Significance
UV-induced DNA lesions are an important contributor to melanomas and other skin cancers. To understand how UV damage leads to cancer-associated mutations, it is important to know how the chromosomal landscape influences initial UV damage formation and repair. We have developed a UV damage mapping procedure to precisely map UV damage throughout the genome. We used this method to map the genome-wide distribution of UV lesions in yeast, a model eukaryote. We found that UV damage is not uniformly distributed, but that damage formation is significantly modulated in a predictable way by nucleosomes and DNA-bound transcription factors. Additionally, genome-wide analysis of removal of UV lesions indicates that repair is significantly inhibited near the center of strongly positioned nucleosomes.
Keywords: DNA repair, DNA damage, nucleosome, chromatin, transcription factor
Abstract
UV-induced DNA lesions are important contributors to mutagenesis and cancer, but it is not fully understood how the chromosomal landscape influences UV lesion formation and repair. Genome-wide profiling of repair activity in UV irradiated cells has revealed significant variations in repair kinetics across the genome, not only among large chromatin domains, but also at individual transcription factor binding sites. Here we report that there is also a striking but predictable variation in initial UV damage levels across a eukaryotic genome. We used a new high-throughput sequencing method, known as CPD-seq, to precisely map UV-induced cyclobutane pyrimidine dimers (CPDs) at single-nucleotide resolution throughout the yeast genome. This analysis revealed that individual nucleosomes significantly alter CPD formation, protecting nucleosomal DNA with an inward rotational setting, even though such DNA is, on average, more intrinsically prone to form CPD lesions. CPD formation is also inhibited by DNA-bound transcription factors, in effect shielding important DNA elements from UV damage. Analysis of CPD repair revealed that initial differences in CPD damage formation often persist, even at later repair time points. Furthermore, our high-resolution data demonstrate, to our knowledge for the first time, that CPD repair is significantly less efficient at translational positions near the dyad of strongly positioned nucleosomes in the yeast genome. These findings define the global roles of nucleosomes and transcription factors in both UV damage formation and repair, and have important implications for our understanding of UV-induced mutagenesis in human cancers.
Ultraviolet (UV) light causes extensive damage to DNA by inducing the formation of cyclobutane pyrimidine dimers (CPDs) and, to a lesser extent, 6-4 pyrimidine-pyrimidone photoproducts (6-4PPs). If unrepaired, these DNA lesions block normal DNA replication and are major contributors to mutagenesis in skin cancers (1). CPDs and 6-4PPs are primarily repaired in cells by the nucleotide excision repair (NER) pathway (1). CPD lesions in actively transcribed strands (TS) of DNA are repaired rapidly by the transcription coupled-NER (TC-NER) branch of this repair pathway, which is triggered by RNA polymerase II stalling at UV damage (2, 3). In contrast, CPD lesions in the remainder of the genome are repaired by the global genome NER (GG-NER) subpathway. Differences in repair rates between transcribed and nontranscribed DNA, and between accessible and inaccessible chromatin domains, have been invoked to explain the mutational heterogeneity in many cancer genomes (4–6). To gain new insight into the mutational processes that lead to human cancer, however, a more detailed understanding is needed of the complex interplay of UV damage formation and repair across the genome.
Our understanding of how NER operates in different sequence and chromatin contexts has been aided by two recent genome-wide surveys of NER activity in UV-irradiated human fibroblasts (7, 8). In these studies, a high-throughput sequencing method, known as excision repair-sequencing (XR-seq), was used to analyze oligonucleotide fragments excised during NER of CPDs or 6-4PPs. These studies found increased repair activity associated with TC-NER of the TS of actively expressed genes and noncoding transcripts (7, 8). There was also more rapid repair in active chromatin domains (i.e., enhancers and promoters) and DNase I hypersensitivity regions, and slower repair associated with repressed and heterochromatin regions (8). Reanalysis of these data by other groups indicated that there is decreased repair activity associated with transcription factor binding sites (9) and transcription initiation sites (10), suggesting that NER may be inhibited by the binding of transcription factors and the transcription machinery to DNA. These differences in repair activity may contribute to cancer-associated mutagenesis, as genomic regions with decreased repair activity (e.g., heterochromatin domains, transcription factor binding sites, and so forth) were found to be associated with increased mutation density in human melanomas (8–10).
The implicit assumption in these and other studies is that initial UV damage levels across the genome are relatively uniform (e.g., ref. 8); however, past biochemical and cellular studies have suggested that nucleosomes and DNA-binding proteins may significantly alter the formation of UV damage (11–14). Previous attempts to measure initial CPD levels using microarray-based methods (15–17) suggested that UV damage formation is primarily influenced by the DNA sequence. For example, CPDs occur most frequently at TT dipyrimidines, followed by TC, CT, and CC. However, the low resolution of these studies precluded analysis of how chromatin or transcription factors affect CPD formation or repair throughout the genome. More recently, a high-throughput sequencing method called Excision-seq was used to map CPDs and 6-4PPs in the yeast genome at single-nucleotide resolution (18). This method did confirm the expected DNA sequence preferences for CPD and 6-4PP formation (18), but there was no reported effect of chromatin or DNA-binding proteins on UV damage formation. Moreover, Excision-seq requires very high, nonphysiological doses of UV light (∼10,000 J/m2), which could bias such analyses (17).
To investigate how the chromosomal landscape affects the initial formation and subsequent repair of UV-induced CPD lesions on a genome-wide scale, we have developed CPD-seq, a high-throughput sequencing method to map CPD lesions at single-nucleotide resolution. In this study, we used CPD-seq to map the initial formation and subsequent removal of CPD lesions in the yeast genome. We found that nucleosomes and DNA-bound transcription factors significantly modulate the formation of UV damage. Furthermore, we show that differences in initial UV damage levels persist during repair. High-resolution analysis of CPD repair revealed that NER is significantly inhibited at translational positions near the nucleosome dyad.
Results
Genome-Wide Map of CPD Lesions at Single-Nucleotide Resolution.
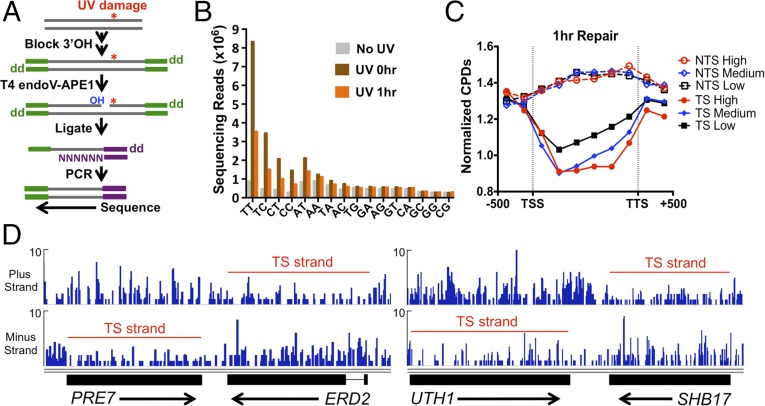
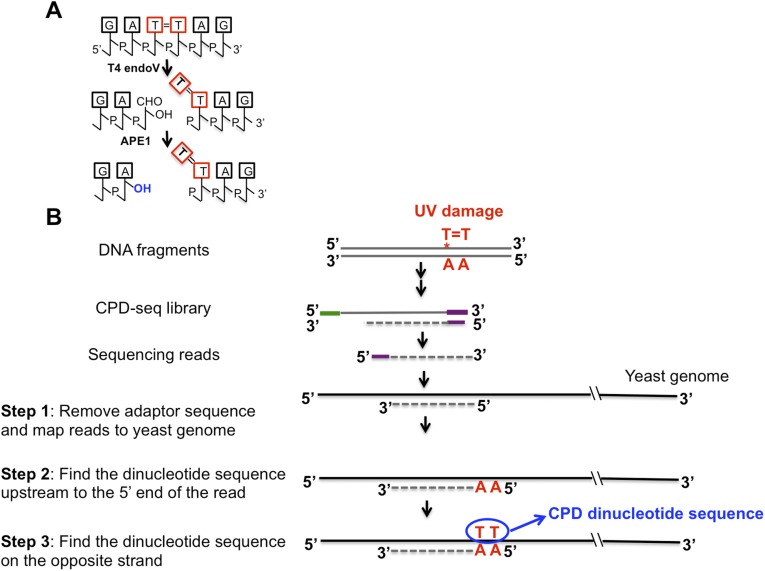
To examine how the chromosomal landscape affects UV damage formation and repair, we developed a high-throughput sequencing method, which we call “CPD-seq,” to map CPD lesions throughout the yeast genome at single-nucleotide resolution (Fig. 1A). CPD-seq is adapted from a previous method used to map ribonucleotide lesions in yeast (19). Briefly, isolated genomic DNA was sonicated, and free 3′ hydroxyls (3′OHs) at the ends of the sonicated fragments are blocked by ligation with the trP1 adapter (Fig. 1A, green), and by incubation with terminal transferase and dideoxyATP (ddATP). The DNA was cleaved at CPD lesions using the repair enzymes T4 endonuclease V (T4 endoV) and an apurinic/apyrimidinic (AP) endonuclease, APE1, to generate a ligatable free 3′OH immediately upstream of the CPD lesion (Fig. S1A). A second adapter (adapter A) (Fig. 1A, purple) was ligated to the free 3′OH generated by T4 endoV/APE1 cleavage, and the ligated DNA fragments were purified, PCR-amplified (six cycles), and sequenced. CPD-seq was used to map CPD lesions in yeast cells irradiated with 125 J/m2 UV light (primarily 254 nm) and allowed the cells to repair for 0 or 1 h. For each sequencing read, we identified the adjacent dinucleotide sequence on the opposite strand corresponding to the putative CPD site (Fig. S1B).
Fig. 1.
CPD-seq method for mapping UV damage formation and repair in the yeast genome. (A) Experimental strategy for the CPD-seq method. The trP1 adapter is colored green, and the A adapter is in purple. “OH” indicates a free 3′OH; “dd” indicates dideoxy (i.e., 3′H). (B) Analysis of dinucleotide sequences associated with CPD-seq sequencing reads. In UV-treated samples, there is an enrichment of sequencing reads at dipyrimidine sequences. (C) Significantly fewer CPDs remain in the TS following 1 h of repair than the NTS. The number of CPD reads normalized by the total number of nucleotides in dipyrimidine sequences for each DNA strand was calculated for the promoter, coding region, and termination site for high (>10 mRNAs per hour), medium (1–10 mRNAs per hour), low (<1 mRNAs per hour) transcribed genes (20). TSS is the transcription start site; TTS is the transcription termination site. (D) Snapshot of the distribution of CPD reads following 1 h of repair for the PRE7, ERD2, UTH1, and SHB17 genes. The red line indicates the transcribed strand [gene coordinates and image drawn using the Integrative Genomics Viewer (33)]. y axis depicts the number of CPD reads.
Fig. S1.
(A) T4 endonuclease V and APE1 cleave at CPD lesions, generating a free 3′OH immediately upstream of the CPD damage site. (B) Bioinformatics procedure to extract CPD dinucleotide sequences. Green, trP1 adaptor; purple, A adaptor.
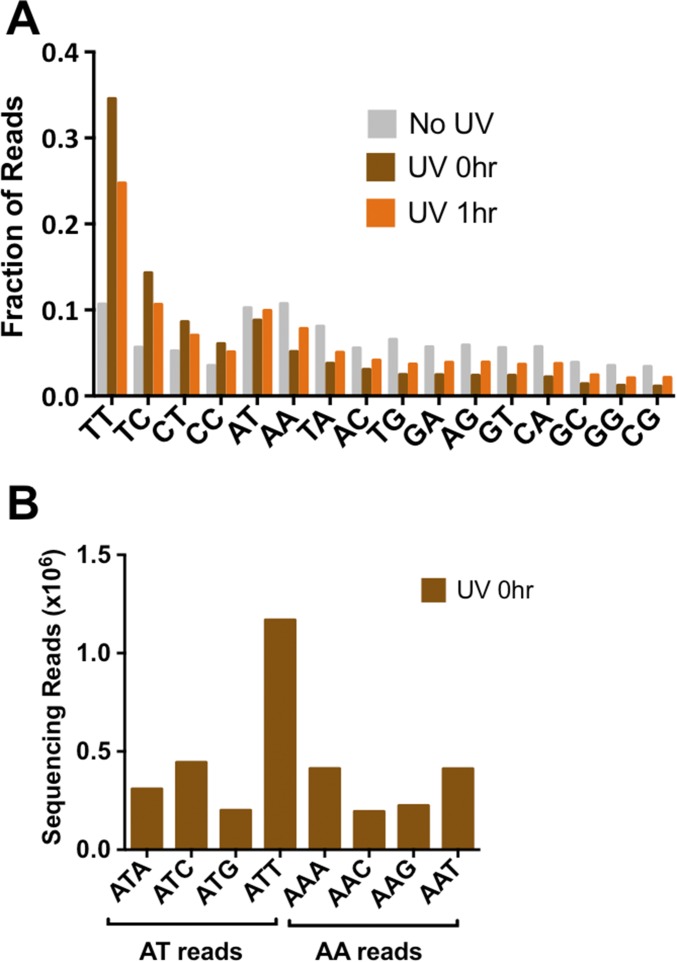
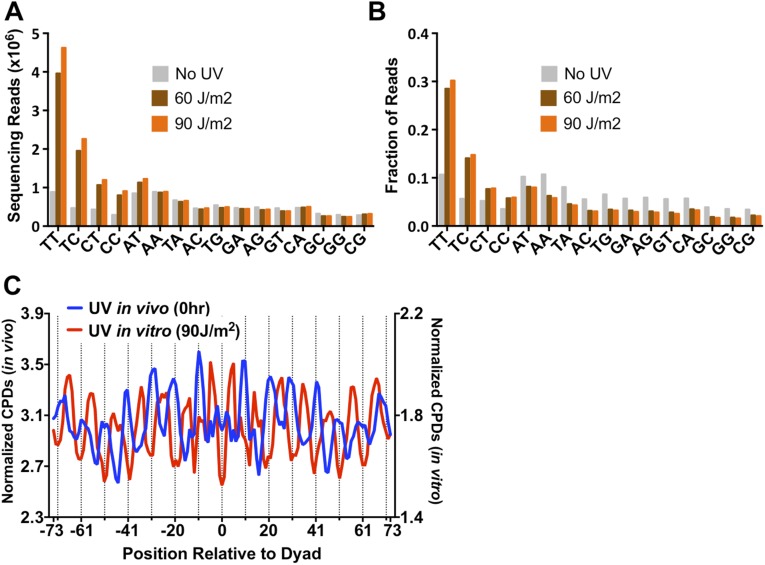
Analysis of the UV-treated samples revealed a clear enrichment of sequencing reads associated with dipyrimidine sequences (Fig. 1B and Fig. S2A). The majority of these reads occurred at TT sequences, followed by TC, CT, and CC, with a relative ratio of 54:22:14:10 in the UV 0-h sample. Much smaller numbers of reads were associated with each of the nondipyrimidine sequences (Fig. 1B and Fig. S2A). Presumably, these reads represent background because of unblocked 3′OHs, or could reflect cleavage by APE1 at abasic sites present in the isolated genomic DNA. A fairly large number of reads were associated with AT dinucleotides (Fig. 1B). However, these were primarily associated with ATT and ATC sequences (Fig. S2B), suggesting that they may be derived from CPD lesions at the adjacent TT or TC sequence. Comparison of the 0-h and 1-h UV samples revealed a significant decrease in the enrichment of dipyrimidine-associated reads in the 1-h UV sample (Fig. 1B and Fig. S2A), representing removal of CPD lesions during the 1-h repair incubation. There was no significant enrichment of sequencing reads associated with dipyrimidine sequences in the No UV sample (Fig. 1B and Fig. S2A), indicating that the enrichment of dipyrimidines in the UV-treated samples reflects UV-induced DNA damage. In all subsequent analysis, we excluded “background” reads that mapped to nondipyrimidine sequences.
Fig. S2.
(A) Fraction of dinucleotide sequences associated with CPD-seq sequencing reads after normalizing by the total number of reads for each sample. (B) Frequency of trinucleotide sequences associated with AT and AA reads. The high frequency of ATC- and ATT-associated reads suggests that the enrichment of AT reads in the UV-treated samples is likely derived from lesions associated with adjacent TT and TC sequences, potentially because of the weak exonuclease activity of APE1. In contrast, there was no enrichment of AAC- or AAT-associated reads.
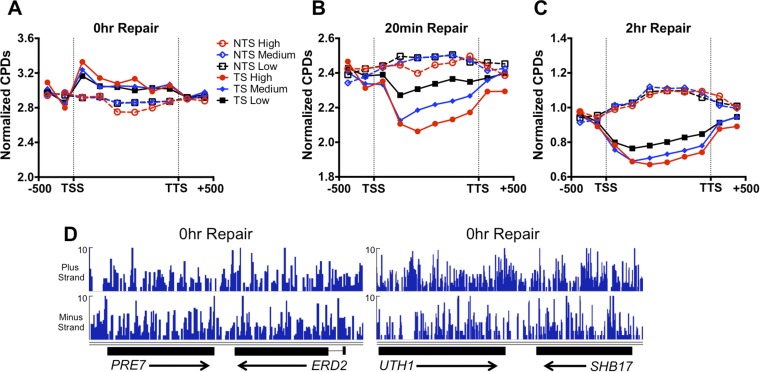
Analysis of CPDs in the 1-h repair sample revealed a clear signature of TC-NER (Fig. 1C). Following 1 h of repair, the number of CPDs remaining in the TS was significantly decreased relative to the nontranscribed strand (NTS). There were also fewer unrepaired CPDs in the TS than in adjacent intergenic regions upstream of the transcription start site or downstream of the transcription termination site; however, the opposite trend (i.e., more CPDs remaining in the coding region than adjacent intergenic regions) was apparent in the NTS (Fig. 1C). Analysis of the 0-h UV sample revealed that there was on average more initial damage in the TS than the NTS (Fig. S3A), indicating that data shown in Fig. 1C underestimate the difference in CPD removal between the TS and NTS. CPD removal appeared to be correlated with transcription rate (Fig. 1C), but a bias for CPD removal from the TS relative to the NTS was apparent even for low expressed genes [<1 mRNA per hour (20)]. We also used CPD-seq to map CPD lesions at earlier (20 min) and later (2 h) repair times. The TS strand also showed more CPD removal following only 20 min of repair, particularly for high expressed genes (Fig. S3B), and the difference in CPD removal between the TS and NTS strands was still detectable after 2 h of repair (Fig. S3C). Strand-specific differences in CPD removal were apparent even at individual genes (Fig. 1D). In contrast, there were no obvious differences in initial CPD levels in the TS and NTS for these genes in the 0-h UV sample (Fig. S3D).
Fig. S3.
(A) More UV damage is initially present on the TS than the NTS in the UV 0-h sample. (B) TC-NER of TS strand is apparent after 20 min of repair. (C) Preferential repair of TS strand is apparent even after 2 h of repair. (D) No significant difference in CPD reads between the TS and NTS strands of the PRE7, ERD2, UTH1, and SHB17 genes in the UV 0-h sample.
CPD Formation Is Modulated by the Rotational Setting of Nucleosomal DNA.
To investigate how chromatin packaging influences UV damage formation and repair in the yeast genome, we analyzed the distribution of CPD lesions within nucleosomes. CPD reads in the UV 0-h sample were aligned with a published, high-resolution map of yeast nucleosome positions (21). To account for potential sequence biases in the nucleosome sequences, the frequency of CPD reads was normalized by the number of nucleotides in dipyrimidine sequences at each position along the nucleosomal DNA. The normalized CPD reads were plotted for both DNA strands at positions relative to the nucleosome dyad (e.g., −73 bp to +73 bp).
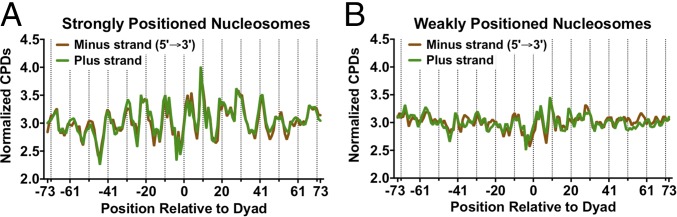
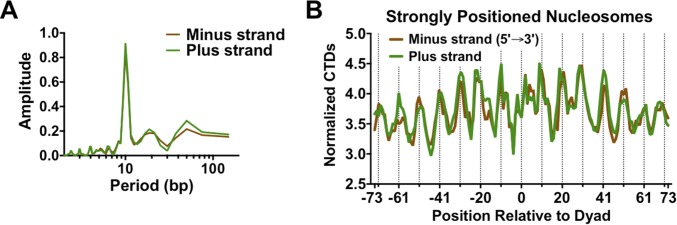
The distribution of normalized CPD reads for the UV 0-h sample showed significant periodicity within strongly positioned nucleosomes (Fig. 2A). The distribution of normalized CPD reads on each strand was analyzed for ∼10,000 strongly positioned nucleosomes, defined as having a nucleosome center positioning (NCP) score greater than 5 (21). When the plus and minus strands were aligned in the same orientation (5′ to 3′), they showed very similar patterns of CPD formation (Fig. 2A). The peaks of CPD formation coincided with outward rotational settings in the nucleosomes (Fig. 2A, dashed lines), exhibiting a clear UV “photo-footprint.” Analysis of the normalized CPD distribution revealed a periodicity of ∼10.0–10.7 bp (Fig. S4A), depending upon which smoothing method was used. However, among ∼7,500 weakly positioned nucleosomes (NCP score < 1) (21), there was little if any UV photo-footprint (Fig. 2B). Because the majority of UV damage was associated with TT dipyrimidines, we also analyzed the distribution of TT lesions [i.e., cyclobutane thymine dimers (CTDs)] among strongly positioned nucleosomes. Analysis of the CTD lesion data revealed an even more prominent UV photo-footprint at strongly positioned nucleosomes (Fig. S4B).
Fig. 2.
Strongly positioned nucleosomes in yeast cause an ∼10-bp UV photo-footprint. (A) CPD damage in strongly positioned nucleosomes (nucleosome score > 5) is higher at outward rotational settings (dashed lines). The normalized CPD distribution for the 0-h UV sample was analyzed along the plus and minus strands of ∼10,000 strongly positioned nucleosomes. Both plus and minus strands were oriented in the 5′ to 3′ direction. (B) CPD damage in weakly positioned nucleosomes (nucleosome score < 1) does not show a significant UV photo-footprint.
Fig. S4.
(A) Periodogram of nucleosome UV photo-footprint reveals a periodicity of CPD formation of ∼10 bp. (B) CTD show a particularly prominent UV photo-footprint in strongly positioned nucleosomes. The number of sequencing reads associated with TT dinucleotides in the UV 0-h sample was normalized by the number of TT dinucleotides at positions relative to the nucleosome dyad.
The Nucleosome Photo-Footprint Overrides Intrinsic DNA Sequence Preferences for CPD formation and Persists During Repair.
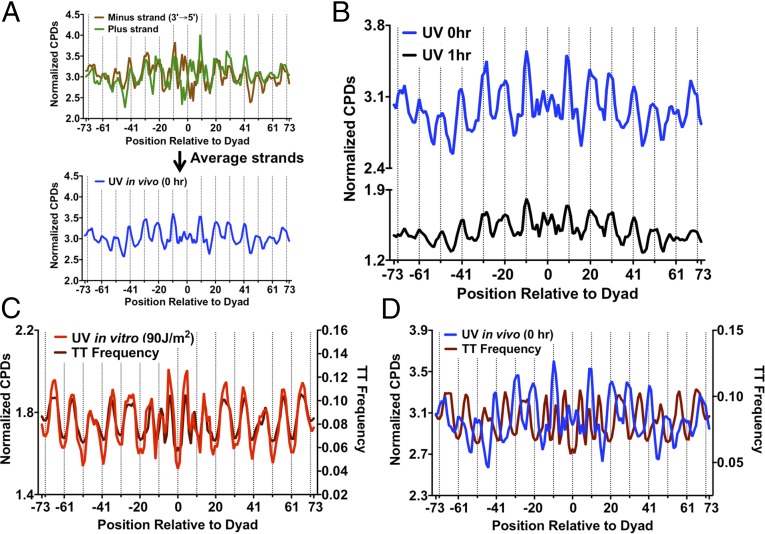
Nucleosomal DNA is known to have a biased sequence composition that favors DNA bending around the histone octamer. The pyrimidine frequency in the two DNA strands is negatively correlated when the strands are aligned in their normal antiparallel orientation (i.e., minus strand 3′ to 5′) (Fig. 3A). Hence, to eliminate the effects of DNA strand bias on CPD formation in the nucleosome, we averaged the normalized CPD values between the two DNA strands (Fig. 3A). Analysis of the strand average of CPD formation again revealed a very clear nucleosome photo-footprint, with peaks of damage formation at outward rotational settings (Fig. 3B). Importantly, a similar photo-footprint was still apparent after 1 h (Fig. 3B) or 2 h of repair (see, for example, Fig. 5B, below). These data suggest that the differences in initial damage levels between inward and outward rotational settings persists during repair, even though a large fraction of the CPDs have been removed by these time points.
Fig. 3.
The nucleosome UV photo-footprint protects T-rich DNA sequences from UV damage. (A) Method for averaging normalized CPD counts between plus and minus DNA strands in strongly positioned nucleosomes. The DNA strands were aligned in their normal antiparallel orientation to remove strand-specific sequence biases, and the normalized CPD data were calculated as a weighted average. (B) The nucleosome UV photo-footprint persists at 1-h repair. DNA regions that fell within the transcribed strand of a gene were excluded from this analysis. (C) In vitro CPD formation in the absence of nucleosomes is strongly correlated with the TT frequency in DNA. Both in vitro CPD formation and TT frequency peak at inward rotational positions. (D) In vivo CPD formation in the presence of strongly positioned nucleosomes is anticorrelated with TT frequency in nucleosomal DNA.
Fig. 5.
Analysis of repair of CPD lesions. (A) Comparison of normalized CPDs at high-occupancy Reb1 binding sites at 0-h and 1-h repair time points. The weighted average of normalized CPDs of both strands is depicted. (B) Plot of normalized CPD reads in strongly positioned (nucleosome score > 5) and weakly positioned (nucleosome score < 5) nucleosomes after 2 h of repair. Dashed lines indicate outward rotational settings (dashed lines). The weighted average of normalized CPDs of both strands is shown. (C) More CPDs remain unrepaired adjacent to the nucleosome dyad in strongly positioned nucleosomes. The fraction of CPDs remaining was calculated by comparing the 2-h repair sample to its matched 0-h control. (D) Telomere regions show slower CPD removal following 1 h of repair. The fraction of CPDs remaining was calculated by comparing the 1-h repair sample to its matched 0-h control.
To examine the influence of nucleosomal DNA sequence composition on CPD formation, we irradiated purified yeast genomic DNA with 60- and 90-J/m2 UVC light and mapped CPD lesions using CPD-seq. These UV doses were chosen because they yielded roughly similar damage levels in naked DNA in vitro as when irradiating yeast cells with 125 J/m2. Analysis of the sequencing data for the 60- and 90-J/m2 samples revealed a significant enrichment of dipyrimidine associated reads, as expected (Fig. S5 A and B).
Fig. S5.
(A) Counts of dinucleotide sequences associated with CPD-seq reads for naked DNA irradiated in vitro with 60 or 90 J/m2 UVC light. The “No UV” data are the same as in Fig. 1B. (B) Fraction of dinucleotide sequences associated with CPD-seq sequencing reads after normalizing by the total number of reads for each sample. (C) When yeast genomic DNA was irradiated in vitro (UV 90J sample), CPD formation occurs more frequently in DNA that adopts an inward rotational setting in vivo. Inward rotational settings occur at the midpoints between the dashed lines. CPD formation in vivo (0-h UV sample) shows the opposite trend.
Surprisingly, the pattern of CPD formation in nucleosomal DNA was very different when the DNA was irradiated in vitro in the absence of nucleosomes (Fig. S5C). Whereas the peaks of CPD formation for nucleosomal DNA in vivo coincided with outward rotational settings (Fig. S5C, dashed lines), the same DNA regions had the lowest levels of CPD formation in naked DNA irradiated in vitro. The highest levels of CPD formation in naked DNA were associated with DNA regions that have inward rotational settings in nucleosomes (i.e., ∼5 bp offset from the outward rotational settings). Nucleosomal DNA at inward rotational settings tends to be A-T rich (22). Because CPDs preferentially form at TT dinucleotides, we hypothesized that A-T–rich sequences in inward rotational settings in nucleosomal DNA were responsible for the observed CPD peaks in the irradiated naked DNA. Indeed, analysis of the TT frequency in nucleosomal DNA supported this hypothesis. Peaks in TT composition overlapped with peaks of CPD formation in naked DNA, and both coincided with inward rotational settings (Fig. 3C). In contrast, there was essentially no overlap between peaks of TT composition and CPD formation in vivo, but they instead showed opposite trends (inward versus outward) (Fig. 3D). These data suggest that nucleosomes strongly inhibit CPD formation in nucleosomal DNA with an inward rotation setting, even though such A-T–rich DNA is more intrinsically prone to form CPD lesions.
DNA-Bound Transcription Factors Inhibit CPD Formation.
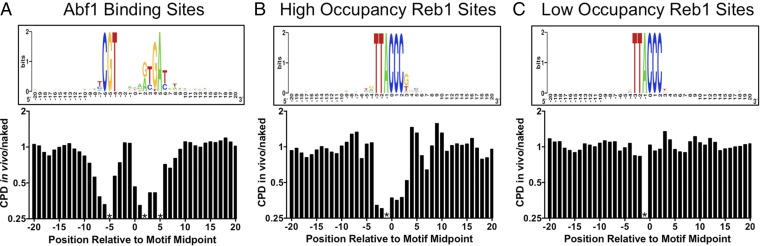
We analyzed CPD formation at experimentally mapped binding sites for two well-studied yeast transcription factors: Abf1 and Reb1. These particular factors were chosen because they bind many sites in the yeast genome and because high-resolution in vivo binding profiles were available for analysis (23). CPD formation was analyzed at 661 sites that were bound by Abf1 (23) and contained a canonical Abf1 sequence motif (Fig. 4A). To account for potential biases in DNA sequence composition, relative differences in CPD formation were measured using the ratio of CPDs in the 0-h UV sample (in vivo) relative to naked DNA (UV 90 J/m2). This ratio was scaled to 1 and analyzed along the aligned Abf1 binding sites.
Fig. 4.
The yeast transcription factors Abf1 and Reb1 induce a significant UV photo-footprint at their DNA binding sites. (A) Abf1-bound DNA sites show altered CPD formation. (Upper) The DNA consensus sequence of 661 Abf1 binding sites [generated using WebLogo (34)], including DNA flanking each binding site. (Lower) The scaled ratio of normalized CPDs in the UV 0-h sample (in vivo) relative to the UV 90-J/m2 sample (naked) for the plus strand of the Abf1 binding sites. Asterisks indicate that the indicated position in the motif cannot form CPD lesions because of DNA sequence constraints. (B) Same as A, except the plus strand of high-occupancy Reb1 binding sites (Reb1 occupancy > 10) were analyzed. (C) Same as A, except a plus strand of low-occupancy Reb1 binding sites (Reb1 occupancy < 10) were analyzed.
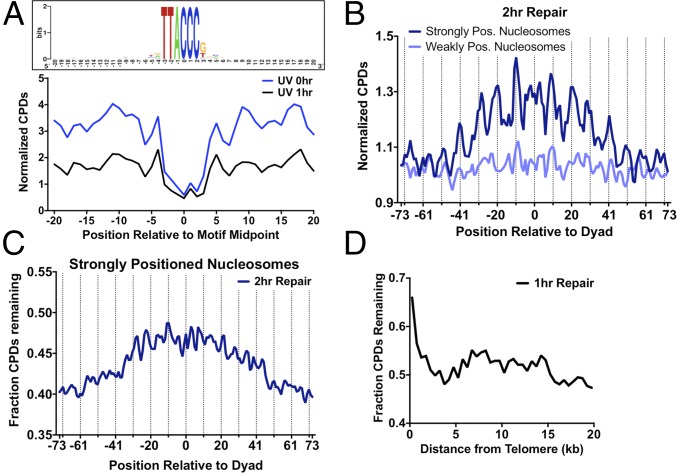
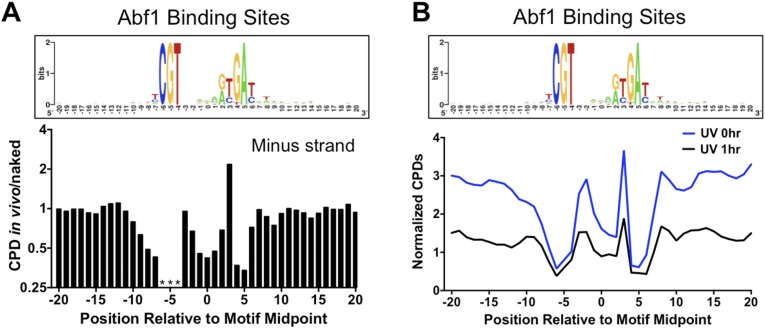
Although there were roughly similar levels of damage (i.e., CPD in vivo/naked ratio ∼1) in regions flanking Abf1 binding sites, there was considerably less CPD formation in vivo within Abf1 binding sites, particularly in the highly conserved regions of the Abf1 motif (compare Upper and Lower panels in Fig. 4A). The magnitude of inhibition ranged from a twofold to threefold decrease in CPD formation in vivo relative to naked DNA at positions in both the plus and minus strands (Fig. 4A and Fig. S6A). The CPD inhibition was primarily associated with nucleotide positions important for Abf1 binding (24). Although CPD formation was generally inhibited at Abf1 binding sites, there was a strong enhancement of CPD formation in the minus strand at position 3 of the Abf1 binding site (Fig. S6A).
Fig. S6.
(A) Abf1-bound DNA sites show altered CPD formation. (Upper) The DNA consensus sequence of 661 Abf1 binding sites [generated using WebLogo (34)], including DNA flanking each binding site. (Lower) The scaled ratio of normalized CPDs in the UV 0-h sample (in vivo) relative to the UV 90J sample (naked) for the minus strands of Abf1 binding sites. Asterisks indicate that the indicated position in the motif cannot form CPD lesions because of DNA sequence constraints. (B) Comparison of normalized CPDs at Abf1 binding sites at 0-h and 1-h repair time points. The weighted average of normalized CPDs of both strands is depicted.
Analysis of CPD formation at binding sites for the yeast Reb1 transcription factor showed similar results (Fig. 4 B and C). The Reb1 binding sites were stratified into “high-occupancy” (Reb1 occupancy > 10) and “low-occupancy” (Reb1 occupancy < 10) binding sites based on in vivo binding data (23). CPD formation was analyzed at 784 high-occupancy and 472 low-occupancy Reb1 binding sites that contained a canonical Reb1 sequence motif (TTACCC). CPD formation was significantly inhibited in vivo at high-occupancy Reb1 binding sites, but not in flanking DNA (Fig. 4B). In contrast, CPD formation was not affected at low-occupancy binding sites (Fig. 4C), even though these sites have nearly identical DNA sequence motifs (compare Fig. 4 B and C, Upper). These data indicate that transcription factor binding alters UV damage formation at binding sites in vivo. Importantly, the transcription factor UV photo-footprint persists during repair, as normalized damage levels at high-occupancy Reb1 (Fig. 5A) and Abf1 binding sites (Fig. S6B) remain low relative to flanking DNA following 1 h of repair.
Location of CPD Lesions near a Nucleosome Dyad or in Heterochromatin Inhibits Repair.
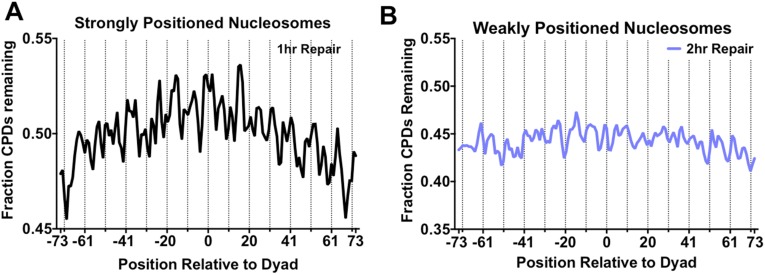
We analyzed how positioning of CPD lesions within well-positioned nucleosomes affected repair. Transcribed DNA was eliminated from this analysis to analyze the effects of nucleosomes specifically on GG-NER. Although the number of CPD lesions decreased following 1 h of repair, the distribution of lesions was similar to the initial 0-h time point, with peaks of CPD lesions remaining at outward rotational settings (Fig. 3B). However, there was a subtle trend of slower removal of CPD lesions in regions near the nucleosome dyad. Analysis of the fraction of CPDs remaining following 1 h of repair revealed that a slightly higher fraction of unrepaired damage was present adjacent to the dyad relative to more distal regions of nucleosomal DNA (Fig. S7A). After 2 h of repair, there was an even more pronounced trend of higher CPD levels near the dyad of strongly positioned nucleosomes; however, this trend was not observed among weakly positioned nucleosomes (Fig. 5B). Analysis of the fraction of CPDs remaining following 2 h of repair, relative to the matched 0-h sample, confirmed these results. Significantly more CPD lesions remained after 2 h of repair near the dyad of strongly positioned nucleosomes (Fig. 5C), but not at weakly positioned nucleosomes (Fig. S7B). Although translational positioning affected repair, there was no obvious effect of rotational positioning on CPD removal (Fig. 5C and Fig. S7).
Fig. S7.
(A) Fraction of CPDs remaining following 1 h of repair relative to the dyad of strongly positioned nucleosomes. (B) Positioning relative to the dyad does not affect CPD removal in weakly positioned nucleosomes. The fraction of CPDs remaining was calculated by comparing the 2-h repair sample to its matched 0-h control.
Highly compact heterochromatin, such as that found in yeast telomeres or silent mating loci, is also thought to impede NER (8). Analysis of the 1-h repair data revealed a much higher fraction of CPDs remained unrepaired in regions adjacent to yeast telomeres than elsewhere in yeast chromosomes (Fig. 5D), indicating that heterochromatin inhibits NER.
Discussion
Previous genomic studies have suggested that NER activity is affected by chromatin state and inhibited by transcription factor binding (7–10). However, it is not known how these and other features of the chromosomal landscape affect initial UV damage formation. In this study, we addressed this question using a new method to map CPD lesions throughout the yeast genome at nucleotide resolution. Using this method, we discovered that initial CPD formation is significantly altered at nucleosomes and transcription factor binding sites throughout the yeast genome. Importantly, we show that differences in initial UV damage formation persist during repair, and thus may affect the genome-wide distribution of UV-induced mutations.
Nucleosomes Significantly Modulate UV Damage Formation and Repair.
Analysis of the CPD distribution immediately after UV irradiation revealed that nucleosomes dramatically alter the formation of CPD lesions. CPD formation was lower at DNA located at inward rotational settings within nucleosomes, whereas CPD formation was higher at DNA at outward rotational settings, yielding a readily discernible nucleosome photo-footprint with ∼10-bp periodicity (Fig. S8). Such a nucleosome photo-footprint was identified in mammalian chromatin nearly 30 y ago (11), but had been missed by previous genome-wide studies of CPD-formation (15, 16, 18, 25). Nucleosomal DNA with an inward rotational setting is likely protected from UV damage as a result of constraints on DNA bending and flexibility imposed by the nucleosome structure (22). Formation of CPDs in DNA depends not only on the absorption of UV photons, but also the flexibility and bending of adjacent pyrimidine bases to allow the photo-induced [2 + 2] cycloaddition reaction to occur (26, 27). At inward rotational settings in the nucleosome, the decrease of DNA flexibility and compression of the minor groove is likely responsible for suppressing CPD formation at such sites (27).
Fig. S8.
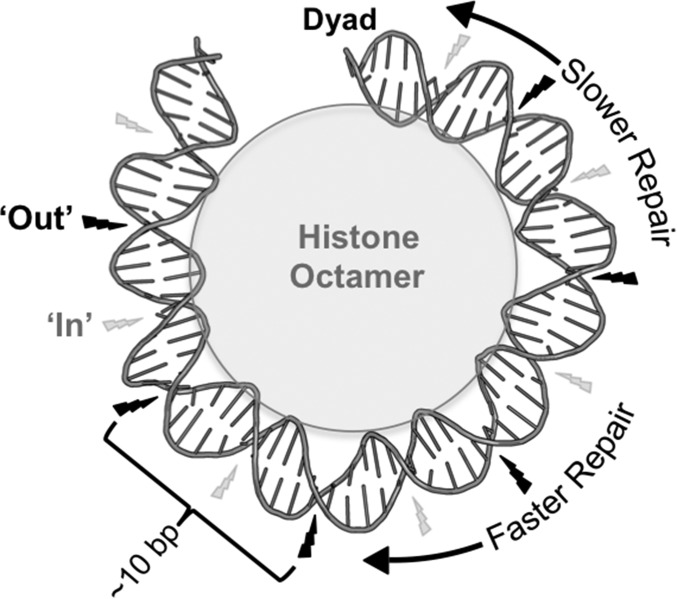
Model of the how strongly positioned nucleosomes affect UV damage formation and repair. In strongly positioned nucleosomes, CPD damage formation is highest at outward (“Out”) rotational settings (indicated by large black lightning bolt), which gives rise to the ∼10-bp periodicity. Inward (“In”) rotational settings have the lowest CPD damage formation (indicated by small gray lightning bolt), even though such regions are TT rich. NER is inhibited at translational positions near the nucleosome dyad. DNA structure was depicted using Pymol to visualize 1ID3.
Our data indicate that among strongly positioned nucleosomes (corresponding to ∼15% of yeast nucleosomes), normalized CPD formation can vary up to 50% between inward and outward rotational settings. However, the actual magnitude of the nucleosome photo-footprint is likely greater than this estimate, as a result of biases in the DNA sequence composition of nucleosomal DNA. Within nucleosomes, the more compressed minor grooves of short segments of A-T–rich DNA tend to adopt inward rotational settings to facilitate DNA bending around the histone octamer (22) and to strengthen histone interactions with the DNA minor groove (28). However, A-T–rich DNA also contains more TT dipyrimidines, which are more prone to form CPD lesions than other dyprimidine sequences (1). Hence, in UV-irradiated naked DNA there is much higher CPD formation in A-T–rich DNA regions, which adopt inward rotational settings in nucleosomes. However, the opposite pattern is detected in vivo in the presence of well-positioned nucleosomes, indicating that the nucleosome photo-footprint can override even intrinsic DNA sequence preferences for CPD formation. Taken together, these findings suggest that nucleosomes may act to protect short A-T–rich DNA sequences from UV damage by orienting such sequences inward toward the histone octamer.
In addition to modulating CPD formation, our data indicate that strongly positioned nucleosomes influence excision repair of CPD lesions. Interestingly, translational positioning of the lesion plays a more important role in CPD repair than rotational settings, as shown by faster repair in more distal regions of strongly positioned nucleosomal DNA relative to the central dyad (Fig. S8). This result is supported by previous studies, which did not observe an influence of rotational positioning on repair (29, 30). In contrast, translational positioning did not correlate with repair in weakly positioned nucleosomes; this may be because of resident chromatin remodeling enzymes in the cell, which are actively involved in NER to partially disrupt nucleosome structure and allow access of NER enzymes to bulky lesions (30, 31). It is likely that DNA regions near the dyad of strongly positioned nucleosomes are more refractory to chromatin remodeling activity and less accessible to the NER machinery.
UV Damage Formation Is Significantly Inhibited at Transcription Factor Binding Sites.
Our analysis revealed that DNA-bound transcription factors significantly affect CPD formation at their binding sites. At binding sites for the yeast transcription factors Abf1 and Reb1, there were up to threefold fewer CPD lesions than expected in naked DNA. This effect was particularly apparent at highly conserved regions of the binding motifs, and was associated with transcription factor binding. Although transcription factor binding largely inhibited CPD formation, at one conserved position in the Abf1 motif there was ∼twofold more CPD damage than expected. Because this position coincides with a known DNase I hypersensitivity site in the Abf1 motif (24), it is likely that in the Abf1–DNA complex this position is constrained in such a way as to facilitate CPD formation. Such UV hypersensitive sites have been detected in previous biochemical studies of UV damage in protein-bound DNA (e.g., refs. 13 and 32). In contrast, the remainder of the DNA motif is likely bent in an unfavorable manner so as to inhibit CPD formation, thus generating the observed transcription factor photo-footprint. Our study suggests that these important functional DNA elements are partially protected from UV damage by ongoing transcription factor binding.
A recent study reported that NER is inhibited by transcription factor binding in the human genome (9). This conclusion was based on analysis of XR-seq data, which essentially counts the number of excised DNA fragments during repair. However, low numbers of excised fragments could also be a result of low initial damage levels. Analysis of our repair data suggests that NER may occur more slowly at yeast transcription factor binding sites (Fig. 5A), but this is difficult to determine precisely with relatively low initial damage levels. Hence, more data are needed to adequately evaluate this claim. CPD-seq should be applicable to mapping CPD lesions in the human genome, although the sensitivity of the method may need to be improved, particularly for lower UV doses.
In summary, this study demonstrates that UV damage formation in a eukaryotic genome is not uniform, but instead is significantly modulated by nucleosomes and other DNA-binding proteins. Similarly, genome sequencing of clinical melanomas has revealed significant heterogeneity in UV-induced mutation density throughout the genome (4–6). Such differences in mutation occurrence are often attributed to variations in DNA repair efficiency between different regions of the genome (8–10). We propose that differences in initial UV damage formation, because of the photo-footprints of bound transcription factors and nucleosomes, also play an important role in the heterogeneity in mutation density in human melanomas and potentially other human cancers.
Materials and Methods
Wild-type yeast (BY4741) were grown to midlog phase and UV irradiated with 125 J/m2 UVC light. Genomic DNA extraction protocol is described in SI Materials and Methods. The CPD-seq procedure is adopted from the recently published emRiboSeq method (19), with a major modification at the enzymatic digestion step to allow specific cutting at CPD sites and generate ligatable 3′OH groups (Fig. 1A). Detailed descriptions of the CPD-seq method, validation, and data analysis are given in SI Materials and Methods and Fig. S9.
Fig. S9.
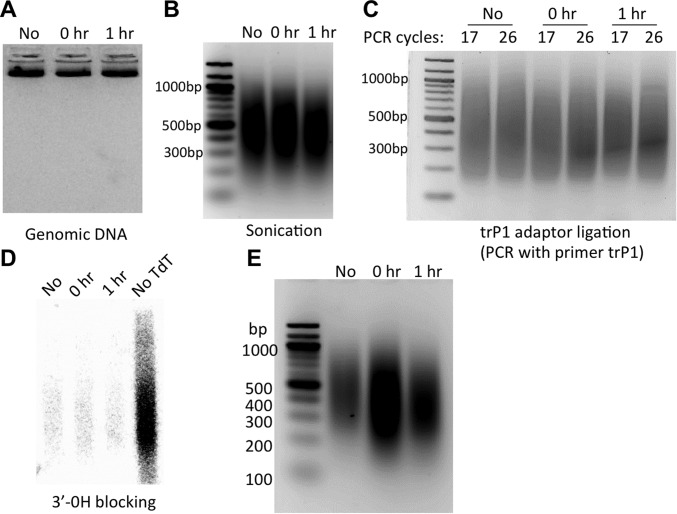
Results at intermediate steps in CPD-seq procedure. Yeast cells were irradiated with 125 J/m2 UVC light. Samples were taken before UV treatment (“No”), immediately after UV (0 h), and after 1 h of repair (1 h). (A) Isolated genomic DNA. (B) Sonicated genomic DNA. (C) PCR confirmation of trP1 adaptor ligation. (D) PCR confirmation of 3′OH blocking by TdT and ddATP (see SI Materials and Methods). (E) Gel verification of CPD-seq libraries.
SI Materials and Methods
UV Irradiation and Yeast Genomic DNA Isolation.
Yeast cultures (BY4741) were grown to OD600 ∼ 0.8. Cells were collected and resuspended in sterile deionized H2O and irradiated with 125 J/m2 UVC. After UV irradiation, cells were spun down and resuspended in prewarmed YPD medium (yeast extract, peptone, dextrose) and grown in the dark for repair experiments. Aliquots (10 mL of yeast culture) were taken before UV irradiation (“no UV”), immediately after UV irradiation (0 h), and at different repair time points as indicated. Cells were spun down and kept at −80 °C until genomic DNA isolation.
To isolate genomic DNA, cell pellets were mixed with 250 μL of DNA lysis buffer [2% (vol/vol) Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris⋅HCl, pH 8.0, 1 mM EDTA], 250 μL of PCI (phenol:chloroform:isoamyl alcohol = 25:24:1), and 150 μL of acid-washed glass beads, and vortexed vigorously twice, 2 min each time. After adding 200 μL of TE (10 mM Tris⋅HCl, pH 7.5, 1 mM EDTA) buffer, cell lysates were centrifuged and the supernatant was mixed with 1 mL of ethanol at room temperature for 5 min to precipitate DNA. Following centrifugation and washing with 70% (vol/vol) ethanol, the DNA pellet was dissolved in 200 μL of TE and incubated with 2 μL of RNase A/T1 (ThermoFisher Scientific) at 37 °C for 1 h to remove RNA. DNA was subsequently purified by extraction with PCI, ethanol-precipitated, and dissolved in sterile deionized H2O. The purified genomic DNA was quantified and 12 μg of DNA was used for each sequencing library preparation.
UV Irradiation of Naked DNA.
Yeast genomic DNA was isolated from unirradiated yeast cells. For UV radiation of naked DNA, a total of 15 μg of DNA (120 μL) was spotted on a clean microscope cover glass, 10 μL in each spot. The cover glass with DNA spots was irradiated with 60 J/m2 or 90 J/m2 UVC light. After UV treatment, the irradiated DNA collected into a microcentrifuge tube and 12 μg of DNA was used for sonication. The sonicated DNA was used for CPD-seq library preparation.
Oligonucleotides for CPD-Seq Library Preparation.
The following oligonucleotides were used for the first ligation: trP1-top (5′-CCTCTCTATGGGCAGTCGGTGAT-phosphorothioate-T-3′), and trP1-bottom (5′-phosphate-ATCACCGACTGCCCATAGAGAGGC-dideoxy-3′). The following barcoded oligonucleotides (A1, A2, or A3) were used for the second ligation: A1-top (5′-phosphate-ATCCTCTTCTGAGTCGGAGACACGCAGGGATGAGATGGC-dideoxy-3′), A1-bottom (5′-biotin-CCATCTCATCCCTGCGTGTCTCCGACTCAGAAGAGGATNNNNNN-C3 phosphoramidite-3′); A2-top (5′-phosphate-ATCACGAACTGAGTCGGAGACACGCAGGGATGAGATGGC-dideoxy-3′), A2-bottom (5′-biotin-CCATCTCATCCCTGCGTGTCTCCGACTCAGTTCGTGATNNNNNN-C3 phosphoramidite-3′); A3-top (5′-phosphate-ATCTCAGGCTGAGTCGGAGACACGCAGGGATGAGATGGC-dideoxy-3′), A3-bottom (5′-biotin-CCATCTCATCCCTGCGTGTCTCCGACTCAGCCTGAGATNNNNNN-C3 phosphoramidite-3′). The oligonucleotides used for PCR confirmation and library amplification were Primer A (5′-CCATCTCATCCCTGCGTGTCTCCGAC-3′), and Primer trP1 (5′-CCTCTCTATGGGCAGTCGGTGATT-3′).
CPD-Seq Validation.
To validate the CPD-seq method, we mapped CPD lesions in yeast cells irradiated with 125 J/m2 UV light (254 nm) and allowed to repair for 0 or 1 h. As a control, we also included yeast DNA that was not irradiated with UV (no UV) in our experiments. The isolated genomic DNA, sonication, and trP1 adaptor ligation were monitored by gel electrophoresis (Fig. S9). A critical step in CPD-seq is the blockage of free 3′OH before T4 endoV digestion, so we analyzed the efficiency of blocking free 3′OH. Following the terminal transferase step, 10% of DNA was separated and denatured to ligate any remaining free 3′OHs to the A adapter. Ligation was analyzed by PCR using radiolabeled primer A and cold primer trP1. The PCR data indicate that the terminal transferase step efficiently blocked adapter A ligation, as shown by the significant reduction of PCR signal in the terminal transferase-treated DNA (Fig. S9D). Subsequent treatment of the remaining DNA (90%) with T4 endoV and APE1 generated many free 3′OHs in the UV 0-h and 1-h samples, which could be efficiently ligated to adapter A and PCR-amplified (Fig. S9E).
CPD-Seq Library Preparation and Sequencing.
Yeast genomic DNA was sonicated to an average size of ∼400 bp using a Bioruptor 300 sonicator (15 cycles, 30-s ON and 30-s OFF). After ethanol precipitation and size selection with 1.2 volumes of AMpure XP beads (Beckman Coulter), ∼5 μg DNA was retained. The sonicated DNA was sequentially end-repaired and dA-tailed, using NEBNext End Repair Module and NEBNext dA-Tailing Module (New England Biolabs), respectively. DNA was then ligated to the double-stranded adaptor DNA trP1, which contains a T overhang at the 3′ end of one strand and a 5′ phosphate group on the other strand. Ligation was performed at 16 °C for 16 h, using NEBNext Quick Ligation Module. DNA was purified with AMpure XP beads and subsequently incubated with Terminal Transferase (TdT, New England Biolabs) and dideoxy ATP (Roche) for 2 h at 37 °C to block all free 3′ ends. After purification with AMPure XP beads, DNA (∼2 μg) was incubated with 30 units of CPD-specific T4 endonuclease V (T4 endoV; Epicentre Biotechnologies) at 37 °C for 2 h to generate single-strand DNA breaks at CPD sites. T4 endoV is known to be specific for CPD lesions and does not cleave at 6-4PPs (35). A relatively high concentration of T4 endoV was used to ensure that CPDs in different sequence contexts were fully cleaved. DNA was purified and subsequently digested with 15 units of apurinic/apyrimidinic (AP) endonuclease APE1 (New England Biolabs) at 37 °C for 1.5 h to create ligatable 3′-OH groups. DNA was purified and incubated with 5 units of shrimp alkaline phosphatase (Affymetrix) at 37 °C for 1 h to remove 5′ phosphates. The remaining phosphatase activity was eliminated by heating DNA at 65 °C for 15 min. After purification, the DNA was denatured by heating at 95 °C for 5 min and immediately cooled on ice. The denatured DNA (∼1.2 μg) was mixed with 240 pmol of double-stranded A adaptor and ligation was performed at 16 °C for 16 h using the NEBNext Quick Ligation Module. One of three different A adaptors (i.e., A1, A2, or A3) that contain distinct barcode sequences was used for ligation and library preparation of each sample. The 5′ biotin label on the A adaptors was used to capture the ligated products with streptavidin beads (Life Technologies). The DNA strand without the biotin label was released from streptavidin beads with 0.15 M NaOH. The resulting single-strand (ss) DNA was used as the template to synthesize double-strand (ds) DNA using primer A. The dsDNA library was purified with AMPure XP beads, and verified by PCR with 32P-labeled primer A and cold primer trP1. The dsDNA library was briefly amplified for six cycles in a standard PCR by adding cold primer A and primer trP1, and the resulting PCR products were purified with AMPure XP beads. Equal volumes of three CPD-seq libraries with different barcodes were mixed and sequenced using the Ion Proton platform (Life Technologies).
CPD-Seq Data Analysis.
The ion proton sequencing reads were separated into distinct libraries using the unique A1, A2, and A3 barcode sequences, and the barcode sequences were trimmed. The trimmed sequencing reads were aligned to the SacCer3 yeast genome using the bowtie2 software (36). In some of the samples, a small amount of UV-irradiated pUC19 DNA was added as a spiked control, but reads mapping to the pUC19 plasmid were excluded from subsequent analysis. The resulting alignment file was converted to a BED file using samtools (37) and BEDtools (38) software. The putative CPD damage site was inferred from the sequencing read by extracting the dinucleotide sequence immediately adjacent to the 5′ end of the sequencing read but on the opposite strand (Fig. S1). Sequencing reads that mapped to nondipyrimidine damage sites were excluded from further analysis. The resulting BED files containing the positions of CPD damage sites were split into plus and minus strands, sorted, and counted using IGVtools (33) to generate WIG files. This method in effect counts a CPD read for both positions in the dinucleotide damage site. For each resulting WIG file, a background value of zero was included at positions of dipyrimidine sequences in the genome that had no mapped CPD reads. The normalized CPD reads value for a bin (e.g., location relative to a set of nucleosome dyads or transcription factor binding sites) was computed by averaging the CPD read values for all nucleotides in dipyrimidine sequences. This represents the average number of CPD reads per nucleotide in a dipyrimidine sequence.
Analysis of TC-NER.
TS and NTS for each yeast gene were divided into six equal sized bins, and the number of CPD reads was tabulated for each bin. Two additional bins were tabulated for the promoter, from −1 to −250 and −251 to −500 nucleotides upstream of the transcription start site. Two additional bins were also tabulated for the regions from +1 to +250 and +251 to +500 downstream of the transcription termination site. To account for strand-specific biases in sequence composition, the total number of nucleotides in dipyrimidine sequences in each bin was used to normalize the frequency of CPD reads. This calculation yielded the number of normalized CPDs, which is in essence the average number of CPD reads per dipyrimidine sequence. Normalized CPD values for each bin were averaged for each group of yeast genes. Transcript boundaries were obtained from ref. 39 and lifted over to the SacCer3 genome.
Analysis of the Nucleosome Photo-Footprint and Repair.
A high-resolution yeast dataset of nucleosome dyad positions (21) was used as a reference map of nucleosomes. A custom perl script was used to map the normalized CPDs data onto the nucleosome dyad coordinates. Strongly positioned nucleosomes were defined as those with a nucleosome score greater than 5, whereas weakly positioned nucleosomes had a nucleosome score less than 1, based on the nucleosome scoring system derived in reference (21). The periodicity analysis for the nucleosome photo-footprint was done using the R package specpgram and a span of 3.
Repair data were analyzed similarly, except DNA regions that fell within the transcribed strand of a gene were excluded from the analysis, to specifically examine the effects of nucleosomes on GG-NER (and not TC-NER). The normalized CPD value from the repair time point was divided by the normalized CPD value from the 0-h time point to yield the “fraction CPDs remaining.” Note, this is a only a relative estimate of repair, as the repair datasets were not normalized to the 0-h dataset.
Analysis of Transcription Factor Photo-Footprints.
Transcription factor binding sites for Abf1 and Reb1 were based on ORGANIC data from experiments using 10-min micrococcal nuclease digestion, 80 mM NaCl, and <100-bp DNA fragments (len50) (23). Binding sites were aligned based on the position of the canonical Abf1 or Reb1 binding site motif (CGTNNNNNRNKA and TTACCC, respectively). Binding sites that did not match the canonical motif were excluded from further analysis. The normalized CPD read data for the UV 0-h (in vivo) and UV 90J (naked DNA) datasets were mapped to the Abf1 and Reb1 binding site positions. The ratio of normalized CPD reads in the UV 0 h relative to the UV 90J naked DNA control (i.e., CPD in vivo/naked) was scaled using the ratio of the total number of CPD reads in each sample. DNA sequence logos of the Abf1 and Reb1 binding site motifs were generated using the WebLogo software (34).
Acknowledgments
We thank Amelia Hodges for helpful comments and suggestions, and Mark Wildung and Wei Wei Du for technical assistance with Ion Proton sequencing. This work was supported by National Institute of Environmental Health Sciences Grants ES002614 (to M.J.S. and J.J.W.) and ES022633 (to S.A.R.); and the Breast Cancer Research Program Breakthrough Award BC141727 from the Department of Defense (to S.A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE79977).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606667113/-/DCSupplemental.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd Ed ASM Press; Washington, DC: 2006. [Google Scholar]
- 2.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 3.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15(7):465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 4.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polak P, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2014;32(1):71–75. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29(9):948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci USA. 2016;113(15):E2124–E2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532(7598):264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- 10.Perera D, et al. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532(7598):259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- 11.Gale JM, Nissen KA, Smerdon MJ. UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc Natl Acad Sci USA. 1987;84(19):6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker MM, Wang JC. Use of light for footprinting DNA in vivo. Nature. 1984;309(5970):682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Conconi A, Smerdon MJ. Strand-specific modulation of UV photoproducts in 5S rDNA by TFIIIA binding and their effect on TFIIIA complex formation. Biochemistry. 1997;36(44):13710–13717. doi: 10.1021/bi9716736. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Mann DB, Suquet C, Springer DL, Smerdon MJ. Ultraviolet damage and nucleosome folding of the 5S ribosomal RNA gene. Biochemistry. 2000;39(3):557–566. doi: 10.1021/bi991771m. [DOI] [PubMed] [Google Scholar]
- 15.Teng Y, et al. A novel method for the genome-wide high resolution analysis of DNA damage. Nucleic Acids Res. 2011;39(2):e10. doi: 10.1093/nar/gkq1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell JR, et al. 3D-DIP-Chip: A microarray-based method to measure genomic DNA damage. Sci Rep. 2015;5:7975. doi: 10.1038/srep07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyrick JJ, Roberts SA. Genomic approaches to DNA repair and mutagenesis. DNA Repair (Amst) 2015;36:146–155. doi: 10.1016/j.dnarep.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryan DS, Ransom M, Adane B, York K, Hesselberth JR. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 2014;24(9):1534–1542. doi: 10.1101/gr.174052.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding J, Taylor MS, Jackson AP, Reijns MA. Genome-wide mapping of embedded ribonucleotides and other noncanonical nucleotides using emRiboSeq and EndoSeq. Nat Protoc. 2015;10(9):1433–1444. doi: 10.1038/nprot.2015.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95(5):717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 21.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486(7404):496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015;115(6):2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods. 2014;11(2):203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBroom LD, Sadowski PD. Contacts of the ABF1 protein of Saccharomyces cerevisiae with a DNA binding site at MATa. J Biol Chem. 1994;269(23):16455–16460. [PubMed] [Google Scholar]
- 25.Zavala AG, Morris RT, Wyrick JJ, Smerdon MJ. High-resolution characterization of CPD hotspot formation in human fibroblasts. Nucleic Acids Res. 2014;42(2):893–905. doi: 10.1093/nar/gkt912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pehrson JR, Cohen LH. Effects of DNA looping on pyrimidine dimer formation. Nucleic Acids Res. 1992;20(6):1321–1324. doi: 10.1093/nar/20.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smerdon MJ, Conconi A. Modulation of DNA damage and DNA repair in chromatin. Prog Nucleic Acid Res Mol Biol. 1999;62:227–255. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 28.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svedruzić ZM, Wang C, Kosmoski JV, Smerdon MJ. Accommodation and repair of a UV photoproduct in DNA at different rotational settings on the nucleosome surface. J Biol Chem. 2005;280(48):40051–40057. doi: 10.1074/jbc.M509478200. [DOI] [PubMed] [Google Scholar]
- 30.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618(1-2):65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters R, van Eijk P, Reed S. Histone modification and chromatin remodeling during NER. DNA Repair (Amst) 2015;36:105–113. doi: 10.1016/j.dnarep.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer GP, Drouin R, Riggs AD, Holmquist GP. Binding of transcription factors creates hot spots for UV photoproducts in vivo. Mol Cell Biol. 1992;12(4):1798–1804. doi: 10.1128/mcb.12.4.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodson ML, Lloyd RS. Structure-function studies of the T4 endonuclease V repair enzyme. Mutat Res. 1989;218(2):49–65. doi: 10.1016/0921-8777(89)90011-6. [DOI] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10(10):R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]