Significance
A substrate-conjugated polyubiquitin chain is accepted as the “canonical” proteasomal degradation signal. Using a cellular (human and yeast) proteomic screen in the exclusive presence of nonpolymerizable ubiquitin, we show that a large group of proteins is degraded by the proteasome following monoubiquitination. The screen also unraveled polyubiquitin-dependent substrates, as they are stabilized in the presence of this ubiquitin mutant. Notably, monoubiquitination- and polyubiquitination-dependent substrates display distinct important characteristics. Monoubiquitinated proteins are of lower molecular mass and of lesser structural disorder. The two groups can be assigned to defined cellular pathways. Furthermore, some of the characteristics are confined to either human or yeast cells, suggesting that the mechanism of action/recognition of the ubiquitin system in the two organisms are different somehow.
Keywords: monoubiquitination, 26S proteasome, protein degradation, ubiquitin replacement
Abstract
The “canonical” proteasomal degradation signal is a substrate-anchored polyubiquitin chain. However, a handful of proteins were shown to be targeted following monoubiquitination. In this study, we established—in both human and yeast cells—a systematic approach for the identification of monoubiquitination-dependent proteasomal substrates. The cellular wild-type polymerizable ubiquitin was replaced with ubiquitin that cannot form chains. Using proteomic analysis, we screened for substrates that are nevertheless degraded under these conditions compared with those that are stabilized, and therefore require polyubiquitination for their degradation. For randomly sampled representative substrates, we confirmed that their cellular stability is in agreement with our screening prediction. Importantly, the two groups display unique features: monoubiquitinated substrates are smaller than the polyubiquitinated ones, are enriched in specific pathways, and, in humans, are structurally less disordered. We suggest that monoubiquitination-dependent degradation is more widespread than assumed previously, and plays key roles in various cellular processes.
Polymers of ubiquitin (Ub) are formed on proteasomal substrates in eukaryotic cells by the concerted action of three enzymes: the Ub-activating enzyme (E1), a Ub-carrier protein [E2; known also as Ub-conjugating enzyme (UBC)], and a Ub ligase (E3), which is the specific substrate-recognizing element of the system. The Ub chains typically consist of multiple moieties linked to one another via an isopeptide bond between the C-terminal Gly residue of the distal moiety and the ε-NH2 group of Lys48 of the proximal one (1).
In addition, the system can also catalyze modification by a single Ub moiety (monoubiquitination) or multiple single Ub moieties (multiple monoubiquitinations), each modifying a distinct lysine residue (2). Generally, monoubiquitination has been conceived as a nondestructive signal. Furthermore, it has been suggested that efficient proteasomal targeting requires a chain with a minimal length of four Ub moieties (3). Monoubiquitination is known to be involved in multiple biological processes. For example, monoubiquitination of proteins containing a ubiquitin-binding domain (UBD) often mediates autoinhibition by a UBD–Ub interaction (4, 5). Signal transduction by membrane receptors, such as the EGFR, is attenuated by monoubiquitination-mediated receptor internalization (6). The subcellular localization of small GTPases is controlled, among other posttranslational modifications, by monoubiquitination (7, 8), and histone monoubiquitination regulates nucleasomal structure, thus affecting gene expression (9). Emerging reports indicate, however, that several substrates can be degraded following monoubiquitination (10–13). Partial degradation/processing of the p105 precursor of NF-κB, which results in release of the p50 active subunit of the transcription factor, is dependent on multiple monoubiquitinations (14). Importantly, these findings demonstrate that the proteasome can recognize a single Ub moiety (moieties) and imply the existence of monoubiquitination vs. polyubiquitination “decision” mechanisms. In this context, a previous study has suggested that the chain length required for proteasomal degradation is determined by the size of the substrate, and possibly other characteristics that affect the affinity of the modified substrate to the proteasome. Specifically, it was suggested that substrates smaller than 150 aa are degraded following monoubiquitination, whereas longer substrates require longer chains. Thus, a dynamic model was proposed, according to which the chain elongates to a point where the affinity to the proteasome is high enough to secure a stable binding of the conjugated substrate, its concomitant detachment from the E3, and its subsequent degradation (15). Another study demonstrated that restricting the number of ubiquitinatable Lys residues can switch the mode of modification necessary for degradation from multiple monoubiquitinations to polyubiquitination, suggesting that, in the cell, the masking of Lys residues by protein–protein interactions or posttranslational modifications can affect the mode of ubiquitination (11).
However, all of these studies have been carried out using specific substrates. Therefore, general conclusions regarding monoubiquitination-dependent degradation mechanisms, the population of substrates that are degraded following this modification, and importantly, whether they have common distinct characteristics, have remained limited.
In this study, we used a systematic proteomic approach for the identification and characterization of monoubiquitination-dependent proteasomal substrates. By silencing the endogenous WT Ub followed by expression of nonpolymerizable lysineless Ub, we identified numerous substrates in both mammalian and yeast cells that are targeted by the proteasome following monoubiquitination or multiple monoubiquitinations. Interestingly, we confirmed a previous hypothesis (15) that there is a correlation between the length of the substrate and its requirement for either monoubiquitination or polyubiquitination. Also, the monoubiquitinated substrates are enriched in specific pathways [e.g., oxidative stress, carbohydrate transport, and components of the ubiquitin–proteasome system (UPS) itself] and, in humans, are structurally less disordered.
Results
Establishing a System for Induction of Monoubiquitination in Cells.
In general, to model monoubiquitination (or multiple monoubiquitinations), we silenced endogenous Ub expression and replaced it with a lysineless Ub (UbK0). This nonpolymerizable Ub species, in which all seven Lys residues were replaced with Arg, can modify each Lys in the target substrate only once and cannot be further ubiquitinated (16–19).
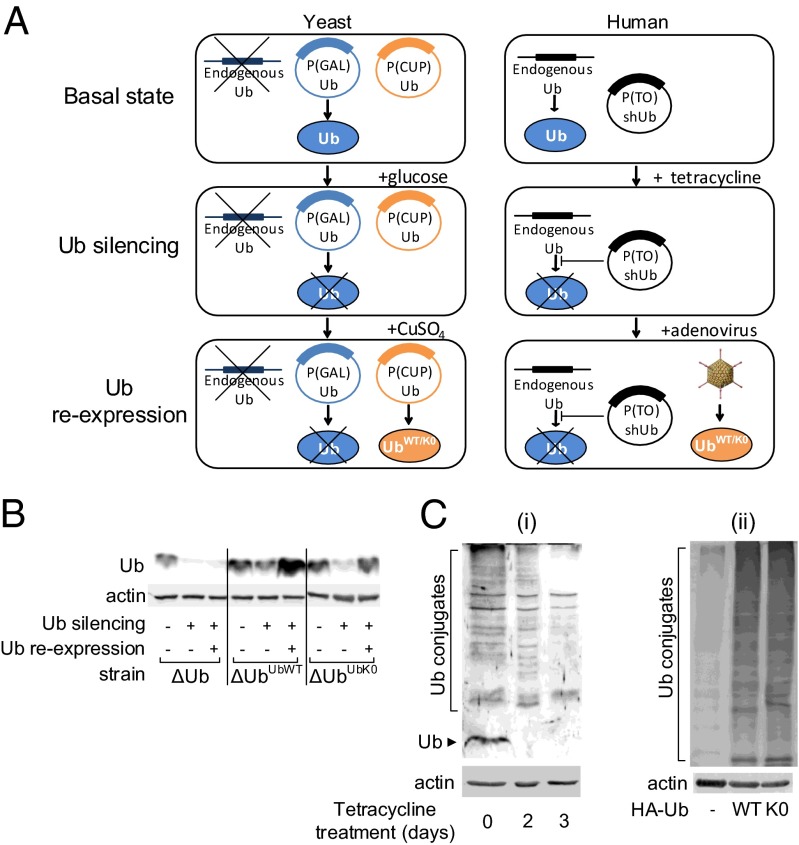
To study monoubiquitination in yeast, we used a modification of a previously described Ub replacement method (20). Briefly, all of the Ub-coding genes were deleted and replaced by galactose-inducible Ub (ΔUb strain), and either copper-inducible UbWT or UbK0 (ΔUbUbWT or ΔUbUbK0, respectively). Thus, Ub expression can be silenced by adding glucose, and Ub reexpression can be induced by adding copper to the growth medium (Fig. 1A). To validate Ub silencing and reexpression, ΔUb, ΔUbUbWT, and ΔUbUbK0 yeast strains were treated with glucose and copper. As shown in Fig. 1B, Ub expression was efficiently suppressed, and both UbWT and UbK0 were markedly expressed.
Fig. 1.
Replacement of endogenous Ub by UbK0 in yeast and mammalian cells. (A) Workflow describing Ub silencing and reexpression (see a detailed description under Experimental Procedures). (B) Ub replacement in yeast cells. ΔUb, ΔUbUbWT, and ΔUbUbK0 yeast cells were treated for Ub silencing and Ub reexpression as indicated. Yeast cells were analyzed by trichloroacetic acid (TCA) lysis followed by SDS/PAGE and Western blotting (WB) using the indicated antibodies. (C) Ub replacement in human cells. (i) Ub silencing. To silence Ub, U2OSshUb cells were treated with tetracycline (1 μg/mL) for the indicated times. (ii) Ub reexpression. Following Ub silencing, cells were infected with viral vectors expressing UbWT or UbK0. In both panels, lysates were analyzed via SDS/PAGE followed by WB using the indicated antibodies.
To assess monoubiquitination in human cells, we used a modification of a previously described Ub replacement model in human cultured cells (21). Briefly, endogenous Ub is silenced in U2OS cells by a Ub-specific tetracycline-induced shRNA (shUb), and either HA-UbWT or HA-UbK0 is expressed following infection with an adenoviral vector (Fig. 1A). To evaluate Ub silencing efficiency, we monitored Ub and Ub conjugates level in U2OSshUb cells following tetracycline treatment. As demonstrated in Fig. 1Ci, the level of both Ub and Ub–protein conjugates were significantly decreased. In the endogenous Ub-silenced cells, both HA-UbWT and HA-UbK0 were efficiently expressed and assembled into high–molecular-mass conjugates following adenoviral expression (Fig. 1Cii). It should be noted that the pattern of conjugation appears similar for both UbWT and UbK0 expression. This is probably due to the numerous substrates with a broad range of molecular mass that are conjugated, and from the possibility that many of them are modified by multiple monoubiquitinations.
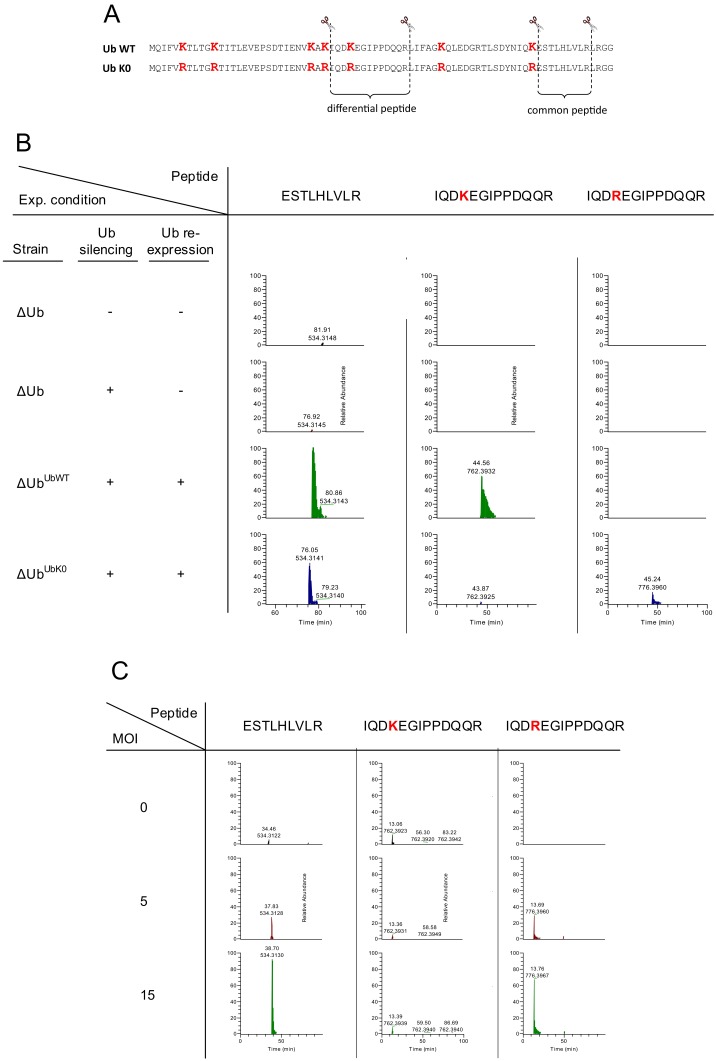
To demonstrate Ub replacement using an additional method, we quantified Ub using mass spectrometry (MS). As shown in Fig. S1A, tryptic digestion of UbWT and UbK0 yields both common and differential MS-detectable peptides. To assess Ub replacement in yeast, we treated ΔUbUbK0 cells with glucose and copper, and quantified Ub-derived peptides by MS. As illustrated in Fig. S1B, UbK0 was markedly more abundant than endogenous Ub. To evaluate UbK0 expression in human cells, we overexpressed HA-UbK0 via adenoviral infection using increasing multiplicities of infection (MOIs). As displayed in Fig. S1C, UbK0 expression level was MOI dependent and significantly exceeded the level of endogenous Ub.
Fig. S1.
Mass-spectrometric verification of replacement of endogenous Ub by UbK0 in yeast and mammalian cells. (A) Ubwt- and UbK0-derived tryptic peptides used for mass-spectrometric quantification of UbK0 expression. Human Ub sequence is shown. The indicated differential and common peptides correspond also to the yeast Ub. (B) ΔUb, ΔUbUbWT, and ΔUbUbK0 yeast cells were treated for Ub replacement as indicated. In all experiments, total Ub abundance (K0 and WT species) was assessed based on the relative MS intensity of the common peptide, and UbWT and UbK0 abundances were assessed based on the relevant differential peptide. Ub replacement and MS analysis were performed as described in Experimental Procedures and SI Experimental Procedures, respectively. MS peaks representing the indicated peptides are shown with values representing column retention times (upper value; in minutes) and m/z ratio (lower value). (C) U2OSshUb cells were either left untreated or treated for Ub replacement with UbK0 using the indicated MOI. Data were processed as described in B.
Taken together, these data demonstrate the effectiveness of our Ub replacement strategies and suggest that our experimental systems are suitable for studying protein monoubiquitination.
Systematic Identification of Monoubiquitination-Dependent Proteasome Substrates.
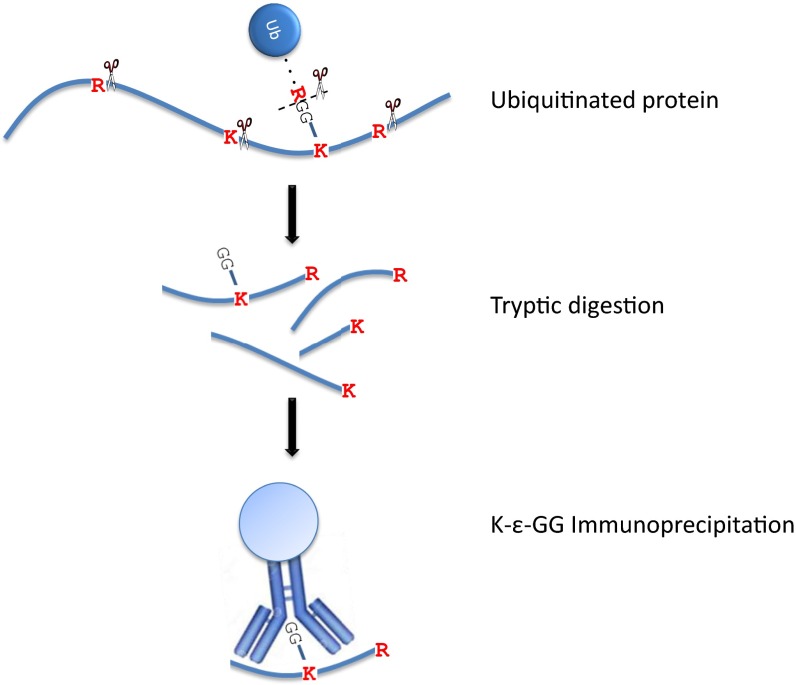
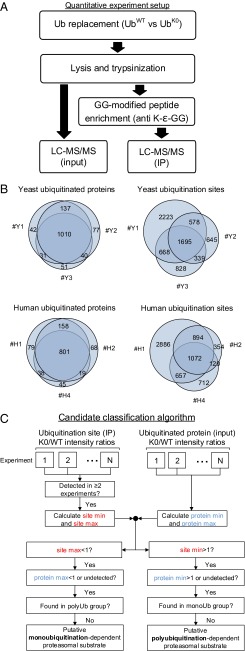
To identify substrates that are degraded following monoubiquitination, we replaced Ub with either UbK0 or UbWT (as a control). We then used anti–K-ε-GG immunoprecipitation (Fig. S2) to enrich and quantify by MS GlyGly-modified peptides derived from tryptic digestion of ubiquitinated proteins (Fig. S2). This method enabled us also to identify ubiquitination sites (Ubsites). To verify that the ubiquitinated proteins serve indeed as proteolytic substrates, we monitored also the level of nonmodified peptides derived from them (Fig. 2A). To ascertain reproducibility, we performed several independent biological replicates for each model organism, using both stable isotope labeling by amino acids in cell culture (SILAC) and label-free quantification (Table S1). As shown in Fig. 2B, identification of the proteins was quite reproducible. Similar to previous data (22), identification of Ubsites within proteins was less reproducible.
Fig. S2.
Ubiquitination sites enrichment. Schematic illustration of isolation of GG-modified peptides using anti–K-ε-GG immunoprecipitation. For detailed description, see Experimental Procedures.
Fig. 2.
Identification of monoubiquitination- and polyubiquitination-dependent proteasome substrates. (A) Experimental workflow. See a detailed description in Results. (B) Ubiquitination sites and ubiquitinated proteins identified in independent experiments. See Table S1 for experiment names. (C) The algorithm used for classification to monoubiquitination- and polyubiquitination-dependent substrates.
Table S1.
Quantitative experiments for the identification of monoubiquitination-dependent proteasomal substrates
| No. experiment | Model organism | Experimental setup | No. of identified peptides |
| Y1 | Yeast | Label free | 5,164 |
| Y2 | Yeast | Label free | 3,257 |
| Y3 | Yeast | Label free | 3,530 |
| H1 | Human | Label free | 5,509 |
| H2 | Human | Label free | 2,440 |
| H3 | Human | SILAC | 1,156 |
| H4 | Human | Label free | 2,561 |
This established experimental setup enabled us to discriminate between proteins degraded following modification by monoubiquitination (or multiple monoubiquitination), and those that are degraded only following polyubiquitination. We calculated the MS signal intensity ratios following either UbK0 or UbWT expression for both proteins and immunoprecipitated ubiquitinated sites (denoted “protein K0/WT intensity ratio” and “site K0/WT intensity ratio,” respectively; see Dataset S1 for raw data). As illustrated in Fig. 2C and based on K0/WT intensity ratios, we classified ubiquitinated proteins to putative monoubiquitination- and polyubiquitination-dependent proteasome substrates as follows.
Monoubiquitination-dependent proteasomal substrates are expected to be unaffected by UbK0 expression. Alternatively, as UbK0 expression renders proteasomes less occupied by polyubiquitination-dependent substrates, increased proteasome availability may result in accelerated degradation of monoubiquitination-dependent substrates. Thus, we required these substrates to (i) have a site K0/WT ratio <1; (ii) have a detectable MS signal in at least two independent experiments; and (iii) have a protein K0/WT ratio <1 (if the protein is detectable).
The degradation of a polyubiquitination-dependent substrate is expected to be inhibited upon UbK0 expression. Consequently, we expect their level to increase. Thus, we require these substrates to (i) have a site K0/WT ratio >1; (ii) have a detectable MS signal in at least two independent experiments; and (iii) have a protein K0/WT ratio >1 (if the protein is detectable).
A small fraction (<3%) of proteins were identified as belonging to the two groups. These proteins were excluded from the survey.
Applying these criteria in both yeast and human cells, we identified 82 and 220 monoubiquitination-dependent and 416 and 303 polyubiquitination-dependent putative proteasomal substrates, respectively (Dataset S2). Samples of each group are presented in Table 1, describing gene names and ubiquitinated Lys positions. As expected, the polyubiquitination-dependent substrate group included several previously suggested proteasomal substrates, e.g., Pdc1p, Ole1p, and Eno1p in yeast, and HIF1A, POLD2, and IER3 in human cells (23, 24).
Table 1.
Putative ubiquitination-mediated proteasomal substrates
| Entry | Protein name | Gene name | Ubiquitination positions | Ubiquitination mode/organism |
| Q00416 | Helicase SEN1 | SEN1 | 464 | Mono/yeast |
| P22943 | 12-kDa heat shock protein | HSP12 | 12, 18, 23, 30, 36, 50, 64, 86, 88, 108 | |
| Q08969 | Protein GRE1 | GRE1 | 7 | |
| Q12734 | Transcription factor CSR2 | CSR2 | 670 | |
| Q92325 | Cullin-associated NEDD8-dissociated protein 1 homolog | LAG2 | 7 | |
| Q04602 | Vacuolar basic amino acid transporter 4 | VBA4 | 368 | |
| P36035 | Carboxylic acid transporter protein homolog | JEN1 | 607 | |
| P01094 | Protease A inhibitor 3 | PAI3 | 16, 18, 31, 32, 50 | |
| Q12358 | α-Ketoglutarate-dependent sulfonate dioxygenase | JLP1 | 114 | Poly/yeast |
| P08679 | Citrate synthase, peroxisomal | CIT2 | 96, 208, 454 | |
| Q05637 | Phosphate metabolism protein 6 | PHM6 | 19 | |
| P40483 | Putative zinc metalloproteinase YIL108W | YIL108W | 5, 348, 524 | |
| P07347 | N-terminal acetyltransferase A complex catalytic subunit ARD1 | ARD1 | 191 | |
| P00924 | Enolase 1 | ENO1 | 195, 409 | |
| P06169 | Pyruvate decarboxylase isozyme 1 | PDC1 | 8 | |
| P21147 | Acyl-CoA desaturase 1 | OLE1 | 457 | |
| Q92890 | Ubiquitin fusion degradation protein 1 homolog | UFD1L | 279 | Mono/human |
| Q9BT67 | NEDD4 family-interacting protein 1 | NDFIP1, N4WBP5, PSEC0192, PSEC0223 | 83 | |
| Q9P2T1 | GMP reductase 2 | GMPR2 | 190 | |
| Q92621 | Nuclear pore complex protein Nup205 | NUP205, C7orf14, KIAA0225 | 69 | |
| Q9H9T3 | Elongator complex protein 3 | ELP3 | 338, 392, 517, 544 | |
| P17174 | Aspartate aminotransferase, cytoplasmic | GOT1 | 97, 99 | |
| Q5JVF3 | PCI domain-containing protein 2 | PCID2, HT004 | 213 | |
| O95302 | Peptidyl-prolyl cis-trans isomerase FKBP9 | FKBP9, FKBP60, FKBP63 | 527 | |
| Q5JW28 | Double-stranded RNA-binding protein Staufen homolog 1 | STAU1 | 127 | Poly/human |
| P36543 | V-type proton ATPase subunit E 1 | ATP6V1E1, ATP6E, ATP6E2 | 10 | |
| Q9NWF9 | E3 ubiquitin-protein ligase RNF216 | RNF216, TRIAD3, UBCE7IP1, ZIN | 487, 584, 773 | |
| Q9NS91 | E3 ubiquitin-protein ligase RAD18 | RAD18, RNF73 | 115, 376 | |
| Q12834 | Cell division cycle protein 20 homolog | CDC20 | 136 | |
| Q16665 | Hypoxia-inducible factor 1α | HIF1A, BHLHE78, MOP1, PASD8 | 377, 389, 674, 709 | |
| P46695 | Radiation-inducible immediate-early gene IEX-1 | IER3, DIF2, IEX1, PRG1 | 84 | |
| P49005 | DNA polymerase δ-subunit 2 | POLD2 | 267 |
Candidate Substrate Validation.
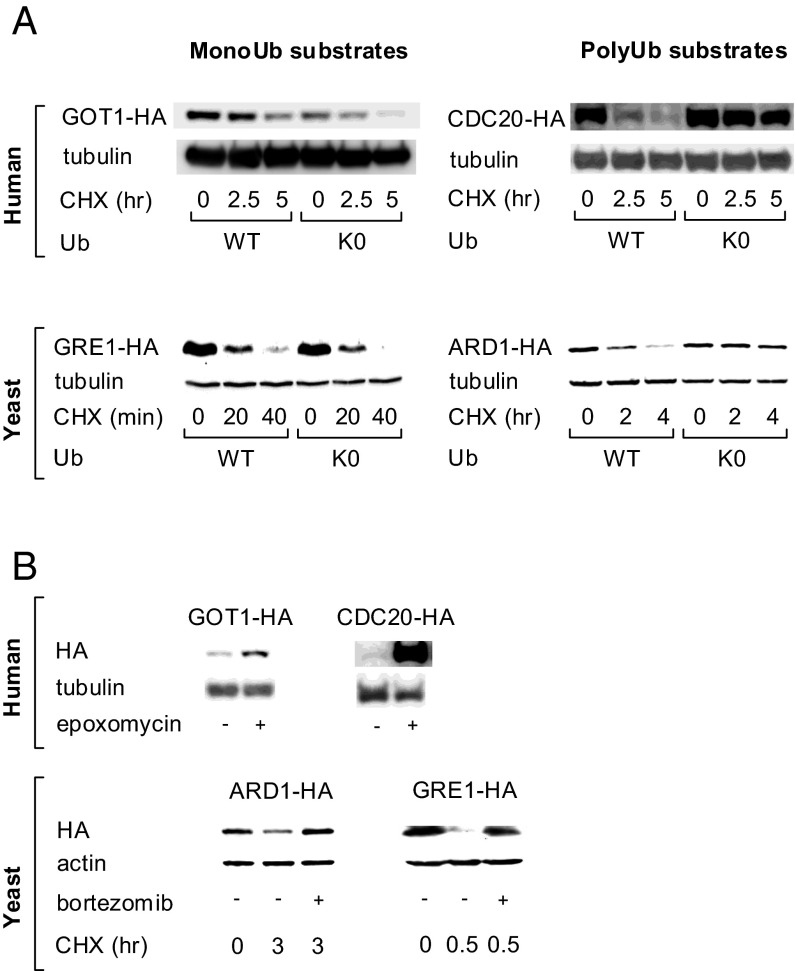
To validate the results of our algorithm and experimental setup, we monitored the cellular stability (using cycloheximide chase) of randomly sampled representative candidate substrates following Ub replacement. As demonstrated in Fig. 3A, replacing UbWT with UbK0 stabilized Ard1p in yeast and CDC20 in human cells (polyubiquitination-dependent substrates; Table 1). In contrast, the predicted monoubiquitination-dependent substrates, Gre1p in yeast and GOT1 in human cells, remained unstable. All four substrates were clearly degraded by the 26S proteasome, as they were stabilized following treatment with a proteasome inhibitor (Fig. 3B). Taken together, these results strongly suggest that our experimental setup is suitable for the systematic identification of monoubiquitination-dependent proteasomal substrates.
Fig. 3.
Validation of representative candidate substrates degraded by the proteasome following monoubiquitination and polyubiquitination. (A, Upper) MCF7 cells were cotransfected with plasmids coding for GOT1-HA or CDC20-HA along with plasmids coding for UbWT or UbK0 as indicated. (Lower) ΔUbUbWT and ΔUbUbK0 yeast cells were transformed with plasmids coding for Gre1p-HA or Ard1p-HA, and Ub replacement was carried out as described in Fig. 1. In all experiments, substrate stability was monitored as described in Experimental Procedures. (B, Upper) MCF7 cells were transfected with either GOT1-HA or CDC20-HA, followed by the treatment with epoxomicin (2 µM, 24 h) as indicated. (Lower) ΔPDR5 yeast cells (strain Y12409 from the EUROSCARF collection) were transformed with plasmids coding for either Ard1p-HA or Gre1p-HA. Strains were treated with cycloheximide (CHX) and bortezomib (100 µM each) as indicated. Samples were collected at the indicated time points. Cell lysates were analyzed via SDS/PAGE followed by WB using the indicated antibodies.
Physical Characteristics of Protein Substrates Play a Role in Their Mode of Ubiquitination: (i) Structural Disorder.
Evidently, a significant number of proteins are degraded following monoubiquitination (and probably also multiple monoubiquitinations) in both yeast and human cells. This observation challenges the prevailing paradigm of polyubiquitination being the prerequisite for protein degradation. Intriguingly, bioinformatics analysis of the data reveals that yeast and human cells significantly differ in their preferences for using the two types of signals. In yeast, the ratio of proteins degraded by polyubiquitination vs. monoubiquitination is 5.07 (416/82), whereas in human cells it is only 1.37 (303/220). Because the identification method was unbiased, the significant difference in the ratios likely points to important differences in the mode of recognition of the UPS in the two species. Yeast apparently operates much more by the traditional signal, polyubiquitin, whereas in humans monoubiquitination is used almost as frequently as polyubiquitination to mark proteins for degradation. The difference may arise from a combination of factors such as using different sets of conjugating enzymes (E2s and E3s), different preferences for local sequence and structural features of substrates, and the recognition elements of the proteasome. Some of these issues are addressed below.
In previous studies, it has been shown that protein ubiquitination and degradation are intimately linked with structural disorder. Intrinsically disordered protein regions lack a well-defined tertiary structure, yet they fulfill important functional roles linked with their highly flexible and adaptable structure (25–27). Structural disorder correlates with all three elements of degradation signals: location of the ubiquitin ligase recognition motif on substrates, the Lys residue(s) to which ubiquitin is attached, and a nearby long disordered region (LDR) (a region of at least 30 consecutive disordered residues) that initiates the unfolding of the substrate engaged with the proteasome (15, 28–30). Structural disorder may also be required for ubiquitin conjugation itself, in two different ways. It is repeatedly reported that the mutation of the Lys residue that is the site of modification does not usually abrogate sensitivity to UPS degradation (11, 31), because flexibility of the substrate enables multiple modifications on neighboring Lys resides. By a similar logic, the buildup of a polyubiquitin chain can also benefit from local structural disorder, because it enables the processive addition of subsequent ubiquitin moieties to the end of the growing polyubiquitin chain.
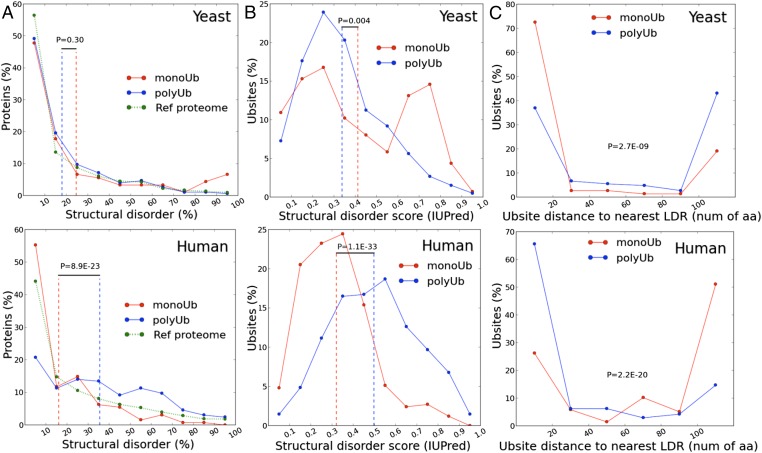
To test whether these disorder features may be related to the use of monoubiquitination vs. polyubiquitination, we tested whether predicted structural disorder of proteins differs in the different datasets (Fig. 4A). We found significant differences between yeast and human proteins: in yeast, the occurrence of structural disorder does not differ between monoubiquitinated and polyubiquitinated substrates, whereas in human cells, structural disorder prevails in polyubiquitinated substrates. Given that polyubiquitin is a stronger signal, we presume that structural disorder is primarily used for the buildup of the polyubiquitin chain. Because yeast relies more heavily on polyubiquitination, yet its polyubiquitinated substrates do not have more disorder, it is possible that their E3 ligases differ from humans in some critical features. These can be their number/redundancy, structural disorder, or binding heterogeneity, the structural disorder of which ensures processive addition of ubiquitin moieties in the ubiquitin chain.
Fig. 4.
Yeast and human substrates demonstrate distinct patterns of structural disorder. (A) Human polyubiquitinated substrates are more disordered than monoubiquitinated substrates. The distribution of overall disorder content in monoubiquitinated and polyubiquitinated substrates is plotted for yeast (Upper) and human (Lower). Overall disorder content is the fraction of predicted disordered residues in a given protein sequence. The distribution for the reference proteome corresponding to each species is shown for comparison. Dotted vertical lines correspond to the average values for the distribution. (B) Local disorder at the ubiquitination sites is more prominent in yeast monoubiquitination substrates and in human polyubiquitination substrates. The distribution of local disorder score is plotted for monoubiquitinated and polyubiquitinated sites from yeast (Upper) and human (Lower). The disorder profile for each sequence is first calculated using IUPred, and then the average disorder score of a 21-residue sequence window centered on each Ubsite is calculated. The distribution for the reference proteome corresponding to each species is shown for comparison. Dotted vertical lines correspond to the average values for the distribution. (C) The distance of Ubsites from their nearest LDR (see Experimental Procedures for definition) is plotted for yeast (Upper) and human (Lower) sites. The averages of the distributions are not shown here for the sake of clarity.
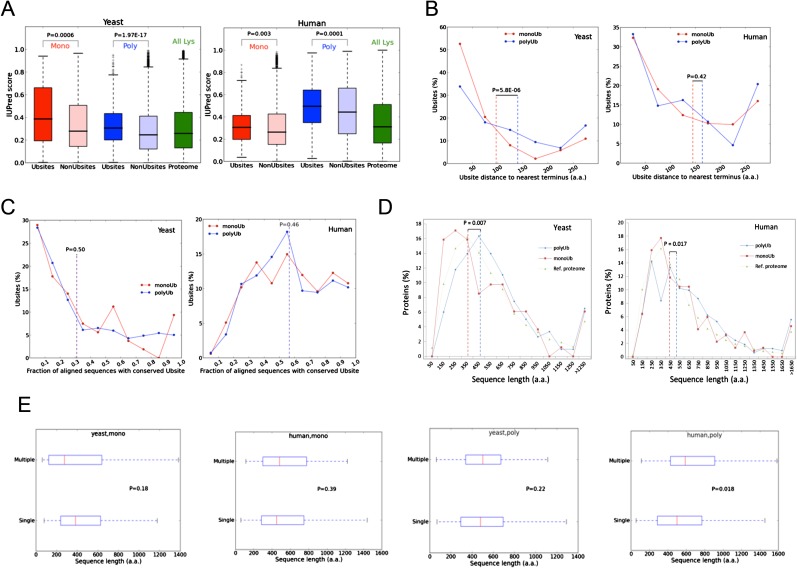
These differences also have to manifest themselves in the local disorder of the protein chain around the ubiquitinated Lys residues. In general, the sites of posttranslational modifications (PTMs) in proteins tend to exhibit local disorder, as studied in detail for phosphorylation (32) and also ubiquitination (28). In our entire dataset, the lysines that are the sites of ubiquitination tend to be locally disordered, but even more importantly, they show a highly characteristic difference between the two species. In yeast, monoubiquitination sites, whereas in human cells polyubiquitination sites, are significantly more disordered (Fig. 4B). Our interpretation, again, is that these differences point to likely differences in the UPS in the two species. In yeast, polyubiquitination is robust, and it is the rare monoubiquitination sites that have to be supported by local disorder, probably more for initiation of degradation than modification (29). In human cells, the sites of polyubiquitination are significantly more disordered, probably as much for the processive buildup of the polyubiquitin chain as for initiation of degradation (as suggested above). These are genuine differences and do not result from the natural tendency of lysines to be locally disordered, which, as disorder-promoting amino acids, tend to be located in disordered regions of proteins (33) (Fig. S3A).
Fig. S3.
Analyses of different characteristics of ubiquitinated substrates. (A) Boxplots showing the local disorder scores for Ubsites, NonUbsites, and all lysines in the reference proteome. Ubsites correspond to the identified ubiquitination sites in each protein, and NonUbsites are the remaining lysines from the proteolytic substrates. The y axis corresponds to the average IUPred score of 21-residue sequence windows flanking each lysine. (B) Plot showing the distance of yeast (Left) and human (Right) Ubsites from their nearer protein chain terminus. Dotted lines represent the average values of each distribution. (C) Plot showing Ubsite residue conservation across orthologous proteins (see SI Experimental Procedures for details). On the x axis, 0.0 corresponds to a completely variable Ubsite, whereas 1.0 indicates a fully conserved Ubsite. Dotted lines represent the average values of each distribution. (D) Length of substrates that undergo monoubiquitination- and polyubiquitination-dependent degradation. The plot is showing the length distribution of monoubiquitinated and polyubiquitinated substrates from yeast (Left) and human (Right). The reference proteome corresponding to each species is shown for comparison. Dotted vertical lines correspond to the median values for the distribution. (E) Boxplots of the distribution of sequence lengths for proteins containing a single or multiple Ubsites. The plots show data from yeast and human cells and from monoubiquitinated and polyubiquitinated proteins. Outliers are not shown.
The distinction between the signaling strength and functionality of monoubiquitin and polyubiquitin chains in yeast and human cells is also reflected in characteristic differences in the proximity of LDRs to Ubsites, which might be the sites of initiation of proteasomal degradation (28–30). The need of such assistance for monoubiquitination sites in yeast is apparent from the larger proportion of such sites that are close to an LDR (Fig. 4C), and probably also by a larger proportion of such sites that are close to the termini of the proteins, which are generally flexible (Fig. S3B).
Conservation in evolution can be an important indicator of the functionality of PTM sites. For example, functional phosphorylation sites (i.e., were shown to have a functional role by direct or indirect evidence, e.g., mutagenesis and/or functional assays) evolve significantly slower than those without evidence for a functional role (34). Interestingly, Ubsites are significantly more conserved in human cells than in yeast (Fig. S3C) (these differences are significant because the phylogenetic coverage of the respective multiple alignments is comparable). These results infer that yeast sites are under a significantly lower evolutionary pressure, which may point to the fact that they are functionally more promiscuous.
Physical Characteristics of the Proteins Play a Role in Their Mode of Ubiquitination: (ii) Size of the Protein.
In a previous study, we demonstrated that substrates degraded following conjugation of a single Ub moiety are usually shorter than 150 aa (15), which suggests that monoubiquitination is a weaker signal. Consistently, we found in this study a significant enrichment of shorter proteins among monoubiquitinated substrates in both yeast and human (Fig. S3D). This difference is not reflected in a preference of multiple modifications in longer proteins: there are no significant differences in the length of substrates that are degraded after the attachment of a single vs. multiple monoubiquitin or polyubiquitin chains (Fig. S3E).
Monoubiquitination- and Polyubiquitination-Dependent Substrates Are Differentially Enriched in Specific Biological Processes.
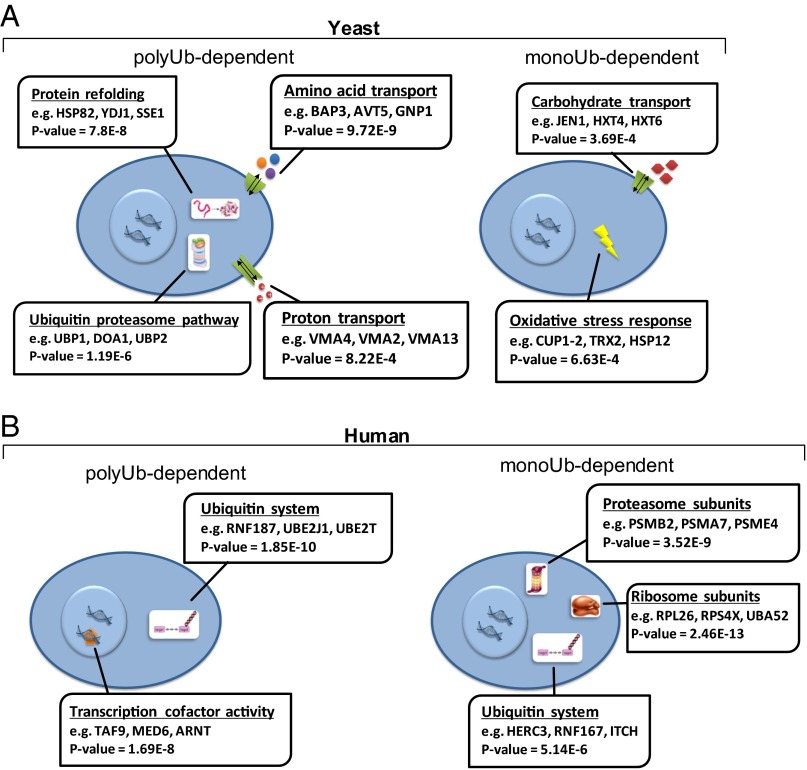
To characterize the two different modes of ubiquitination in a cellular function perspective, we searched for enriched gene ontology (GO) terms, using the GOrilla tool (35, 36). As illustrated in Fig. 5A, it appears that monoubiquitination-dependent proteasomal substrates are enriched in oxidative stress response and carbohydrate transport pathways. As presented in Fig. 5B, molecular-function–specific GO terms were also identified. For example, monoubiquitination-dependent substrates were found enriched among ribosomal and proteasomal subunits. Similarities between the monoubiquitinated and polyubiquitinated substrates were also observed, as Ub system components were enriched in both groups. Interestingly, however, monoubiquitinated substrates consisted of mostly Ub ligases, E3s (e.g., HERC3, ITCH, XIAP), whereas polyubiquitinated substrates were enriched with Ub-conjugating enzymes, E2s (e.g., UBE2J1, UBE2T, and UBE2L6).
Fig. 5.
Analysis of molecular functions and involvement in different cellular processes for monoubiquitination and polyubiquitination-dependent substrates. Monoubiquitination- and polyubiquitination-dependent substrate groups were analyzed for gene ontology (GO) term enrichment using the GOrilla tool, as described under Experimental Procedures. Representative proteins are shown for each enriched term, and the full list is shown in Table S2. (A) Biological-process GO term analysis for yeast substrates. (B) Molecular-function GO term analysis for human substrates.
Table S2.
Analysis of molecular functions and involvement in different cellular processes for monoubiquitination- and polyubiquitination-dependent substrates (related to Fig. 5)
| Organism | Ubiquitination type | Enriched GO term | Protein names | P value |
| Yeast | MonoUb | Carbohydrate transport | JEN1, HXT4, HXT6, HXT7, HXT5 | 3.69E-4 |
| Yeast | MonoUb | Oxidative stress response | CUP1-2, TRX2, HSP12, GRX7, SOD1, GRX2, YJR096W | 6.63E-4 |
| Yeast | PolyUb | Amino acid transport | BAP3, AVT5, GNP1, AGP2, AGP1, HNM1, HIP1, GAP1, MUP3, YCT1, AVT2, CAN1, MUP1, TAT2, VBA1, TAT1, FEN2, LYP1, BAP2 | 9.72E-9 |
| Yeast | PolyUb | Protein refolding | HSP82, YDJ1, SSE1, SSC1, SSE2, HSP60, SSA1, HSP104, CPR6, HSC82 | 7.80E-8 |
| Yeast | PolyUb | Proton transport | VMA4, VMA2, VMA13, ATP2, VMA1, VMA6 | 8.22E-4 |
| Yeast | PolyUb | Ubiquitin proteasome pathway | UBP1, DOA1, UBP2, UBX2, PRE3, UBP6, CDC48, PRE10, CDC53, UBR1, PRE5, RPT1, UFD2, YDR333C, SCL1, PRE2, PRE9, RPT2, PUP3, RPT5, RPT4, RPT3, CDC34, PRE6, RPN12 | 1.19E-6 |
| Human | MonoUb | Proteasome subunits | PSMB2, PSMA7, PSME4, PSMA3, PSMB10, PSMD1, PSMC2, PSMA2, PSMD3 | 3.52E-9 |
| Human | MonoUb | Ribosome subunits | RPL26, RPS4X, UBA52, RPS3, RPL19, RPL14, RPL18, RPS15A, RPS20, RPL11, RPS11, RPL10, RPL7A, RPS14, RPL6, RPL7, RPL4, RPS6, RPL3 | 2.46E-13 |
| Human | MonoUb | Ubiquitin system | HERC3, RNF167, ITCH, XIAP, SMURF2, RNF10, UBOX5, WWP2, NEDD4, SIAH1, MIB2, HECTD3, MIB1, TRIM5 | 5.14E-6 |
| Human | PolyUb | Transcription cofactor activity | TAF9, MED6, ARNT, TAF7, AEBP1, N4BP2L2, RYBP, ARID5B, ELK3, TFDP1, EID1, GMNN, ATF3, MED14, LMCD1, ID3, SKIL, RB1, MKL1, RNF20, ANKRD1, HTATIP2, ZNF281, TDG, CBX4, DMAP1, MED10, NCOA4 | 1.69E-8 |
| Human | PolyUb | Ubiquitin system | RNF187, UBE2J1, UBE2T, UBE2L6, UBE2E3, UBE2D1, RNF20, HERC4, RNF25, CDC34, TRIM37, ANAPC10, UBE2G2, UBE2J2, AMFR, PJA1, UBE2E1, BTRC, BIRC6, UBE2Q1, BRAP | 1.85E-10 |
Ubiquitination Site Sequence Analysis Reveals Unique Patterns.
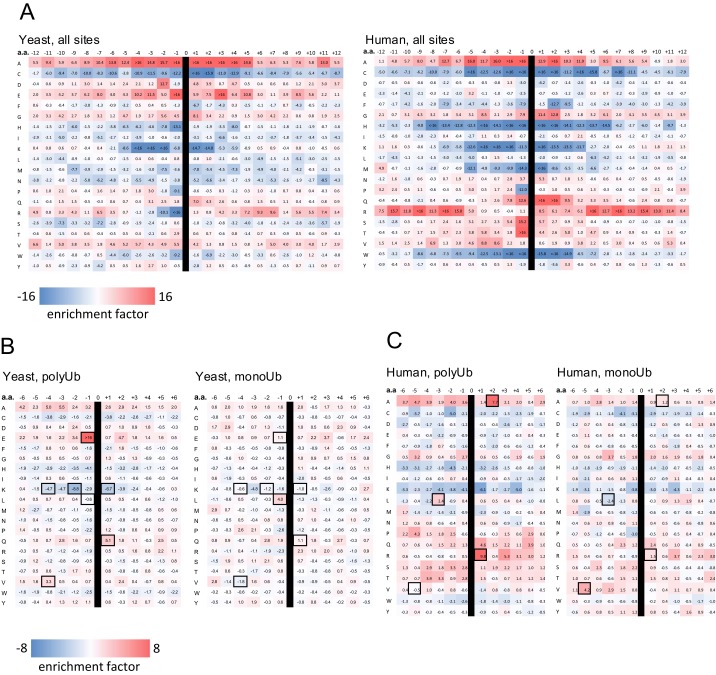
Previous studies have demonstrated motifs and sequence patterns specific for protein posttranslational modifications such as phosphorylation, acetylation, and SUMOylation (29, 30, 37, 38). To identify Ubsite motifs, we analyzed our data using the Motif-X algorithm (39). Consistent with previous findings (24), no specific motif was found when analyzing all identified Ubsites in either yeast or human cells. To analyze Ubsite amino acid composition, all identified Ubsites were examined. We constructed an alignment of peptides that are surrounding each modified Lys residue. Residue- and position-specific amino acid occurrences were calculated and were compared with the corresponding proteomic occurrence in a lysine-centered peptide. As depicted in Fig. S4A, ubiquitination sites demonstrated residues-specific enrichment (e.g., Ala, Gly, Gln) and underrepresentation (e.g., Cys, His, Lys, Met, Trp) for both yeast and human cells. Enrichment of Glu, and to a lesser extent Asp, was specific to yeast sites. Importantly, for several amino acids, the enrichment factor depended on the proximity to the modified Lys, suggesting that these residues can affect conjugation mechanisms.
Fig. S4.
Analysis of amino acids occurrences around ubiquitination sites. Analysis of sequence windows with ubiquitinated lysines in their center. Residue- and position-specific enrichment factors (EF) were calculated as described in SI Experimental Procedures. (A) All identified ubiquitination sites in yeast (Left) and in human (Right). (B) Yeast ubiquitination sites of polyubiquitination-dependent (Left), and monoubiquitination-dependent (Right) substrates. The highlighted coordinates represent major pattern differences between monoubiquitinated and polyubiquitinated substrates in which . (C) Same as in B, but for human substrates.
To compare the ubiquitination site composition of monoubiquitination- vs. polyubiquitination-dependent substrates, we performed the above analysis for each group separately. As shown in Fig. S4 B and C, each group displays a unique pattern of enrichment factors, and several differences can be observed between the patterns of monoubiquitination- and polyubiquitination-dependent substrates.
Discussion
In this study, we identified 82 yeast and 220 mammalian proteins that are degraded by the proteasome following monoubiquitination. These significant numbers point to a much broader phenomenon to what was considered until recently as an exception. Therefore, it allows for analysis that sheds light on the mechanisms that underlie the different modes of ubiquitination. Because we did not use proteasome inhibitors, it is likely that some low level, rapidly degrading proteins were below the detection threshold, and therefore the numbers are probably larger.
Experimentally, we replaced UbWT with UbK0 in cells to enforce monoubiquitination. This strategy has been used successfully to inhibit polyubiquitination-dependent proteasomal degradation (14, 17–19). Other methods to study monoubiquitination have been reported, such as inhibition of polyubiquitin chain formation by methylated Ub (40), or detection by Western blotting of specific substrates that appear to be monoubiquitinated (10). However, using methylated Ub is limited to cell-free systems, and using endogenous Ub can identify only individual substrates. Therefore, these methods limit the ability to identify and characterize the broad population of target substrates degraded by the proteasome following monoubiquitination and polyubiquitination. Thus, UbWT-to-UbK0 replacement in cells seemed to be the most suitable strategy for our objective of carrying out a proteome-wide screen to identify these two distinct populations.
Notably, although efficient UbK0 expression inhibits polyubiquitination, it may still support the conjugation of several UbK0 molecules to a protein substrate, resulting in multiple monoubiquitinations. In our study, we included monoubiquitinated and multiple-monoubiquitinated substrates in the same group, as discriminating between the two is complicated experimentally.
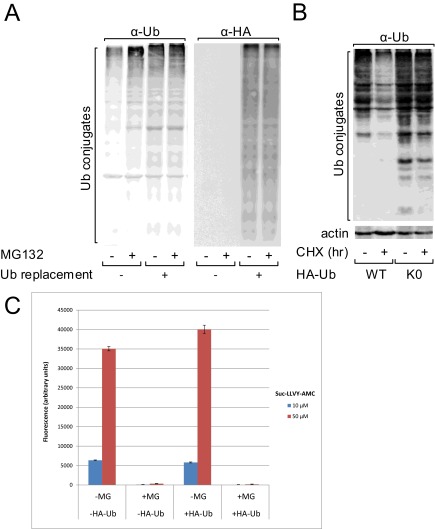
Importantly, in our screen, we decided not to use proteasome inhibitors as a tool to identify substrates that are nevertheless degraded following Ub replacement; the reason being that, in human cells infected with adenoviral HA-UbWT, we could not observe up-regulation of Ub conjugates following treatment with a proteasome inhibitor (Fig. S5A). It should be noted that the proteasome in the control cells was active (Fig. S5B), and its inhibition was efficient (Fig. S5C). This finding is consistent with partial proteasome inhibition by free Ub chains that are accumulated due to the high level of HA-UbWT (41). Also, the use of proteasome inhibitors is challenging in yeast cells, as low permeability results in low cellular concentration of the drugs (42). Notably, in human cells, it was reported that proteasome inhibitors impair protein synthesis in a selective manner, thus affecting the proteome in more than one mode. Therefore, interpretation of proteomic data of experiments carried out in their presence is not straightforward (43). Accordingly, we have adapted alternative classification criteria to identify monoubiquitinated vs. polyubiquitinated proteasomal substrates based on MS analyses of the proteome and ubiquitome under normal conditions and where only monoUb is available (Results and Fig. 2B). To confirm our results and to nevertheless relate them to proteasomal degradation, we integrated data from previous studies (23, 24, 44) and constructed a reference list of known ubiquitin–proteasome substrates (Dataset S3). Compared with this list, our polyubiquitination-dependent substrates were highly enriched with known proteasomal substrates (P valueyeast = 6.90 × 10−23 and P valuehuman = 5.06 × 10−5, by hypergeometric test). Furthermore, our monoubiquitination-dependent candidates were also highly enriched with proteasomal substrates (P valuehuman = 1.10 × 10−36). These findings strongly suggest that our experimental model is faithful and offers a reliable method for the identification of UPS substrates. Furthermore, we validated biochemically that several randomly sampled monoubiquitination and polyubiquitination-dependent candidates that emerged from the screen do indeed belong to their respective predicted categories (Fig. 3).
Fig. S5.
Measurement of ubiquitin–proteasome system activity following Ub replacement in U2OSshUb cells. (A) Where indicated, endogenous Ub was replaced by HA-UbWT in U2OSshUb cells as described in Experimental Procedures. Cells were further treated as indicated with MG132 at 20 µM for 4 h. Lysates were analyzed by SDS/PAGE followed by WB using the indicated antibodies. (B) Endogenous Ub was replaced by HA-UbWT in U2OSshUb cells. The stability of Ub conjugates was monitored following addition of cycloheximide for 6 h as described in SI Experimental Procedures. (C) U2OSshUb cells were treated for Ub replacement by HA-UbWT and for proteasome inhibition with MG132 as indicated. Proteasome activity was measured by a fluorescence-based assay as described in SI Experimental Procedures.
Reportedly, membrane proteins such as receptor tyrosine kinases (RTKs) (45) are subjected to monoubiquitination-dependent lysosomal degradation. To specifically identify proteasomal substrates in the absence of proteasome inhibitors, plasma membrane proteins were excluded from our screen.
It is interesting to refer to specific proteins that were identified in previous studies as targeted by monoubiquitination. For example, Syndecan-4 (12) did exhibit site K0/WT ratio <1 (for Lys105) but was detected in one replicate only. Cks-2 (15) displayed protein K0/WT ratio <1 at the protein level but had conflicting ratios at site level. Other substrates including Pax-3 (13) and α-synuclein (10) could not be detected, probably due to low abundance in the bone-derived U2OS cells (46). Taken together, it seems that our results are in agreement with random previous data but should be further substantiated by experiments in cells from different tissues.
From the bioinformatics analyses of the substrates, several important and intriguing conclusions can be drawn. First, in agreement with previous studies in which it was shown that substrates of up to 150 residues can be degraded following monoubiquitination (15), we found that the distribution of monoubiquitination-dependent substrates is shifted toward shorter proteins (Fig. S3D).
Furthermore, a difference between yeast and human is also apparent when comparing the ratio of monoubiquitination- and polyubiquitination-dependent substrates. Yeast relies more heavily on polyubiquitination, whereas human cells use both monoubiquitination and polyubiquitination with a similar frequency. This difference can be interpreted if structural disorder is considered as shown by our own data (Fig. 4 and Fig. S3), and if we assume that a single ubiquitin moiety is a weaker signal for degradation than polyubiquitin. As suggested, local structural disorder is involved in various steps of the UPS cascade, from recognition motifs of E3 ligases through local disorder of ubiquitination sites to an LDR initiation site of substrate unfolding (28–30).
We have screened our monoubiquitination-dependent substrates for enriched biological-process–related GO terms and found a highly significant overrepresentation of genes associated with carbohydrate transport and oxidative stress response pathways (Fig. 5A). Because carbohydrate transporters are plasma membrane proteins, this finding is consistent with previous studies that demonstrated membrane receptors down-regulation via monoubiquitination-mediated endocytosis (47). Notably, as this pathway results in lysosomal/vacuolar rather than proteasomal degradation, this finding highlights the challenge in distinguishing between these two degradation modes using our experimental system. Oxidative stress was shown to activate cellular signal transduction cascades, and to result in gene expression modulation (48). The enrichment of oxidative stress response proteins in our monoubiquitination-dependent substrates may suggest that they are regulated by a common monoubiquitinating E3 ligase(s). Thus, oxidative stress-mediated down-regulation of this putative E3 may result in up-regulation of oxidative stress pathway components and activate the respective cellular response. Consistently, microarray experiments have shown that the expression of the E3s UBR1 and HUL4, and the E2s CDC43, RAD6, and UBC11, is decreased following exposure to oxidative stress (49).
UPS components were enriched in both groups (Fig. 5B). This finding most likely represents the previously reported autoubiquitination of E2s (50–52) and E3s (53–55). Our findings show that E3s are preferably classified as monoubiquitination-dependent substrates.
In this study, we have determined the sequence positions of thousands of ubiquitinated lysines. The unsuccessful attempts to identify a ubiquitination-site motif in this study and in others (24, 56) reflect site-level promiscuity, which is supported by low ubiquitination sites conservation across eukaryotic species (56), and by the flexible selection of the ubiquitinated lysines within a given substrate (11, 31). Additionally, we used position-specific analysis of relative amino acid abundance to characterize ubiquitination sites (Fig. S4). This method yields a more thorough representation of the amino acid composition and reflects both enrichment and underrepresentation trends. According to the notion of promiscuity, a ubiquitination site should merely provide a sterically available ε-amino group of a Lys residue. Consistently, we found enrichment of small residues (e.g., Ala and Gly) and decrease in bulky (e.g., Trp) residues in the proximity of the ubiquitinated Lys. The depletion of Pro at position −1 also supports this concept, because Pro disrupts the protein’s secondary structure and might impair the solvent accessibility of a following Lys residue.
Experimental Procedures
Adenovirus-Mediated Ub Replacement in Human Cells.
For Ub silencing, U2OSshUb (described in ref. 21) cells were treated with 1 µg/mL tetracycline for 24 h. Fresh tetracycline and adenoviruses encoding either HA-UbWT or HA-UbK0 were added, and cells were incubated for additional 24 h.
Ub Replacement in Yeast.
The construction of ΔUb strain was described previously [SUB328 (57)]. Briefly, endogenous Ub genes were deleted and replaced with a Ub gene expressed under a Gal promoter. To construct ΔUbUbWT and ΔUbUbK0 strains, ΔUb yeast cells were transformed with pUb39 UbWT and pUb39 UbK0, respectively (both genes are under the Cup1 promoter). To replace the Gal-induced Ub, ΔUbUbWT and ΔUbUbK0 yeast cells were grown to 1.0 OD600nm in standard Hartwell's complete medium (HC) without glucose, supplemented with 2% (wt/vol) galactose and 2% (wt/vol) raffinose. Cells were then washed in double-distilled water (DDW) to remove galactose and raffinose, and resuspended in HC medium with glucose and 50 µM CuSO4. Cells were incubated for 16 h at 30 °C, collected by centrifugation, washed in DDW, and frozen in liquid N2.
Ubiquitination Sites Detection Using GG-Modified Peptide Enrichment.
Dried peptides (SI Experimental Procedures, Sample Preparation for MS) were resuspended in immunoaffinity purification (IAP) buffer (50 mM Mops/NaOH, pH 7.2, 10 mM Na2HPO4, and 50 mM NaCl), and cleared by centrifugation. Supernatants were adjusted to pH 7.0 with NaOH and incubated with immobilized anti–K-ε-GG antibody (Cell Signaling Technology) at 4 °C for 3 h. Beads were washed with IAP buffer and then with a wash buffer (500 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.1% octyl glucoside, pH 7.4). GG-modified peptides were eluted with 0.2% TFA, desalted on C18 tips, and eluted in two fractions of 20% and 80% (vol/vol) acetonitrile. Peptides were analyzed as described in SI Experimental Procedures, MS.
For additional experimental procedures, see SI Experimental Procedures.
SI Experimental Procedures
Materials.
Anti-Ub was described previously (58), anti-HA (16B12) was from Covance, and anti-actin and anti-tubulin were from Millipore. Anti–K-ε-GG Immunoaffinity Beads were from Cell Signaling Technology. Suc-Leu-Leu-Val-Tyr-AMC (Suc-LLVY-AMC) was from Boston Biochem. Materials for SDS/PAGE and Bradford reagent were from Bio-Rad. Tissue culture sera, media, and supplements were from Biological Industries. Peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories. N-carbobenzoxy-l-leucyl-l-leucyl-leucinal (MG132) and epoxomicin were from Calbiochem. Reagents for ECL were from Pierce. JetPEI cell transfection reagent was from Polyplus. Restriction and modifying enzymes were from New England Biolabs. Oligonucleotides were synthesized by Sigma. All other reagents were of high analytical grade.
Plasmids.
All of the plasmids were constructed and manipulated using standard molecular biological techniques. For liposome-mediated transient cell transfection, Flag-UbWT and Flag-UbK6,11,27,29,33,48,63R (Flag-UbK0) were subcloned into pCAGGS (59), and GOT1-HA and CDC20-HA were subcloned into pCS2. For adenovirus preparation, HA-UbWT and HA-UbK0 were subcloned into pAd/CMV/V5-DEST. For yeast transformation, UbWT and UbK0 were subcloned into pUb39 (20), and ARD1-HA and GRE1-HA were subcloned into pCM190.
Cultured Cells.
U2OSshUb cells (21) were grown at 37 °C in DMEM supplemented with 10% (vol/vol) FCS and antibiotics (penicillin–streptomycin). MCF7 cells were grown 37 °C in the same medium supplemented with insulin (100 units/mL).
Protein Concentration Measurement.
Protein concentration was measured by the Bradford method (60) using BSA as the standard.
Protein Detection.
For Western blotting (WB), proteins were resolved via SDS/PAGE, transferred to nitrocellulose (or PVDF) membrane, and incubated with the appropriate antibodies.
Yeast Transformation.
The different strains of Saccharomyces cerevisiae were transformed with plasmids coding for the proteins of interest using the poly(ethylene glycol)/lithium acetate (PEG/LiAc) method (61), followed by appropriate selection.
Mammalian Cell Transfection.
Transient transfections of mammalian cells with plasmids containing cDNAs coding for proteins of interest were carried out using the jetPEI transfection reagent according to the manufacturer’s instructions.
Monitoring Protein Stability.
Protein synthesis was inhibited by adding 100 µg/mL cycloheximide. Samples were collected at the indicated times, and proteins of interest analyzed by SDS/PAGE followed by WB using the indicated antibodies.
Preparation of Adenoviral Vectors.
HEK293 cells were transfected with pAd/CMV/V5-DEST (AdEasy Vector System) encoding the genes of interest. Small- and large-scale virus amplifications were performed according to the manufacturer’s instructions, followed by purification of the viral particles on iodixanol gradient as described previously (62). Viral titer was determined by MOI test according to the manufacturer’s instructions.
Fluorescence-Based Proteasome Activity Assay.
Cells were lysed in 0.3% CHAPS buffer (20 mM Hepes, 100 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, 0.3% CHAPS, 1 mM DTT, 2.5 mM ATP, and protease inhibitor mixture), and proteasomes were immobilized onto agarose to which anti-α6 was bound. Beads to which the proteasomes were not immobilized were used as control. Beads were washed twice with 0.03% CHAPS buffer. Where indicated, MG132 (100 µM) was added to the immobilized proteasomes, and the mixture was incubated at 37 °C for 10 min. Next, proteasomes were incubated at 37 °C for 30 min with the indicated concentrations of Suc-LLVY-AMC in a reaction buffer (40 mM Tris⋅HCl, pH 7.2, 2 mM DTT, 5 mM MgCl2, 10 mM creatine phosphate, 0.1 mg/mL creatine phosphate kinase, 5 mM ATP). Reaction was stopped by adding 1% SDS, and fluorescence was measured at excitation 360 nm/emission 460 nm.
Cell Lysis.
Yeast cell lysates for WB were prepared by trichloroacetic acid (TCA) precipitation as described (14). Yeast lysates for MS analysis and anti–K-ε-GG IP were prepared by suspending cell pellets in urea buffer (8 M urea, 100 mM Tris⋅HCl, pH 8.0, 10 mM iodoacetamide), followed by shaking with 0.5-mm glass beads for 25 min at room temperature. The resulting lysates were cleared by centrifugation. Human cells were lysed by adding urea buffer, followed by brief sonication and clearing by centrifugation.
Sample Preparation for MS.
Cells were harvested and lysed as described in Cell Lysis. Two to 3 mg of protein in 8 M urea and 100 mM ammonium bicarbonate, were incubated with DTT (2.8 mM; 30 min at 60 °C), modified with iodoacetamide (8.8 mM; 30 min at room temperature in the dark), and digested (overnight at 37 °C) with modified trypsin (Promega; 1:50 enzyme-to-substrate ratio) in 2 M urea and 25 mM ammonium bicarbonate. Additional second trypsinization was carried out for 4 h. The tryptic peptides were desalted using Sep-Pak C18 (Waters) and dried. Ten micrograms of the total protein were used for proteome analysis as described in MS, and the remaining material was used for ubiquitination sites analysis as described in Experimental Procedures, Ubiquitination Sites Detection Using GG-Modified Peptide Enrichment.
MS.
Tryptic peptides were analyzed by liquid chromatography (LC)-MS/MS using a Q Exactive plus mass spectrometer (Thermo Fisher Scientific) fitted with a capillary HPLC (easy nLC 1000; Thermo). The peptides were loaded onto a C18 trap column (0.3 × 5 mm; LC-Packings) connected on-line to a homemade capillary column (20 cm; internal diameter, 75 μm) packed with Reprosil C18-Aqua (Dr. Maisch GmbH) in solvent A (0.1% formic acid in water). The peptides mixture was resolved with a 5–28% linear gradient of solvent B [95% (vol/vol) acetonitrile with 0.1% formic acid] for 180 min followed by a 5-min gradient of 28–95% and 25 min at 95% (vol/vol) acetonitrile with 0.1% formic acid in water at a flow rate of 0.15 μL/min. MS was performed in a positive mode (m/z 350–1,800; resolution, 70,000) using repetitively full MS scan followed by collision-induced dissociation (higher-energy collisional dissociation at 35 normalized collision energy) of the 10 most dominant ions (>1 charges) selected from the first MS scan. A dynamic exclusion list was enabled with exclusion duration of 20 s.
Data Analysis.
The MS raw data were analyzed by the MaxQuant software (version 1.4.1.2; www.coxdocs.org/doku.php?id=maxquant:start) for peak picking and quantification. This was followed by identification of the proteins using the Andromeda engine, searching against the human or the yeast UniProt database with mass tolerance of 20 ppm for the precursor masses and for the fragment ions. Met oxidation, N-terminal acetylation, N-ethylmaleimide and carbamidomethyl on Cys, GlyGly on Lys, and phosphorylation on Ser, Thr and Tyr residues, were set as variable posttranslational modifications. Minimal peptide length was set to 6 aa and a maximum of two miscleavages was allowed. Peptide- and protein-level false-discovery rates were filtered to 1% using the target-decoy strategy. Protein tables were filtered to eliminate the identifications from the reverse database and from common contaminants. The MaxQuant software was used for label-free semiquantitative analysis [based on extracted ion currents (XICs) of peptides], enabling quantification from each LC/MS run for each peptide identified in any of the experiments. In samples that were SILAC-labeled, quantification was also done using the MaxQuant 1.4.1.2 software. Data merging and statistical tests were done by the Perseus 1.4 software.
GO Term Enrichment Analysis (GOrilla).
GO term analysis was performed by the GOrilla web tool (35, 36), using the two unranked lists running mode. We used the monoubiquitination- or polyubiquitination-dependent proteasome substrate groups as target lists, and the complete proteome as a background list. Both yeast and human data were analyzed.
Statistical Analysis.
To compare protein sequence lengths of monoubiquitination- and polyubiquitination-dependent proteasomal substrates, length distribution histograms were generated. Statistical significance of differences was determined by the Mann–Whitney U test. To analyze ubiquitination sites amino acid composition, peptides containing GG-modified lysines in their center were aligned, and residue- and position-specific amino acid occurrences (OC) were calculated. As a control, the corresponding proteome’s amino acid occurrences in lysine-centered peptides were estimated (OCcontrol), using lysines from 1,000 random proteins. The statistical significance of enrichment or depletion for each amino acid in every position was determined by the binomial test, and the enrichment factor (EF) was calculated as follows:
Thus, increasing positive EF values represent a more significant enrichment, and increasing negative EF values represent a more significant depletion.
Prediction of Structural Disorder.
IUPred (63) software was used to predict structural disorder using amino acid sequences as input. IUPred output scores were determined for each residue (ranging between 0 and 1); scores greater than 0.5 indicate disordered residues (the “long” default mode of IUPred was used). Using the IUPred scores, long disordered regions (LDRs) were defined as consecutive stretches of at least 30 disordered residues (breaks of up to three consecutive ordered residues within an LDR were permitted). Statistical tests for calculating P values were carried out using the Mann–Whitney U test.
Ortholog Alignments and Calculation of Ubiquitination Site Conservation.
Precomputed multiple sequence alignments of orthologs were obtained from Discovery@Bioware (bioware.ucd.ie/∼compass/biowareweb/) and used to calculate the fraction of aligned sequences in which lysines were retained in orthologous sequences for identified Ubsites. Statistical tests for calculating P values were carried out using the Mann–Whitney U test.
Supplementary Material
Acknowledgments
We thank Drs. Zhijian James Chen (University of Texas Southwestern) and Daniel Finley (Harvard Medical School) for providing us with the mammalian and yeast systems for deleting endogenous Ub genes, respectively. Research in the laboratory of A.C. is supported by grants from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Israel Science Foundation (ISF), the I-CORE Program of the Planning and Budgeting Committee and the ISF (Grant 1775/12), and the Deutsch–Israelische Projektkooperation. I.L. is supported by the Foulkes Fellowship. A.C. is an Israel Cancer Research Fund USA Professor. A.G. is supported by NIH Grant R01 GM101457. This work was supported by Odysseus Grant G.0029.12 from Research Foundation Flanders (to P.T.) and a VIB/Marie Curie COFUND Postdoctoral (omics@VIB) fellowship (to M.G.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 8894.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608644113/-/DCSupplemental.
References
- 1.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37(Pt 5):937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 3.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19(1):94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6(8):610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 5.Hoeller D, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8(2):163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 6.Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5(5):a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: Post-translational modification of RAS. Nat Rev Mol Cell Biol. 2011;13(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21(5):679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA. 2009;106(39):16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rott R, et al. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci USA. 2011;108(46):18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimova NV, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14(2):168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvallo L, et al. Non-canonical Wnt signaling induces ubiquitination and degradation of Syndecan4. J Biol Chem. 2010;285(38):29546–29555. doi: 10.1074/jbc.M110.155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130(2):349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 14.Kravtsova-Ivantsiv Y, Cohen S, Ciechanover A. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol Cell. 2009;33(4):496–504. doi: 10.1016/j.molcel.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48(1):87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Braten O, Shabek N, Kravtsova-Ivantsiv Y, Ciechanover A. Generation of free ubiquitin chains is up-regulated in stress and facilitated by the HECT domain ubiquitin ligases UFD4 and HUL5. Biochem J. 2012;444(3):611–617. doi: 10.1042/BJ20111840. [DOI] [PubMed] [Google Scholar]
- 17.Hospenthal MK, Freund SM, Komander D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat Struct Mol Biol. 2013;20(5):555–565. doi: 10.1038/nsmb.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T, et al. The role of monoubiquitination in endocytic degradation of human ether-a-go-go-related gene (hERG) channels under low K+ conditions. J Biol Chem. 2011;286(8):6751–6759. doi: 10.1074/jbc.M110.198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83(1):121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 20.Finley D, et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14(8):5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36(2):302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emanuele MJ, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147(2):459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6(11):1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21(3):419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 26.van der Lee R, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guharoy M, Bhowmick P, Tompa P. Design principles involving protein disorder facilitate specific substrate selection and degradation by the ubiquitin-proteasome system. J Biol Chem. 2016;291(13):6723–6731. doi: 10.1074/jbc.R115.692665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guharoy M, Bhowmick P, Sallam M, Tompa P. Tripartite degrons confer diversity and specificity on regulated protein degradation in the ubiquitin-proteasome system. Nat Commun. 2016;7:10239. doi: 10.1038/ncomms10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7(3):161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9(9):679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7(9):1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32(3):1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 34.Landry CR, Levy ED, Michnick SW. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25(5):193–197. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007;3(3):e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amanchy R, et al. A curated compendium of phosphorylation motifs. Nat Biotechnol. 2007;25(3):285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 38.Basu A, et al. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci USA. 2009;106(33):13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou MF, Schwartz D. 2011. Biological sequence motif discovery using Motif-X. Curr Protoc Bioinformatics Chap 13:Unit 13.15–24.
- 40.Hershko A, Ganoth D, Pehrson J, Palazzo RE, Cohen LH. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J Biol Chem. 1991;266(25):16376–16379. [PubMed] [Google Scholar]
- 41.Amerik AYu, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16(16):4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271(44):27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 43.Milner E, et al. The effect of proteasome inhibition on the generation of the human leukocyte antigen (HLA) peptidome. Mol Cell Proteomics. 2013;12(7):1853–1864. doi: 10.1074/mcp.M112.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Udeshi ND, et al. Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics. 2013;12(3):825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haglund K, et al. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5(5):461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: A database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 48.Ikner A, Shiozaki K. Yeast signaling pathways in the oxidative stress response. Mutat Res. 2005;569(1-2):13–27. doi: 10.1016/j.mrfmmm.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 49.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285(12):8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432(7017):588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 52.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9(4):422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- 53.Amemiya Y, Azmi P, Seth A. Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol Cancer Res. 2008;6(9):1385–1396. doi: 10.1158/1541-7786.MCR-08-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96(20):11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noels H, et al. Auto-ubiquitination-induced degradation of MALT1-API2 prevents BCL10 destabilization in t(11;18)(q21;q21)-positive MALT lymphoma. PLoS One. 2009;4(3):e4822. doi: 10.1371/journal.pone.0004822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danielsen JM, et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics. 2011;10(3):M110.003590. doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15(3):1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hershko A, Eytan E, Ciechanover A, Haas AL. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982;257(23):13964–13970. [PubMed] [Google Scholar]
- 59.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 60.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 61.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 62.Dormond E, et al. An efficient process for the purification of helper-dependent adenoviral vector and removal of helper virus by iodixanol ultracentrifugation. J Virol Methods. 2010;165(1):83–89. doi: 10.1016/j.jviromet.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 63.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.