Significance
Successful response to stress requires that an organism rapidly direct its energy toward an appropriate survival response. The brain is central to successful survival decisions, and therefore its ability to allocate energetic resources precisely in response to stress is paramount. Glucocorticoid stress hormones have long been known to assist in the liberation of energy during stress via their ability to regulate the activity of the nuclear genome. The cellular powerhouse, the mitochondria, also contains a genome; herein we show that glucocorticoids, acting through their receptors, regulate the expression of mitochondrial genes in the brain. These findings demonstrate a direct molecular linkage between stress and mitochondrial function.
Keywords: nuclear receptors, allostasis, mitochondrial plasticity, brain metabolism, mitochondrial transcription
Abstract
Glucocorticoids (GCs) are involved in stress and circadian regulation, and produce many actions via the GC receptor (GR), which is classically understood to function as a nuclear transcription factor. However, the nuclear genome is not the only genome in eukaryotic cells. The mitochondria also contain a small circular genome, the mitochondrial DNA (mtDNA), that encodes 13 polypeptides. Recent work has established that, in the brain and other systems, the GR is translocated from the cytosol to the mitochondria and that stress and corticosteroids have a direct influence on mtDNA transcription and mitochondrial physiology. To determine if stress affects mitochondrially transcribed mRNA (mtRNA) expression, we exposed adult male rats to both acute and chronic immobilization stress and examined mtRNA expression using quantitative RT-PCR. We found that acute stress had a main effect on mtRNA expression and that expression of NADH dehydrogenase 1, 3, and 6 (ND-1, ND-3, ND-6) and ATP synthase 6 (ATP-6) genes was significantly down-regulated. Chronic stress induced a significant up-regulation of ND-6 expression. Adrenalectomy abolished acute stress-induced mtRNA regulation, demonstrating GC dependence. ChIP sequencing of GR showed that corticosterone treatment induced a dose-dependent association of the GR with the control region of the mitochondrial genome. These findings demonstrate GR and stress-dependent transcriptional regulation of the mitochondrial genome in vivo and are consistent with previous work linking stress and GCs with changes in the function of brain mitochondria.
Stress places energetic demands on an organism that require rapid, coordinated physiologic and energetic responses to ensure the successful response of an organism to environmental challenges. This response requires that energetic resources be shunted from one tissue to serve the needs of another to permit the organism to surmount whatever obstacle the stress represents. The stress response is coordinated to a large part by glucocorticoids (GCs), a class of adrenal steroids released during and after stress that also mediate diurnal variations in physiology. GCs act as regulators of a number of physiologic processes, notably the liberation of energy stores via gluconeogenesis and other means and by suppressing glucose uptake and activity in some tissues, such as adipose- and immune tissue (1, 2) that may be unnecessary to overcome an immediate threat or to reduce the negative impact of excess glucose in neurons (3).
GCs are also important factors in the orchestration of whole-organism behavioral and physiological responses such as the sleep/wake cycle and other circadian rhythms, which also can be disrupted by stress. In the brain, GCs mediate what may be regarded as both the adaptive and maladaptive effects of stress, although it should be emphasized that what is regarded as “adaptive” is context dependent. For example, moderate GC levels improve performance on spatial memory tasks and adaptive immunity, but high, sustained levels of the steroids can impair performance (4, 5). This biphasic pattern, in which acute and chronic stress or corticosteroid exposure have different or even opposed effects, holds for other effects in the nervous system, such as dendritic plasticity and neurogenesis (6, 7), and may serve to adapt the nervous system to the energetic demands of environment in many cases, although it may induce pathology in others (8).
GCs are classically understood to act via the mineralocorticoid receptors (MRs) and GC receptors (GRs). These nuclear receptors, like others in their class, act as ligand-activated transcription factors, which translocate from the cytoplasm to the nucleus to exert their effects by binding to response elements in the genome. It has become apparent that this view of corticosteroid action is incomplete, because a number of rapid, nongenomic effects of GCs and other steroid hormones have been observed (9). Given the effects of GCs on metabolism and energetics across tissues, as well as their impact on the structure and function of individual neurons and synapses, it is logical to look to the organelle responsible for cellular energy generation, the mitochondria, for evidence of GC action.
The mitochondria are present in neurons and glia in numbers varying from hundreds to thousands per cell. Mitochondria are highly dynamic within neurons, undergoing fission and fusion in response to local conditions and being trafficked to regions of high metabolic demand such as the synapse, where they sit at the nexus of multiple biochemical pathways that regulate neural plasticity (10). The mammalian mitochondria contains a 16.6-kb circular chromosome known as mitochondrial DNA (mtDNA), which encodes 37 genes in total, 13 of which code for proteins essential to the function of the mitochondrial electron transport chain. Mutations in the mtDNA have been linked to serious diseases that show a marked effect on the central nervous system in both humans and mouse model systems (11, 12). Steroid hormone receptors, including estrogen receptors and GRs, translocate into brain mitochondria. Studies in cultured cells suggest that mtDNA contains GC response elements (GREs) and that these may have a functional role in regulating mtDNA transcription (13–18). In both cultured neurons and in prefrontal cortex tissue, GC treatment showed a biphasic effect on mitochondrial oxidation, membrane potential, and calcium capacity, with lower doses improving these measures and higher or chronic doses and treatments causing deficits. These effects were correlated with the neuroprotective effects of the GC corticosterone (15). Previous studies have established that GRs can alter the transcription of mitochondrial genes in cultured cells, but GR regulation of mtDNA transcription has not yet been confirmed as occurring in vivo (13). These findings raise the questions: Does GR act to influence transcription in brain mitochondria? Furthermore, do stress or corticosteroids regulate expression of the genes of mtDNA?
Although our previous work focused on the translocation of GR into the mitochondria in the prefrontal cortex, we chose to examine the hippocampus in this study because of its sensitivity to stress. We addressed questions about the capacity of stress, and corticosterone, acting via the GR to alter expression of mRNAs derived from the mitochondrial genome (mtRNA) in the presence or absence of endogenous corticosteroids, and we performed ChIP-sequencing (ChIP-seq) to demonstrate GR interaction with the mitochondrial genome in the rat hippocampus. However, we believe that the findings reported here likely generalize beyond that brain region.
Results
Acute Stress Decreases mtRNA Expression in the Rat Hippocampus.
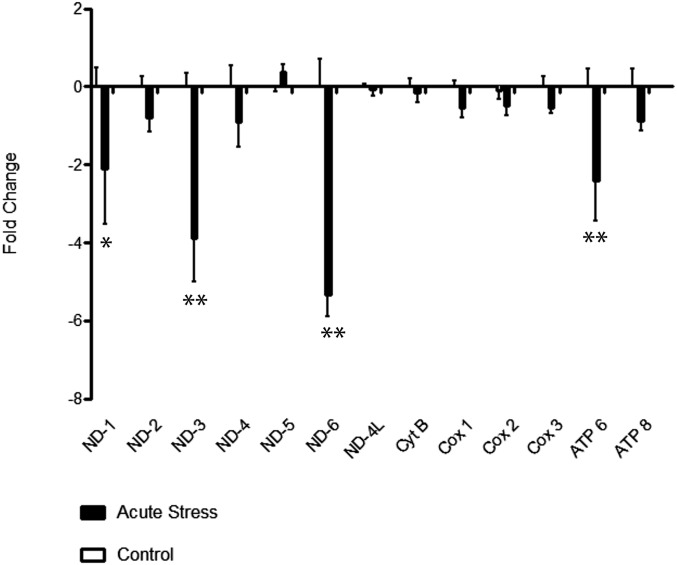
We first determined whether a single acute stressor had an effect on the expression of mtRNA in the hippocampus. We chose a time point 90 min after stress onset to fit with our previous observations of GR translocation into mitochondria (15). We subjected adult male Sprague–Dawley rats to acute immobilization stress and examined whether stress-induced changes in mitochondrial gene expression, as measured with RT-PCR using primers designed in the B.S.M. laboratory (SI Methods, Fig. S1, and Table S1), in the hippocampus, a brain region that expresses high levels of GR and is particularly stress-sensitive. Rats (n = 8) stressed acutely for 30 min and allowed to recover for 1 h showed a main effect of stress on overall mitochondrial gene expression (n = 8, F = 40.3, P < 0.0001) (Fig. 1): four mRNAs, NADH dehydrogenase 1, 3, and 6 (ND-1, ND-3, ND-6) and ATP synthase 6 (ATP-6), showed significant decreases of 50% or more (ND-1: P < 0.05; ND-3, ND-6, and ATP6: P < 0.001). (For mtDNA gene nomenclature, see Table 1.)
Fig. S1.
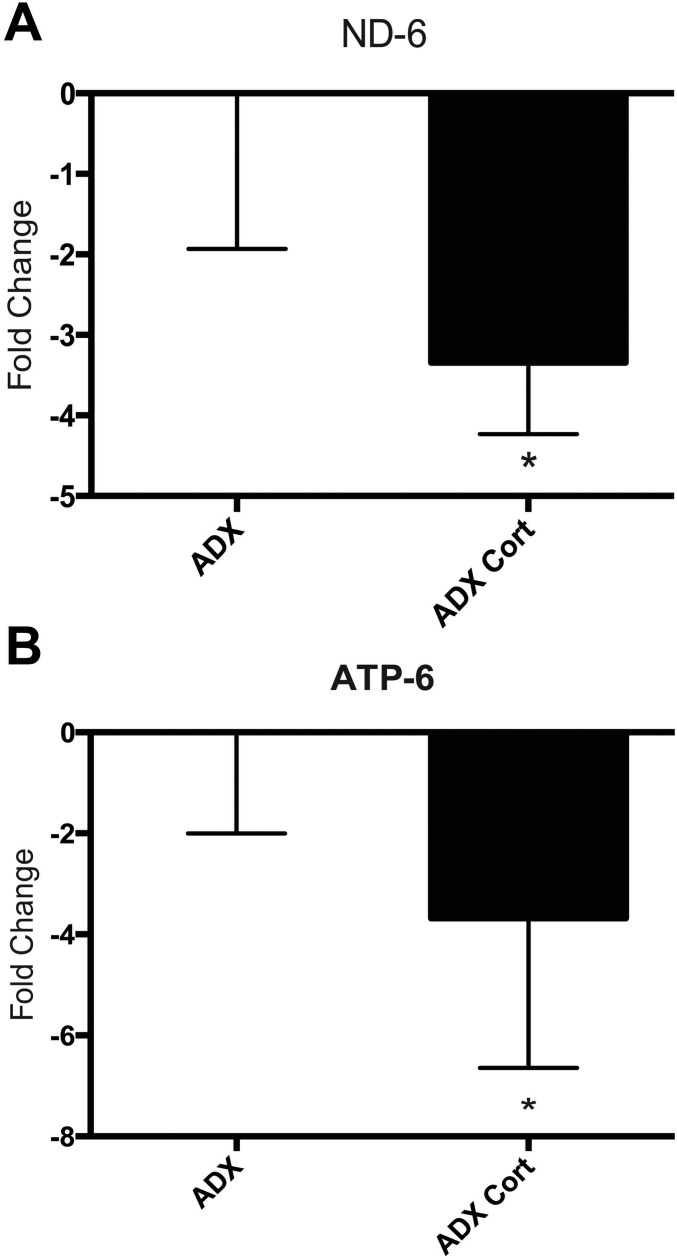
Acute administration of 300 μg corticosterone to adrenalectomized rats recapitulates the effects of acute stress on the expression of the mitochondrial genes ND-6 (A) and ATP-6 (B) (n = 6; *P < 0.03).
Table S1.
Primer sequences for rat mtRNA RT-PCR analyses
| Gene product | Forward | Reverse |
| mtDNA | ||
| ND-1 | CACCCCCTTATCAACCTCAA | ATTTGTTTCTGCGAGGGTTG |
| ND-2 | AACCCAAGCTACAGCCTCAA | GAAATTGCGAGAATGGTGGT |
| ND-3 | TAACATCACCTTATCCTTTATCCTC | GGCAGTTGCTATTATTGTAGTGG |
| ND-4 | TCCCACTCTTAATTGCCCTC | GAGGATGATGAATGGGTAGG |
| ND-4L | CTCCAACTCCATAATCTCCATAAC | GGCTAAACCTACTGCTGCTTC |
| ND-5 | ATCGAAGCCATCAACACGTG | GCAGTTATGGATGTGGCGATT |
| ND-6 | AGCCTTCACCTATTTATG | CACCCAGCCACCACTATC |
| Cytochrome B | CGGCTGACTAATCCGATACC | TGGGAGTACATAGCCCATGA |
| COX-1 | GACACCCGAGCCTACTTTAC | GCTATGATGGCGAATACTGC |
| COX-2 | GCTTACAAGACGCCACATCACC | CGTAGGGAGGGAAGGGCAAT |
| COX-3 | CAGCCTAGTTCCTACCCACGAC | CCCGTTGCTATGAAGAATGTTG |
| ATP-6 | CGAACCTGAGCCCTAATA | GTAGCTCCTCCGATTAGA |
| ATP-8 | TGCCACAACTAGACACATCCA | TGTGGGGGTAATGAAAGAGG |
Fig. 1.
Change in the expression of mitochondrial mRNA after acute immobilization stress and brief (1-h) recovery. There was a main effect of stress on mtRNA expression (n = 8, F = 40.3, P < 0.0001) and were significant decreases in ND-1 (*P < 0.05) and ND-3, ND-6, and ATP-6 expression (**P < 0.001).
Table 1.
mtDNA gene nomenclature
| Electron transport chain complex | Abbreviation | Gene name |
| Complex I NADH dehydrogenase | ND-1 | NADH dehydrogenase 1 |
| ND-2 | NADH dehydrogenase 2 | |
| ND-3 | NADH dehydrogenase 3 | |
| ND-4 | NADH dehydrogenase 4 | |
| ND-4L | NADH dehydrogenase 4L | |
| ND-5 | NADH dehydrogenase 5 | |
| ND-6 | NADH dehydrogenase 6 | |
| Complex III cytochrome B | Cytb | Cytochrome B |
| Complex IV cytochrome C oxidase | COX I | Cytochrome C oxidase I |
| COX II | Cytochrome C oxidase II | |
| COX III | Cytochrome C oxidase III | |
| Complex V ATP synthase | ATP-6 | ATP synthase 6 |
| ATP-8 | ATP synthase 8 |
Chronic Stress Increases Expression of ND-6 mRNA.
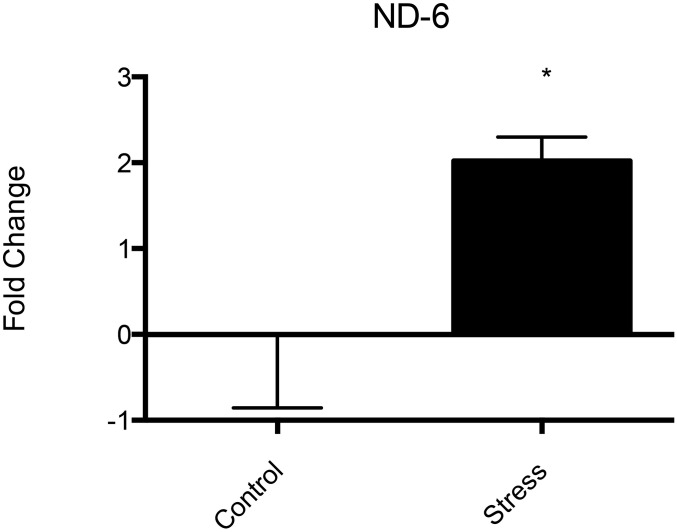
To assess the effect of chronic stress on mtRNA expression, we subjected rats to 21 d of immobilization stress for 1 h/d during the light phase of their cycle. Quantitative RT-PCR analysis of mtRNA expression revealed significant changes in the expression of only one gene, ND-6, which showed a significant (twofold ± 28%; P < 0.05; n = 8) increase in expression over controls (Fig. 2), a direction opposite that seen with acute mobilization stress.
Fig. 2.
Chronic (21-d) immobilization stress increased the expression of the mitochondrial ND-6 gene (n = 8; *P < 0.05) but had no significant effect on the expression of other mitochondrial genes.
Adrenalectomy Blocks the Stress-Induced Down-Regulation of mtRNA.
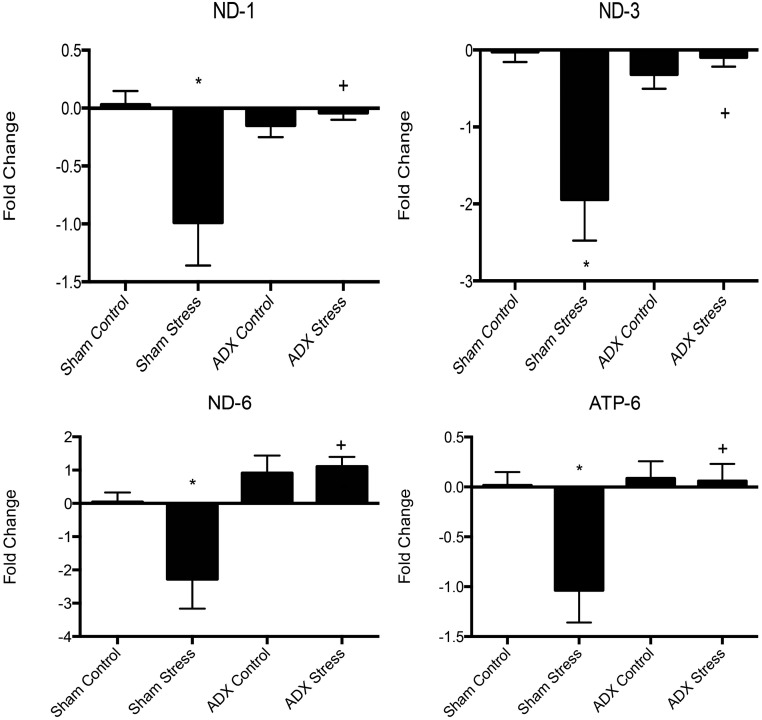
To determine if the acute stress-induced down-regulation of mtRNA was corticosteroid dependent, we removed the adrenal glands from adult male rats or performed sham surgery with an incision in the same region of the flanks. These animals then were subjected to either a single 30-min episode of acute stress, followed by 1 h of recovery or were allowed to pass the time in their home cage (controls). Sham-operated animals subjected to acute stress showed significant decreases in the expression of ND-1, ND-3, ND-6, and ATP-6, as had been observed previously (n = 5–8; P < 0.05 versus sham control), whereas neither the adrenal-excised control rats nor the adrenal-excised rats subjected to stress conditions showed significant changes in mtRNA expression relative to sham controls (Fig. 3). Thus, the acute stress effect is dependent upon adrenal secretions.
Fig. 3.
Acute immobilization stress in adrenalectomized rats did not have an effect on the expression of mtRNA (n = 5–8; *P < 0.05 versus sham controls; +P < 0.05 versus sham stress, no difference from sham control). ADX, adrenalectomized.
The GR Binds the Control Region of the Mitochondrial Genome in the Presence of Corticosterone.
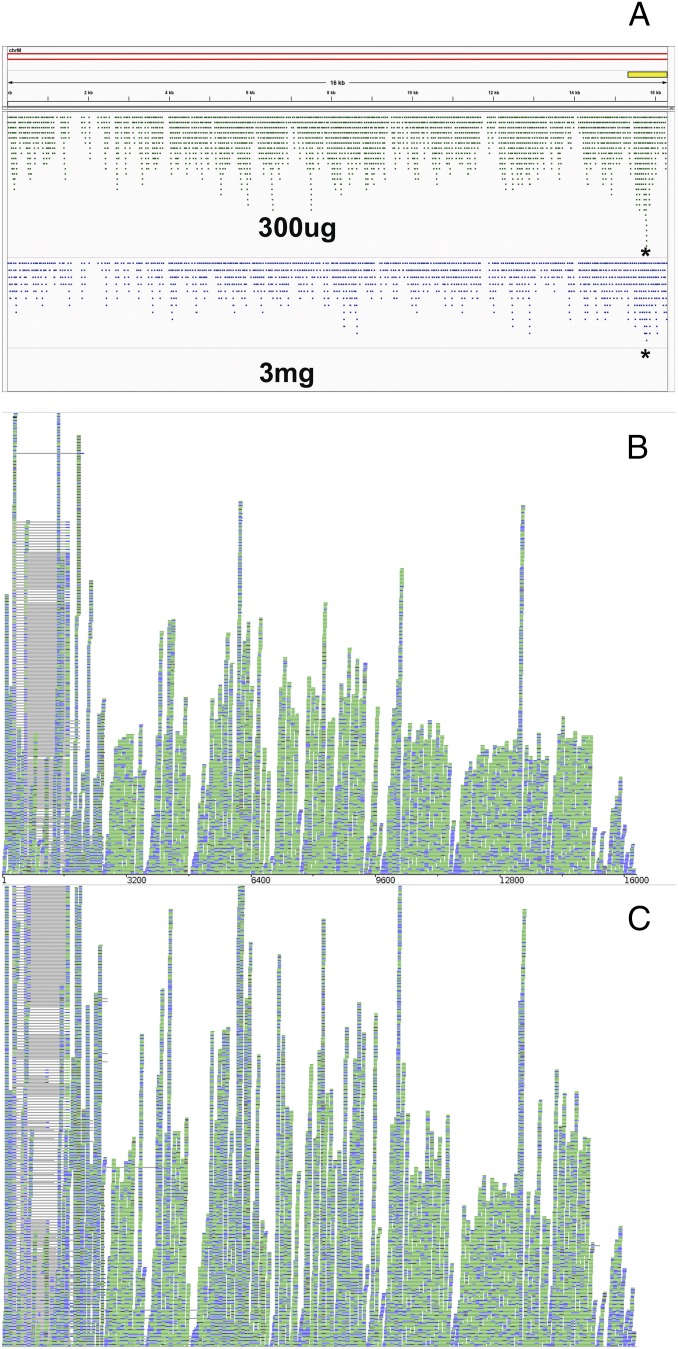
Although in previous studies we have established that GR translocates into mitochondria under conditions of stress or elevated corticosteroid levels (15), and others have shown that GR binding to mtDNA occurs in liver extracts (19), it has not been established whether the GR actually binds to the brain mitochondrial genome as it does to genes in the cell nucleus. To determine if corticosteroid-induced GR binding to mtDNA occurs in the brain, we treated rats with a moderate (300 μg/kg) or high (3,000 μg/kg) dose of corticosterone or performed ChIP-seq with anti-GR antibodies on hippocampal chromatin from those animals. We then examined the resulting data for evidence of significant GR-binding peaks in the mitochondrial genome. We found a significant [fivefold; n = 6; P < 1 × 10−20; false-discovery rate (FDR) < 1 × 10−11%] increase in GR binding to the D-loop control region of the mitochondrial genome in animals treated with the 300-μg/kg dose and a similar but less pronounced fourfold increase in GR binding to the same region in animals treated with the 3,000 μg/kg dose (n = 6; P < 1 × 10−9; FDR < 0.001%) (Fig. 4). In contrast to the response to acute stress seen in intact rats, injection of 300 μg/kg corticosterone also caused a moderate but significant up-regulation of the expression of most mitochondrial genes as measured by RNA-seq (ND-3, ND-4, Cox-2, ND-4L, ATP-6, ATP-8, ND-5, Cox-3, Cox-1, and Cytb; P < 5.0 × 10−5; n = 6) and a moderate down-regulation of ND1 and ND2 (P < 5.0 × 10−5; n = 6) (Fig. 4 B and C). These results also were confirmed by RT-PCR (SI Methods).
Fig. 4.
(A) GR interaction with the mitochondrial chromosome after a dose of either 300 μg/kg (green) or 3,000 μg/kg (blue) [n = 6; *P < 1 × 10−9; FDR < 0.001%; visualization produced using IGV (48)]. The yellow bar denotes the location of the D-loop control region. (B) Representative Bamtools image of mt-mRNA RNA-seq reads from vehicle-treated adrenalectomized rat hippocampus. (C) mt-mRNA RNA-seq reads from acute (300 μg/kg) corticosterone-treated adrenalectomized rat hippocampus. Corticosterone treatment resulted in significantly higher expression of 10 of 13 mtDNA genes (n = 6; P < 0.00005; q < 0.0008).
SI Methods
RT-PCR.
RNA was extracted from dissected and flash-frozen hippocampal tissue using a Qiagen RNeasy lipid tissue extraction kit. The RNA was reverse transcribed using a Roche Transcriptor reverse transcriptase kit, and the cDNA (1 μg per reaction) was used to quantify transcript levels on an Applied Biosciences 7900 RT-PCR system. Quantification was via ΔΔCT to control GAPDH primers.
ChIP-Seq.
DNA for ChIP-seq was prepared from hippocampi dissected and fixed with 1% formaldehyde. Six hippocampi per group were pooled into two technical replicates per group, sheared, and immunoprecipitated as described in ref. 43. They then were ligated and amplified for sequencing on an Illumina Genome Analyzer GA1, which yielded 1.9 × 107 total GR reads and 1.1 × 106 unique alignments to the rat genome (rn4).
mtRNA RT-PCR Primers.
Primers for quantitative RT-PCR of mtRNA were synthesized by Integrated DNA Technologies. Sequences are shown in Table S1.
Discussion
It recently has been proposed that mitochondrial allostatic load, the mitochondrial analog of allostatic load in the broader context of stress biology (20), represents a potentially significant pathogenic agent in stress-related disease and cellular aging (21). Mitochondrial allostatic load can be defined as the sum of the potentially damaging changes to mitochondrial structure and function as a consequence of stress exposure. It has been demonstrated previously that stress and corticosteroids induce translocation of the GR into brain mitochondria and that this translocation alters mitochondrial physiology (15). The GR is a well-described ligand-activated nuclear transcription factor, and the mtDNA contains GREs that can regulate mtDNA transcription in cultured human cell lines (13, 14). These observations and the fact that GR translocates to the mitochondria in response to treatment with its endogenous ligand suggested that it might regulate transcription of the brain mitochondrial genome as well. Here we have shown that stress regulates mitochondrial gene transcription in a dynamic, corticosteroid-dependent manner and that the GR interacts intimately with the mtDNA in response to corticosterone treatment. Interestingly, GR binding to mtDNA was maximal at the 300-μg/kg dose but was lower at the 3 mg/kg dose, suggesting the descending limb of the inverted-U dose–response relationship often observed with corticosteroids (22). Furthermore, the GR binding or interaction site we describe coincides closely with the GRα GRE identified previously by the Sekeris laboratory in human cell lines and mouse hepatocytes (14, 19). It is worth noting that, although our ChIP-Seq data show an interaction with a putative GRE, the method does not demonstrate that a direct GR–DNA binding event has occurred, because other elements of the transcriptional machinery may be involved. Although the GR, androgen, estrogen, and thyroid hormone receptors have been shown to be present in mitochondria (reviewed in ref. 23), the MR has not been found there, and our previous work in neurons failed to show translocation of MR to mitochondria under either basal or corticosteroid-treated conditions (15), suggesting that the effects observed here are GR rather than MR mediated.
Acute vs. Chronic Stress Effects.
In the nuclear genome, acute stress and GC treatment regulate a larger number of genes than chronic stress (24–26), and this pattern holds true for our observations of mitochondrial gene expression as well. Acute stress caused a significant main effect on mitochondrial gene expression and a significant down-regulation of 4 of 13 mitochondrial genes examined, although all but one gene showed reduced expression. Three of these genes (ND-1, ND-3, and ND-6) are subunits of complex I of the respiratory chain, the first step in which electron flow is initiated, and one (ATP-6) is a component of the ATP-synthase, the last step in which mitochondrial ATP is synthesized.
Chronic stress had a significant effect only on the expression of the ND-6 subunit of complex I of the electron transport chain. In contrast to acute stress, chronic stress induced an up-regulation of ND-6 mRNA. ND-6 mRNA also showed the largest fold down-regulation by acute stress. These findings fit well with the body of literature examining the differing effects of acute versus chronic stress on gene expression. Acute stress often shows a pattern of broad, larger-magnitude changes in gene expression, whereas chronic stress shows a less pronounced change, often in a different direction from the response evoked by acute stress or corticosteroid treatment (e.g., refs. 24, 27, and 28).
Role of Adrenal Steroids.
Adrenalectomy followed by acute corticosterone treatment produced increases in most mitochondrial RNAs in the present study, suggesting that the interactions of GR with mtDNA are complex and context specific and that adrenal steroids actually may oppose the effects of acute stress on some of the mitochondrial genes. Previous work has shown similarly complex relations between corticosteroid treatment, acute and chronic stress, and gene expression for a number of individual nuclear genes (e.g., refs. 28–31). More recently, global nuclear gene-expression analyses have shown that the three manipulations have largely nonoverlapping effects on mRNA expression patterns in the rodent hippocampus (24, 32). Thus, our observations of the regulation of the mitochondrial genome by stress and corticosteroids are consonant with previous observations of nuclear gene expression. The time course of hypothalamic–pituitary–adrenal axis activation and adaptation as a result of stress is significant, and future work will be needed to examine the role of the timing and duration of stress in GR–mtDNA interactions.
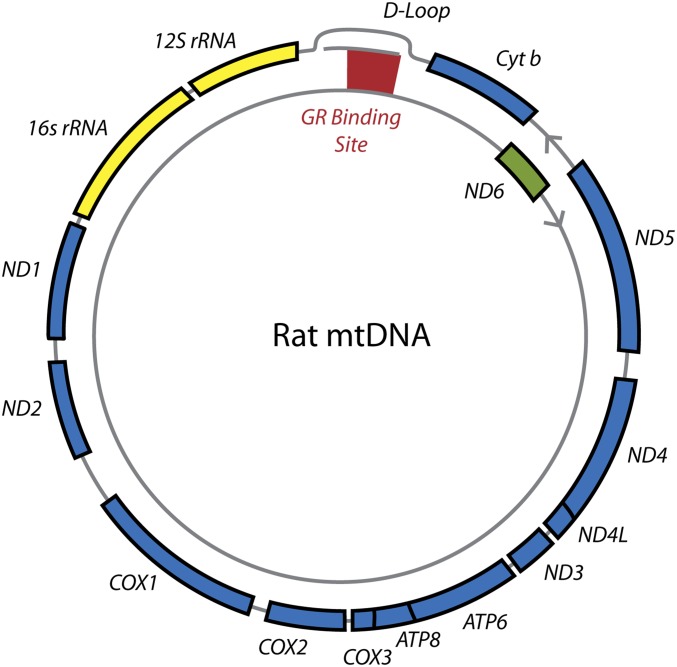
Acute stress produced a global down-regulation in mtRNA expression, although only four genes showed statistically significant changes in expression in our analysis. The reasons for this differential down-regulation are not entirely clear but likely have to do with the polycistronic nature of the mitochondrial genome. In addition, in the rat mitochondrial genome, ND-6 is located close to the GR-binding peak we describe here, as is ND-1 (Fig. 5). ND-3 and ATP-6 also are relatively close to GREs (14). Although we did not observe significant GR interaction with the presumptive GRE near ND-3 and ATP-6, such interaction may have occurred at an earlier time point than was captured in our ChIP-seq analysis. ND-6 is the only protein-coding gene to be oriented in the opposite direction, on the light strand, rather than on the heavy strand with the rest of the protein-coding genes (33); this position may explain why it is more labile in response to GC stimulation and suggests a directional effect of GR binding on transcription. ND-6 also appears to be expressed at levels several fold lower than the other mtDNA genes across all the analyses we performed.
Fig. 5.
Relation of corticosterone-induced GR binding in rat hippocampus to the structure of the rat mitochondrial genome. Heavy-strand mitochondrial genes are shown in blue, light-strand genes are shown in green, and the two mt-rRNA genes are shown in yellow. The D-loop binding site observed at both 300-μg and 3-mg doses of corticosterone is shown as a red box in the D-loop control region.
Functional Implications.
The ND-6 gene codes for NADH-ubiquinone oxidoreductase chain subunit 6, which is a subunit of complex I of the electron transport chain. Exposure to high doses of corticosterone reduce complex I activity and increase reactive oxygen species (ROS) production and consequent molecular damage in rat PC12 cells (34). Consistent with our findings, complex III, which is the other principal source of mitochondrial ROS, did not show a change in activity (34). Chronic stress has been shown to produce results similar to the effects of high-dose corticosteroids on mitochondrial function in intact rodent brains (35–37), suggesting that the transcriptional activity of GR in brain mitochondria is a consistent mechanism for the transduction of psychological stress into molecular pathology. Mitochondrial dysfunction in general, and complex I deficits specifically, have been shown to be associated with a number of neurodegenerative and neuropsychiatric disorders, including Parkinson’s disease, Alzheimer’s disease, and bipolar disorder (38–40). It is possible the increase in ND-6 caused by chronic stress is a compensatory response to the mitochondrial allostatic load or overload, an idea supported by previous work showing deficits in mitochondrial function as a consequence of chronic GC exposure (15). If so, then our findings might support the hypothesis that stress, by increasing mitochondrial allostatic load, could account for much of the individual variability in the age of onset and symptom severity seen in many of these disorders.
In conclusion, although our work here has focused on the hippocampus for methodological reasons (higher GR expression), we and our collaborators have shown previously that the GR translocates into mitochondria in neurons of the prefrontal cortex (15), and the available evidence indicates this translocation is a general process in neurons and is likely to apply broadly to all corticosteroid target tissues, including glial cells, which were included in our approach (16). In a similar vein, it is possible that the effects we observed were dampened by the cellular heterogeneity of our sample, generating potential false negatives. Determining the cell-type specificity of these GR–mtDNA interactions will be an important focus for future work. Nonetheless, these findings demonstrate the capacity of nuclear receptors to regulate the transcription of genes located on the mitochondrial genome. Furthermore, we have shown that this GR dependent regulation of mitochondrial mRNA can occur in response to environmental stress, a known factor in a number of neurologic and psychiatric diseases (41). These findings have implications for a variety of phenomena in neuroscience, from synaptic plasticity, by which local control of mitochondrial activity could be altered, to neurodegenerative diseases in which mitochondrial dysfunction is widely implicated and in which stress can be a contributing factor (10, 42).
Methods
Animals.
For the stress experiments, 90-d-old male Sprague–Dawley rats were housed in the Rockefeller University animal facility on a 12-h light/dark cycle. They were housed in pairs with ad libitum access to food and water. ChIP-seq was performed on male Sprague–Dawley rats housed at Leiden University Medical Center under the same husbandry conditions (43). RNA-seq experiments were performed at the University of Massachusetts Boston. All experiments were approved by the University of Massachusetts, Boston or the Rockefeller University Institutional Animal Care and Use Committee, in accordance with the Guide for Care and Use of Laboratory Animals (44), or by the Local Committee for Animal Health, Ethics, and Research of the University of Leiden, in accordance with European Commission Council Directive of November 1986.
Acute Immobilization Stress.
Rats (n = 8) were restrained for 30 min in well-ventilated plastic bag restrainers, released, and allowed to recover in their home cage for 1 h. Then they were killed, and their hippocampi were processed for RT-PCR (see Table S1 for primer sequences).
Chronic Immobilization Stress.
Rats (n = 8) were restrained daily for 2 h in plastic bag restrainers for 3 wk. After the last restraint they were released and allowed to recover in their home cage for 1 h. Then they were killed, and their hippocampi were processed for RT-PCR.
Adrenalectomy and Acute Stress.
Adrenalectomies and sham surgeries were performed as described previously in ref. 31. Rats were allowed to recover for 4 d after surgery and were provided with 0.9% saline to drink. They then were subjected to acute immobilization or were allowed to remain in their home cage (controls). Then they were killed, and their hippocampi were processed for RT-PCR.
RT-PCR.
RT-PCR was performed in accordance with standard procedures. SYBR Green primers designed for the mRNAs of interest were used (SI Methods).
GR ChIP-seq was performed as described in ref. 43. Rats were adrenalectomized and injected with 300 or 3,000 μg/kg of corticosterone 1 h before they were killed and tissue was processed for ChIP. Hippocampi were bilaterally dissected, finely chopped, fixed with 1% formaldehyde, and then flash frozen on dry ice until ready for further processing. ChIP was performed with anti-GR antibody (H-300) or normal rabbit IgG (Santa Cruz Biotechnology). Single-end sequencing of 35-bp reads was performed using an Illumina Genome Analyzer at the Leiden Genome Technology Center, Leiden University Medical Center.
Read Alignment and Peak Analysis.
Reads were aligned using the Burroughs–Wheeler Aligner (45) to the rat rn5 reference genome, and BED files were generated using BEDtools (46). Peak calling was performed using MACS2 (47) with a P value cut-off of 1.0 × 10−5, a fold cutoff of 30, and a λ value of 1,000. FDRs were calculated by MACS2. Integrated Genomics Viewer (IGV) was used to visualize results (48).
RNA-Seq.
RNA-seq was performed on rats (n = 6) adrenalectomized and treated with either 300 μg/kg corticosterone or vehicle (as in the GR ChIP-seq process described above). Rats were killed 1 h after treatment, and their hippocampi were harvested and flash frozen before RNA extraction. RNA was processed for RNA-seq by the Massachusetts General Hospital (MGH) Next-Generation Sequencing Core on an Illumina HiSeq sequencer. Sequenced .fastq files were transferred via File Transfer Protocol from MGH to the R.G.H. laboratory local galaxy server. Quality scores were converted to Sanger format with the Fastq Groomer tool (v1.0.4) (49) using default settings. The rn5 mitochondrial DNA sequence was downloaded from the University of California, Santa Cruz genome table browser and indexed with Bowtie2 (v0.6) (50) and default settings. The paired-end groomed files were mapped to this index using Tophat (v0.9) (51). After read mapping, the accepted-hits.bam files were run through Cufflinks (v2.2.1.0) to identify transcripts, using the ensembl annotation .gtf file to identify mitochondrial genes. The cufflinks transcripts were merged with the cuffmerge tool (v2.2.1.0) with default settings, and Cuffdiff (v2.2.1.3) (52) was used to look for differences between the two groups.
Acknowledgments
We thank Dr. Martin Picard (Center for Mitochondrial and Epigenomic Medicine, Children’s Hospital of Philadelphia Research Institute, now of Columbia University College of Physicians and Surgeons) for his thoughtful comments on this manuscript. This research was supported by a Betz Family National Association for Research on Schizophrenia and Depression Young Investigator Award (to R.G.H.), National Institutes of Health Grant MH41256, and the Hope for Depression Research Foundation (B.S.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The ChIP-seq and RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE84202 and GSE84249).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602185113/-/DCSupplemental.
References
- 1.Scheffler IE. 2008. Mitochondria (Wiley-Liss, Hoboken, NJ), 2nd Ed.
- 2.Dhabhar FS. Effects of stress on immune function: The good, the bad, and the beautiful. Immunol Res. 2014;58(2-3):193–210. doi: 10.1007/s12026-014-8517-0. [DOI] [PubMed] [Google Scholar]
- 3.de Leon MJ, et al. Cortisol reduces hippocampal glucose metabolism in normal elderly, but not in Alzheimer’s disease. J Clin Endocrinol Metab. 1997;82(10):3251–3259. doi: 10.1210/jcem.82.10.4305. [DOI] [PubMed] [Google Scholar]
- 4.McEwen BS. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol Rev. 2007;87(3):873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- 5.Dhabhar FS, Malarkey WB, Neri E, McEwen BS. Stress-induced redistribution of immune cells—From barracks to boulevards to battlefields: A tale of three hormones—Curt Richter Award winner. Psychoneuroendocrinology. 2012;37(9):1345–1368. doi: 10.1016/j.psyneuen.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McEwen BS. Central effects of stress hormones in health and disease: Understanding the protective and damaging effects of stress and stress mediators. Eur J Pharmacol. 2008;583(2-3):174–185. doi: 10.1016/j.ejphar.2007.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picard M, McEwen BS. Mitochondria impact brain function and cognition. Proc Natl Acad Sci USA. 2014;111(1):7–8. doi: 10.1073/pnas.1321881111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers B, McKlveen JM, Herman JP. Glucocorticoid actions on synapses, circuits, and behavior: Implications for the energetics of stress. Front Neuroendocrinol. 2014;35(2):180–196. doi: 10.1016/j.yfrne.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Kloet ER. Functional profile of the binary brain corticosteroid receptor system: Mediating, multitasking, coordinating, integrating. Eur J Pharmacol. 2013;719(1-3):53–62. doi: 10.1016/j.ejphar.2013.04.053. [DOI] [PubMed] [Google Scholar]
- 10.Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60(5):748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharpley MS, et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151(2):333–343. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFarland R, Taylor RW, Turnbull DM. A neurological perspective on mitochondrial disease. Lancet Neurol. 2010;9(8):829–840. doi: 10.1016/S1474-4422(10)70116-2. [DOI] [PubMed] [Google Scholar]
- 13.Psarra AM, Sekeris CE. Glucocorticoids induce mitochondrial gene transcription in HepG2 cells: Role of the mitochondrial glucocorticoid receptor. Biochim Biophys Acta. 2011;1813(10):1814–1821. doi: 10.1016/j.bbamcr.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Psarra AM, Solakidi S, Sekeris CE. The mitochondrion as a primary site of action of steroid and thyroid hormones: Presence and action of steroid and thyroid hormone receptors in mitochondria of animal cells. Mol Cell Endocrinol. 2006;246(1-2):21–33. doi: 10.1016/j.mce.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Du J, et al. Dynamic regulation of mitochondrial function by glucocorticoids. Proc Natl Acad Sci USA. 2009;106(9):3543–3548. doi: 10.1073/pnas.0812671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Psarra AM, Sekeris CE. Glucocorticoid receptors and other nuclear transcription factors in mitochondria and possible functions. Biochim Biophys Acta. 2009;1787(5):431–436. doi: 10.1016/j.bbabio.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Irwin RW, et al. Progesterone and estrogen regulate oxidative metabolism in brain mitochondria. Endocrinology. 2008;149(6):3167–3175. doi: 10.1210/en.2007-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irwin RW, et al. Selective oestrogen receptor modulators differentially potentiate brain mitochondrial function. J Neuroendocrinol. 2012;24(1):236–248. doi: 10.1111/j.1365-2826.2011.02251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demonacos C, Djordjevic-Markovic R, Tsawdaroglou N, Sekeris CE. The mitochondrion as a primary site of action of glucocorticoids: The interaction of the glucocorticoid receptor with mitochondrial DNA sequences showing partial similarity to the nuclear glucocorticoid responsive elements. J Steroid Biochem Mol Biol. 1995;55(1):43–55. doi: 10.1016/0960-0760(95)00159-w. [DOI] [PubMed] [Google Scholar]
- 20.McEwen BS, Wingfield JC. The concept of allostasis in biology and biomedicine. Horm Behav. 2003;43(1):2–15. doi: 10.1016/s0018-506x(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 21.Picard M, Juster RP, McEwen BS. Mitochondrial allostatic load puts the ‘gluc’ back in glucocorticoids. Nat Rev Endocrinol. 2014;10(5):303–310. doi: 10.1038/nrendo.2014.22. [DOI] [PubMed] [Google Scholar]
- 22.Joëls M. Corticosteroid effects in the brain: U-shape it. Trends Pharmacol Sci. 2006;27(5):244–250. doi: 10.1016/j.tips.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Psarra AM, Sekeris CE. Steroid and thyroid hormone receptors in mitochondria. IUBMB Life. 2008;60(4):210–223. doi: 10.1002/iub.37. [DOI] [PubMed] [Google Scholar]
- 24.Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2014;19(11):1171–1178. doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datson NA, et al. Previous history of chronic stress changes the transcriptional response to glucocorticoid challenge in the dentate gyrus region of the male rat hippocampus. Endocrinology. 2013;154(9):3261–3272. doi: 10.1210/en.2012-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datson NA, et al. The transcriptional response to chronic stress and glucocorticoid receptor blockade in the hippocampal dentate gyrus. Hippocampus. 2012;22(2):359–371. doi: 10.1002/hipo.20905. [DOI] [PubMed] [Google Scholar]
- 27.Polman JA, et al. Glucocorticoids modulate the mTOR pathway in the hippocampus: Differential effects depending on stress history. Endocrinology. 2012;153(9):4317–4327. doi: 10.1210/en.2012-1255. [DOI] [PubMed] [Google Scholar]
- 28.Hunter RG, et al. Regulation of CART mRNA by stress and corticosteroids in the hippocampus and amygdala. Brain Res. 2007;1152:234–240. doi: 10.1016/j.brainres.2007.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reagan LP, et al. Chronic restraint stress up-regulates GLT-1 mRNA and protein expression in the rat hippocampus: Reversal by tianeptine. Proc Natl Acad Sci USA. 2004;101(7):2179–2184. doi: 10.1073/pnas.0307294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Autry AE, et al. Glucocorticoid regulation of GLT-1 glutamate transporter isoform expression in the rat hippocampus. Neuroendocrinology. 2006;83(5-6):371–379. doi: 10.1159/000096092. [DOI] [PubMed] [Google Scholar]
- 31.Hunter RG, et al. Regulation of kainate receptor subunit mRNA by stress and corticosteroids in the rat hippocampus. PLoS One. 2009;4(1):e4328. doi: 10.1371/journal.pone.0004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubin TG, Gray JD, McEwen BS. Experience and the ever-changing brain: What the transcriptome can reveal. BioEssays. 2014;36(11):1072–1081. doi: 10.1002/bies.201400095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang VM, Young AH, Tan H, Beasley C, Wang JF. Glucocorticoids increase protein carbonylation and mitochondrial dysfunction. Horm Metab Res. 2013;45(10):709–715. doi: 10.1055/s-0033-1345119. [DOI] [PubMed] [Google Scholar]
- 35.Gong Y, Chai Y, Ding JH, Sun XL, Hu G. Chronic mild stress damages mitochondrial ultrastructure and function in mouse brain. Neurosci Lett. 2011;488(1):76–80. doi: 10.1016/j.neulet.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Rezin GT, et al. Inhibition of mitochondrial respiratory chain in brain of rats subjected to an experimental model of depression. Neurochem Int. 2008;53(6-8):395–400. doi: 10.1016/j.neuint.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Bennett MC, Mlady GW, Fleshner M, Rose GM. Synergy between chronic corticosterone and sodium azide treatments in producing a spatial learning deficit and inhibiting cytochrome oxidase activity. Proc Natl Acad Sci USA. 1996;93(3):1330–1334. doi: 10.1073/pnas.93.3.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manczak M, Park BS, Jung Y, Reddy PH. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: Implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 2004;5(2):147–162. doi: 10.1385/NMM:5:2:147. [DOI] [PubMed] [Google Scholar]
- 39.Schapira AH, et al. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54(3):823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 40.Anglin RE, Mazurek MF, Tarnopolsky MA, Rosebush PI. The mitochondrial genome and psychiatric illness. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(7):749–759. doi: 10.1002/ajmg.b.32086. [DOI] [PubMed] [Google Scholar]
- 41.McEwen BS. Protection and damage from acute and chronic stress: Allostasis and allostatic overload and relevance to the pathophysiology of psychiatric disorders. Ann N Y Acad Sci. 2004;1032:1–7. doi: 10.1196/annals.1314.001. [DOI] [PubMed] [Google Scholar]
- 42.Manji H, et al. Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci. 2012;13(5):293–307. doi: 10.1038/nrn3229. [DOI] [PubMed] [Google Scholar]
- 43.Polman JA, de Kloet ER, Datson NA. Two populations of glucocorticoid receptor-binding sites in the male rat hippocampal genome. Endocrinology. 2013;154(5):1832–1844. doi: 10.1210/en.2012-2187. [DOI] [PubMed] [Google Scholar]
- 44.Committee on Care and Use of Laboratory Animals 2010. Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blankenberg D, et al. Galaxy Team Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26(14):1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]