons RD, Hansen WR, Converse KA, Senne DA. Type A influenza virus surveillance in free-flying, nonmigratory ducks residing on the eastern shore of Maryland. Avian Dis. 2003;47(3) Suppl:1107–1110. doi: 10.1637/0005-2086-47.s3.1107. [DOI] [PubMed] [Google Scholar]
- 18.Stallknecht DE, Brown JD. Ecology of avian influenza in wild birds. In: Swayne DE, editor. Avian Influenza. Blackwell; Ames, IA: 2008. pp. 43–48. [Google Scholar]
- 19.Ramey AM, et al. Dispersal of H9N2 influenza A viruses between East Asia and North America by wild birds. Virology. 2015;482:79–83. doi: 10.1016/j.virol.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 20.Krauss S, et al. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 2007;3(11):e167. doi: 10.1371/journal.ppat.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahl J, Vijaykrishna D, Holmes EC, Smith GJ, Guan Y. Gene flow and competitive exclusion of avian influenza A virus in natural reservoir hosts. Virology. 2009;390(2):289–297. doi: 10.1016/j.virol.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.zu Dohna H, Li J, Cardona CJ, Miller J, Carpenter TE. Invasions by Eurasian avian influenza virus H6 genes and replacement of the virus’ North American clade. Emerg Infect Dis. 2009;15(7):1040–1045. doi: 10.3201/eid1507.090245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinshaw VS, Webster RG, Bean WJ, Sriram G. The ecology of influenza viruses in ducks and analysis of influenza viruses with monoclonal antibodies. Comp Immunol Microbiol Infect Dis. 1980;3(1-2):155–164. doi: 10.1016/0147-9571(80)90051-x. [DOI] [PubMed] [Google Scholar]
- 24.Latorre-Margalef N, et al. Heterosubtypic immunity to influenza A virus infections in mallards may explain existence of multiple virus subtypes. PLoS Pathog. 2013;9(6):e1003443. doi: 10.1371/journal.ppat.1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber MR, Aldridge JR, Jr, Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci USA. 2010;107(13):5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebarbenchon C, Feare CJ, Renaud F, Thomas F, Gauthier-Clerc M. Persistence of highly pathogenic avian influenza viruses in natural ecosystems. Emerg Infect Dis. 2010;16(7):1057–1062. doi: 10.3201/eid1607.090389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436(7048):191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 28.Alexander DJ. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002–2006. Avian Dis. 2007;51(1) Suppl:161–166. doi: 10.1637/7602-041306R.1. [DOI] [PubMed] [Google Scholar]
- 29.Boyce WM, Sandrock C, Kreuder-Johnson C, Kelly T, Cardona C. Avian influenza viruses in wild birds: A moving target. Comp Immunol Microbiol Infect Dis. 2009;32(4):275–286. doi: 10.1016/j.cimid.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Brown IH. Summary of avian influenza activity in Europe, Asia, and Africa, 2006–2009. Avian Dis. 2010;54(1) Suppl:187–193. doi: 10.1637/8949-053109-Reg.1. [DOI] [PubMed] [Google Scholar]
- 31.Globig A, et al. Epidemiological and ornithological aspects of outbreaks of highly pathogenic avian influenza virus H5N1 of Asian lineage in wild birds in Germany, 2006 and 2007. Transbound Emerg Dis. 2009;56(3):57–72. doi: 10.1111/j.1865-1682.2008.01061.x. [DOI] [PubMed] [Google Scholar]
- 32.Marchenko VY, et al. Influenza A(H5N8) virus isolation in Russia, 2014. Arch Virol. 2015;160(11):2857–2860. doi: 10.1007/s00705-015-2570-4. [DOI] [PubMed] [Google Scholar]
- 33.Hanson BA, et al. Is the occurrence of avian influenza virus in Charadriiformes species- and location-dependent? J Wildl Dis. 2008;44(2):351–361. doi: 10.7589/0090-3558-44.2.351. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization 2002 WHO Manual on Animal Influenza Diagnosis and Surveillance. Available at www.who.int/csr/resources/publications/influenza/en/whocdscsrncs20025rev.pdf. Accessed June 13, 2016.
- 35.Bosch FX, Garten W, Klenk HD, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: Primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113(2):725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]