Significance
Parasitism is a proven way of life that brings about extraordinary phenotypic and genetic modifications. Obtaining organic carbon from a host rather than synthesizing it, nonphotosynthetic plants lose unneeded genes for photosynthesis from their plastid genomes, while essential genes in the same subgenome may evolve rapidly. We show that long before the nonphotosynthetic lifestyle is established, losses of functional complexes repeatedly trigger the disruption of evolutionary stasis, resulting in “roller-coaster rate variation” along the transition to full parasitism. Our model of the molecular evolutionary principles of plastid genome degradation under modified selective constraints makes a significant contribution to our understanding of the complexity of genetic switches in relation to lifestyle changes.
Keywords: parasitism, relaxed selection, evolutionary rates, plastid genomes, Orobanchaceae
Abstract
Because novel environmental conditions alter the selection pressure on genes or entire subgenomes, adaptive and nonadaptive changes will leave a measurable signature in the genomes, shaping their molecular evolution. We present herein a model of the trajectory of plastid genome evolution under progressively relaxed functional constraints during the transition from autotrophy to a nonphotosynthetic parasitic lifestyle. We show that relaxed purifying selection in all plastid genes is linked to obligate parasitism, characterized by the parasite’s dependence on a host to fulfill its life cycle, rather than the loss of photosynthesis. Evolutionary rates and selection pressure coevolve with macrostructural and microstructural changes, the extent of functional reduction, and the establishment of the obligate parasitic lifestyle. Inferred bursts of gene losses coincide with periods of relaxed selection, which are followed by phases of intensified selection and rate deceleration in the retained functional complexes. Our findings suggest that the transition to obligate parasitism relaxes functional constraints on plastid genes in a stepwise manner. During the functional reduction process, the elevation of evolutionary rates reaches several new rate equilibria, possibly relating to the modified protein turnover rates in heterotrophic plastids.
Lineages change over time as they adapt to new environments. Novel conditions determine the selection in genes or cellular genomes and shape their functional and structural evolution. A system well suited to study the evolution of genomic traits in the context of altered selective regimes that is also tractable technically (due to its small size and high copy number) is the plastid genome (plastome). The prime function of plastids is photosynthesis, but this essential plant organelle also produces starch, lipids, amino acids, sulfur compounds, and pigments. As a result of the strong selective pressure on plastid gene function, plastid genomes have a conserved gene content (1; but see ref. 2) and their genes functioning in photosynthesis (atp, ndh, pet, psa, psb, ccsA, cemA, ycf3/4, rbcL), transcription, transcript maturation or translation (rpo, matK, rpl, rps, infA), and other pathways (accD, clpP, ycf1, and ycf2) evolve at lower evolutionary rates than nuclear genes (3). However, in eukaryotic lineages such as Apicomplexan pathogens and nongreen plants that independently made the transition from an autotrophic to a parasitic way of life, plastomes have experienced convergent reductions and accelerations of evolutionary rates (4). Although there is a general understanding of the association of the nonphotosynthetic lifestyle with plastome degradation and rate acceleration, the precise trajectory of plastome evolution under progressively reduced function along the way from being a full autotroph to an obligate nonphotosynthetic parasite remains unknown.
Parasitic plants are an excellent system for studying genome evolution under altered selective constraints because of lifestyle changes such as the transition from an autotrophic to a parasitic way of life (5). These plants directly connect to their host plants through a specialized organ to steal water and nutrients. The large majority of the 4,000–4,500 parasitic flowering plant species are photosynthetic parasites (hemiparasites), whereas only 10% are nonphotosynthetic parasites (holoparasites). Whereas some hemiparasites can complete their life cycle without ever connecting to a host plant (facultative parasites), most hemiparasites and all holoparasites require a host at least during certain life stages to fulfill their life cycle (obligate parasites). The transition from autotrophy to parasitism coincides with the loss of both photosynthetic and housekeeping plastid genes (6, 7). Whether a gene is retained or lost in parasites mainly depends on its function, its size (8), and its physical and/or transcriptional association to essential genes (5). Most of the retained genes continue to evolve under purifying selection despite uncorrelated shifts of codon use (5, 6) and changes in evolutionary rates (4). However, severe reconfigurations of the plastid chromosomal architecture such as increases in the amount of recombinogenic DNA sequences in obligate hemiparasites suggest that already the shift from a free-living to an obligate parasitic lifestyle alters the evolution of the plastome in general (5, 9).
Here, we assess the course of reductive plastome evolution and its underlying causes under progressively relaxed functional constraints. Specifically, we examine mutation rate variation, encompassing nucleotide substitutions and microstructural changes across different functional gene classes, and we test for correlations of mutational rates with lifestyle and genomic features, taking into account potential effects of life history. We use complete plastid genome sequences of 17 parasitic and nonparasitic species across all different trophic specializations of the broomrape family (Orobanchaceae) and two closely related autotrophs. Orobanchaceae represent an ideal group for this type of study, because its phylogeny is well understood and it spans the entire range from autotrophy to full parasitism (10). Our data show that the shift to parasitism and the loss of functional gene groups trigger the disruption of evolutionary stasis, resulting in phases of accelerated evolution alternating with phases of deceleration. Our findings provide the basis for a molecular evolutionary model of plastome degradation along the transition to a nonphotosynthetic way of life.
Results
Plastid Genomes in Parasitic Orobanchaceae.
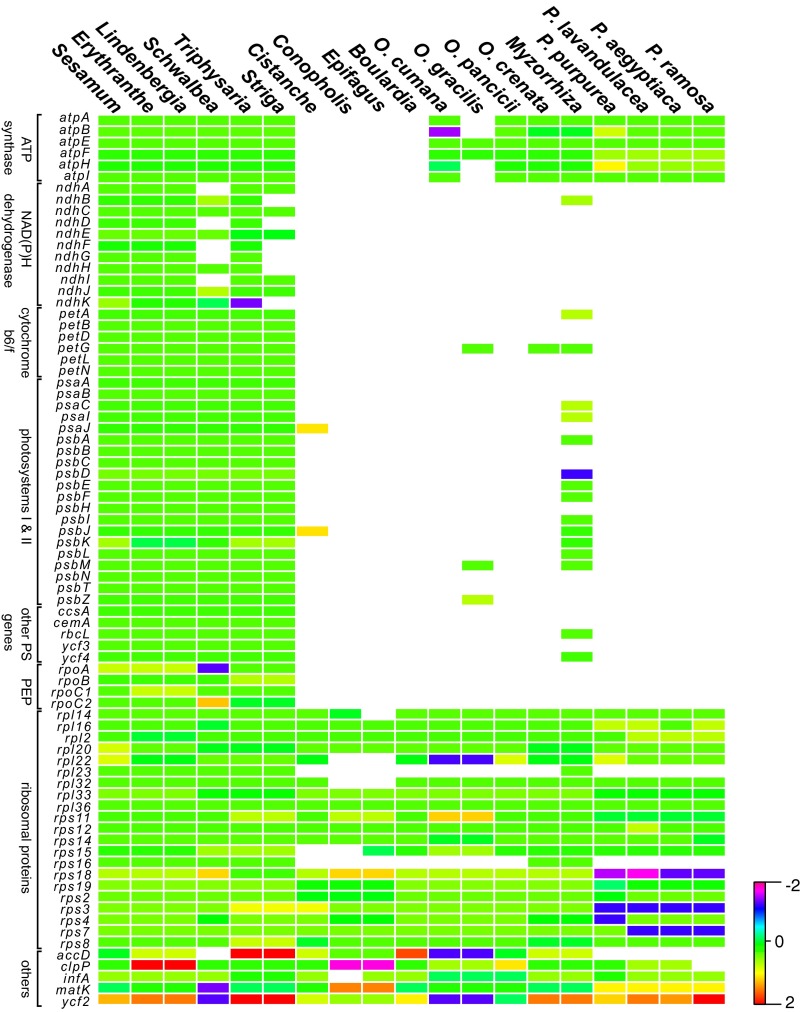
Complete sequencing of 17 species of Orobanchaceae revealed that genes for only 16 proteins, 4 ribosomal RNAs, and 14 transfer RNAs of the 113 unique genes found in the nonparasitic Lindenbergia philippensis (Orobanchaceae), Erythranthe guttata (Phrymaceae), and Sesamum indicum (Pedaliaceae) are commonly present in the plastid genomes of all of the hemiparasitic and holoparasitic plants (Fig. 1). Retaining between 42 and 71 intact genes (Fig. S1), including intact photosynthesis genes (atp genes), the holoparasites are particularly diverse with respect to both gene content and genome structure. Large-scale structural reconfigurations such as inversions, modifications of the large inverted repeat (IR) regions, or their loss characterize the genomes of several parasites, including the obligate hemiparasites Schwalbea americana and Striga hermonthica, as well as the holoparasites Conopholis americana, Orobanche gracilis, Orobanche crenata, and all Phelipanche species (Fig. S2).
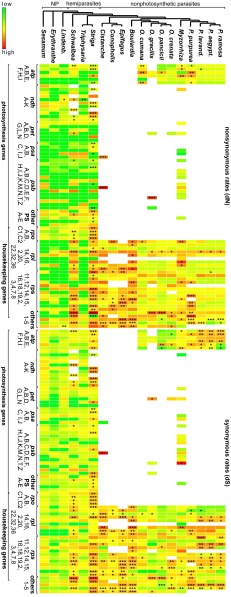
Fig. 1.
Rate variation in Orobanchaceae. Heatmaps illustrate differences in dN and dS for each plastid protein gene (as named from top to bottom per gene class), with low rates shown in green and high rates in red. A phylogenetic tree on top indicates species relationships and the trophic specialization. Asterisks indicate the significance of LRTs of a focal taxon against the non-Orobanchaceae taxa (significance levels: *P < 0.05, **P < 0.01, ***P < 0.001). other PS, other photosynthesis genes (A, ccsA; B, cemA; C, rbcL; D, ycf3; E, ycf4), other HK, housekeeping and metabolic genes (1, matK; 2, infA; 3, ycf2; 4, clpP; 5, accD).
Fig. S1.
Gene content and its evolution. (A) Matrix showing presence (white) or functional or physical absence (black) of genes (names to the right) in Orobanchaceae and the closely related E. guttata and S. indicum. (B) Inferred gene losses in Orobanchaceae, reconstructed using ML. Branch lengths are proportional to the number of (functionally or physically) lost genes, whose names are given along the branches.
Fig. S2.
Plastid genome organization in Orobanchaceae. Physical map of the plastid gene arrangement in L. philippensis with genes being colored according to their functional classes (as in Fig. S1), and locally collinear blocks (LCB) from whole plastome alignments of L. philippensis and 16 parasitic Orobanchaceae. Colored blocks represent distinct LCBs. Colored bars inside the LCBs show the similarity within these regions among the different species, whereby longer bars indicate a greater similarity. One copy of the large inverted repeat (IR, longest purple LCB) region was removed for the plastome alignment. One asterisk indicates that an IR exists in the plastome, but with a considerably reduced length compared with L. philippensis; two asterisks indicate the loss of the IR from the plastome.
Nucleotide Substitution Rates.
We compared nonsynonymous (dN) and synonymous (dS) nucleotide substitution rates of all Orobanchaceae with closely related nonparasites (Fig. 1), building on phylogenetic relationships established earlier (10, 11). In gene-by-gene likelihood ratio tests (LRTs), the facultatively hemiparasitic Triphysaria versicolor shows hardly any significant rate shifts in plastid genes compared with nonparasitic plants (Fig. 1). In contrast, multiple genes evolve at elevated substitution rates in the obligate hemiparasites S. americana and S. hermonthica, mostly with significantly higher dN and dS in the majority of photosynthesis and housekeeping genes. Among holoparasites, dN and dS are highest in the Epifagus virginiana/C. americana/Cistanche phelypaea-clade (Fig. 1). Fewer genes evolve at elevated molecular evolutionary rates in Myzorrhiza californica and most of the Orobanche and Phelipanche species, which all retain genes for the plastid ATP synthase (atp genes) (Fig. 1). However, dN and/or dS of the retained atp genes are mostly accelerated in these holoparasites compared with those of nonparasites (Fig. 1). Despite some disproportional rate accelerations, both dN and dS are highly correlated [Mantel tests (12), all P < 0.001] without apparent lags (Fig. 2A and Fig. S3).
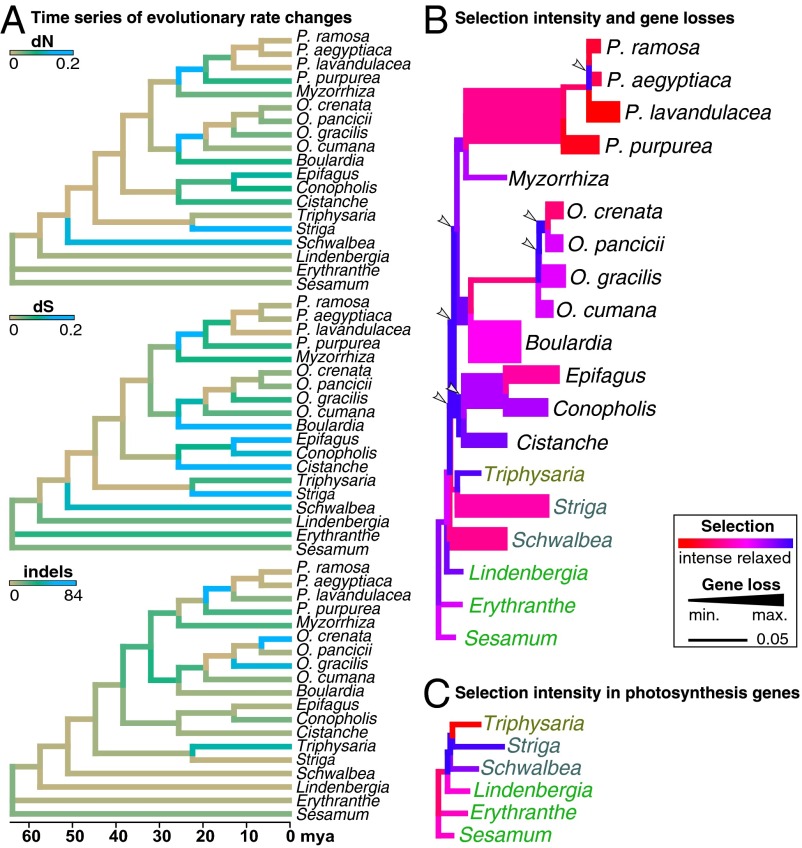
Fig. 2.
Evolution of selectional strength. (A) Evolution of dN, dS, and indels shown on the dated phylogeny. Low rates of nucleotide substitutions and indels are shown in brown and high ones in blue. (B) Selectional changes per branch across all universal protein genes (as in Fig. 1) are color-coded according to the selection strength parameter k, inferred under the general descriptive RELAX model (13). Low k (<1, blue) indicates a relaxation of purifying selection (i.e., a weaker deviation from neutral evolution), whereas high k (>1, red) suggests selection intensification. Branch widths are proportional to the number of gene losses per branch. Arrowheads mark phases of selection relaxations occurring before inferred gene losses. (C) Selectional changes per branch across all photosynthesis genes (as in Fig. 1); colors as in B.
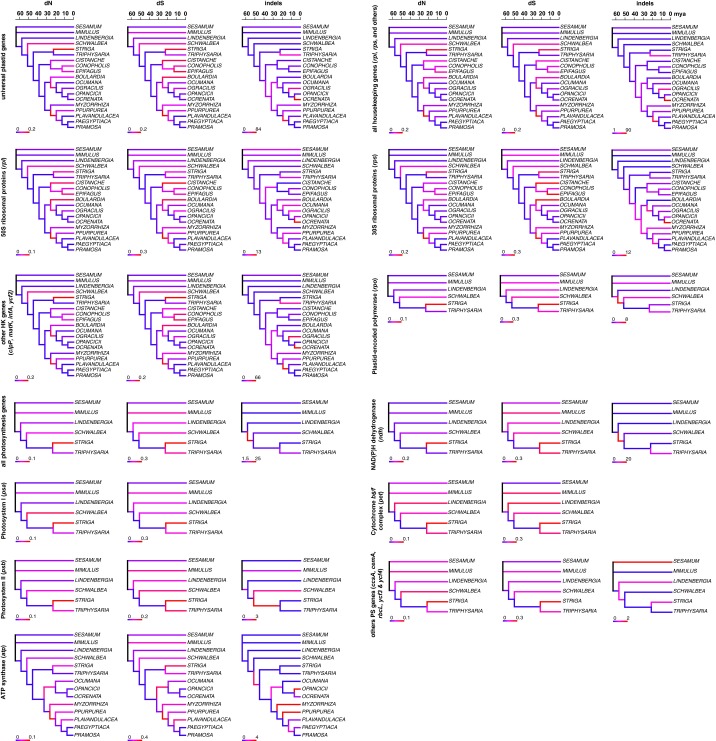
Fig. S3.
Time series of changes in evolutionary rates and indels. Changes in dS, dN, and indels, shown for all universal genes as well as functional classes and gene categories (as indicated), are traced over the dated phylogeny (scales in million years). Low substitution rates and indel numbers are shown in blue, high ones in red; the extent of these changes is indicated by the scale bars at the bottom left of each tree. Because psa and pet genes lack indels, the respective trees are not shown. HK, housekeeping; PS, photosynthesis.
Changes of Selection.
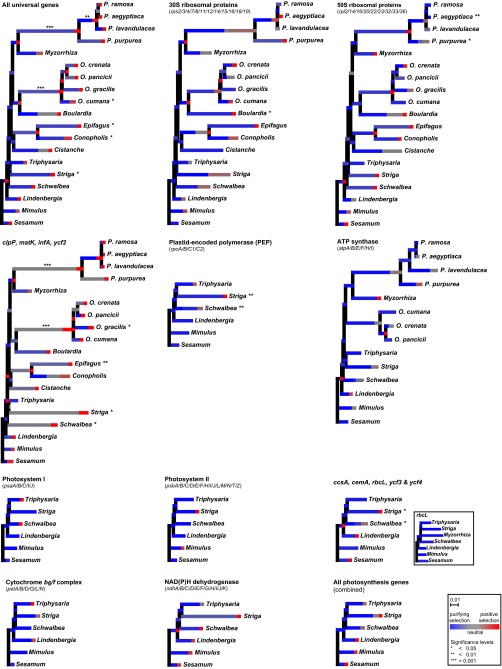
We assessed the direction and the strength of changes of selection across functional gene complexes by ω (ratio of dN to dS) and the selection strength parameter k (according to ref. 13) through branch-site random effects models (13, 14). Alternative branch partitioning and Akaike weights were used to infer the relative contribution of each major shift of lifestyle (i.e., nonparasitism to parasitism to obligate parasitism to holoparasitism and complete loss of photosynthesis) to changes of selection. Plastid genes in general show significant shifts of selection, especially in obligate parasitic Orobanchaceae compared with nonparasitic species and T. versicolor (Fig. 2B). In photosynthesis genes, selectional strength is significantly or—in case of psb genes if analyzed separately—marginally significantly reduced in obligate parasites (Fig. 2C and Table S1). Exceptions are rbcL and pet genes, which all show no significant change in selection (k = 0.64–0.99, LRT P value 0.460–0.879). Ribosomal genes for the small and large ribosome subunit (rps, rpl) show a relaxation of purifying selection in all parasites (i.e., including T. versicolor) compared with nonparasitic species. In contrast, genes for the RNA polymerase (rpo) evolve under lower selectional strength only in obligate parasites (Table 1). All other plastid genes (accD, clpP, ycf2) that are involved in other housekeeping or metabolic processes other than photosynthesis show a slight intensification of purifying selection compared with nonparasites (ω = 0.545 vs. 1.05, LRT P <0.001). Across all universal genes, selectional strength is intensified in obligate parasites (Fig. 2 B and C and Table 1), albeit not evenly. Whereas selection is more relaxed along the backbone (i.e., the selection parameter k is low), it is intensified toward terminal lineages (e.g., S. hermonthica, E. virginiana, Phelipanche) with intermittent phases of again relaxed selection within Orobanche and Phelipanche (Fig. 2 B and C). In housekeeping genes and a few photosynthesis genes (e.g., ccsA, cemA, ycf3, ycf4, but not rbcL), we found evidence for adaptive evolution in a small proportion of sites (Fig. S4).
Table S1.
Lifestyle effects on selection in functional gene complexes
| Gene set | Model statistics | Test statistics | ||||||
| ID | -lnL | AICc | AIC weight | Branch set | mean ω | k | LR | P value |
| ATP | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 16208.55 | 32509.3 | 0.000 | Reference | 0.089 | 0.21 | 25.4 | <0.001 | |
| Test | 0.230 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 16202.58 | 32497.35 | 0.000 | Reference | 0.098 | 0.21 | 37.4 | <0.001 | |
| Test | 0.238 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 16193.03 | 32478.24 | 0.996 | Reference | 0.097 | 0.19 | 56.5 | <0.001 | |
| Test | 0.252 | |||||||
| Reference: nonparasites + hemiparasites; Test: holoparasites | ||||||||
| 16198.56 | 32489.3 | 0.004 | Reference | 0.136 | 0.2 | 45.4 | <0.001 | |
| Test | 0.268 | |||||||
| NDH | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 21040.46 | 42141.01 | 0.000 | Reference | 0.221 | 0.73 | 7.1 | 0.008 | |
| Test | 0.295 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 21036.13 | 42132.35 | 0.000 | Reference | 0.229 | 0.59 | 15.8 | <0.001 | |
| Test | 0.323 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 21008.64 | 42077.36 | 1.000 | Reference | 0.202 | 0.22 | 70.8 | <0.001 | |
| Test | 0.590 | |||||||
| PET | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 4500.8 | 9061.99 | 0.324 | Reference | 0.087 | 2.57 | 0.3 | 0.603 | |
| Test | 0.087 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 4500.84 | 9062.07 | 0.312 | Reference | 0.083 | 0.77 | 0.2 | 0.662 | |
| Test | 0.090 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 4500.68 | 9061.76 | 0.364 | Reference | 0.084 | 0.64 | 0.5 | 0.460 | |
| Test | 0.093 | |||||||
| PSA | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 8930.82 | 17921.83 | 0.022 | Reference | 0.046 | 0.97 | 0.1 | 0.776 | |
| Test | 0.061 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 8929.26 | 17918.71 | 0.106 | Reference | 0.033 | 0.82 | 3.2 | 0.074 | |
| Test | 0.075 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 8927.15 | 17914.49 | 0.872 | Reference | 0.037 | 0.66 | 7.4 | 0.007 | |
| Test | 0.087 | |||||||
| PSB | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 12169.43 | 24399.01 | 0.117 | Reference | 0.069 | 1 | 1 | 0 | |
| Test | 0.068 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 12168.44 | 24397.01 | 0.318 | Reference | 0.055 | 0.88 | 2 | 0.158 | |
| Test | 0.076 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 12167.86 | 24395.86 | 0.565 | Reference | 0.057 | 0.79 | 3.2 | 0.076 | |
| Test | 0.085 | |||||||
| rbcL | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 2862.61 | 5787.82 | 0.545 | Reference | 0.231 | 0.99 | 0.3 | 0.565 | |
| Test | 0.160 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 2862.35 | 5789.34 | 0.255 | Reference | 0.185 | 0.90 | 0.5 | 0.475 | |
| Test | 0.170 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 2862.60 | 5789.83 | 0.199 | Reference | 0.192 | 0.98 | 0.1 | 0.879 | |
| Test | 0.162 | |||||||
| RPL | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 13268.58 | 26649.45 | 0.017 | Reference | 0.332 | 0.92 | 0.2 | 0.683 | |
| Test | 0.389 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 13264.69 | 26641.67 | 0.839 | Reference | 0.249 | 0.46 | 7.9 | 0.005 | |
| Test | 0.403 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 13266.57 | 26645.45 | 0.127 | Reference | 0.255 | 0.76 | 4.2 | 0.041 | |
| Test | 0.414 | |||||||
| Reference: nonparasites + hemiparasites; Test: holoparasites | ||||||||
| 13268.58 | 26649.45 | 0.017 | Reference | 0.332 | 0.83 | 1.7 | 0.191 | |
| Test | 0.389 | |||||||
| RPO | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 22475.51 | 45011.11 | 0.000 | Reference | 0.214 | 0.87 | 2.1 | 0.145 | |
| Test | 0.278 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 22469.13 | 44998.35 | 0.000 | Reference | 0.198 | 0.36 | 14.9 | <0.001 | |
| Test | 0.308 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 22456.37 | 44972.83 | 1.000 | Reference | 0.190 | 0.07 | 40.4 | <0.001 | |
| Test | 0.364 | |||||||
| RPS | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 23774.84 | 47661.86 | 0.004 | Reference | 0.147 | 0.36 | 12.4 | <0.001 | |
| Test | 0.407 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 23769.4 | 47650.98 | 0.995 | Reference | 0.159 | 0.38 | 23.3 | <0.001 | |
| Test | 0.418 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 23777.17 | 47666.53 | 0.000 | Reference | 0.350 | 0.36 | 14.9 | 0.001 | |
| Test | 0.403 | |||||||
| Reference: nonparasites + hemiparasites; Test: holoparasites | ||||||||
| 23777.21 | 47666.6 | 0.000 | Reference | 0.350 | 3.48 | 7.7 | 0.006 | |
| Test | 0.403 | |||||||
| Others | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 46813.3 | 93738.68 | 0.003 | Reference | 0.672 | 1.63 | 10.6 | 0.001 | |
| Test | 0.996 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 46808.23 | 93728.53 | 0.476 | Reference | 0.598 | 2.01 | 20.7 | <0.001 | |
| Test | 1.020 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 46808.14 | 93728.35 | 0.521 | Reference | 0.545 | 1.69 | 22.9 | <0.001 | |
| Test | 1.050 | |||||||
| Reference: nonparasites + hemiparasites; Test: holoparasites | ||||||||
| 46819.26 | 93750.59 | 0.000 | Reference | 1.020 | 1.03 | 0.7 | 0.389 | |
| Test | 0.961 | |||||||
| HK | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 71275.32 | 142662.71 | 0.000 | Reference | 0.346 | 3.29 | 17.5 | <0.001 | |
| Test | 0.581 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 71265.43 | 142642.93 | 0.000 | Reference | 0.308 | 4.95 | 37.3 | <0.001 | |
| Test | 0.598 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 71246.28 | 142604.63 | 1.000 | Reference | 0.279 | 6.71 | 75.7 | <0.001 | |
| Test | 0.620 | |||||||
| Reference: nonparasites + hemiparasites; Test: holoparasites | ||||||||
| 71281.99 | 142676.04 | 0.000 | Reference | 0.592 | 1.12 | 4.2 | 0.040 | |
| Test | 0.559 | |||||||
| PS | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 66017.92 | 132095.86 | 0.000 | Reference | 0.167 | 0.93 | 0.7 | 0.421 | |
| Test | 0.179 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 66014.6 | 132089.23 | 0.000 | Reference | 0.162 | 0.81 | 7.3 | 0.007 | |
| Test | 0.187 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 66004.32 | 132068.66 | 1.000 | Reference | 0.150 | 0.72 | 27.9 | <0.001 | |
| Test | 0.225 | |||||||
| Univ. genes | Reference: Erythranthe, Sesamum; Test: Orobanchaceae | |||||||
| 76618.04 | 153348.13 | 0.000 | Reference | 0.341 | 3.85 | 31.4 | <0.001 | |
| Test | 0.649 | |||||||
| Reference: nonparasites; Test: all parasites | ||||||||
| 76616.47 | 153345 | 0.000 | Reference | 0.351 | 4.13 | 34.6 | <0.001 | |
| Test | 0.657 | |||||||
| Reference: nonparasites + Triphysaria; Test: obligate parasites | ||||||||
| 76600.5 | 153313.05 | 1.000 | Reference | 0.318 | 5.63 | 66.5 | <0.001 | |
| Test | 0.678 | |||||||
| Reference: nonparasites + hemiparasites; Test: holoparasites | ||||||||
| 76723.19 | 153550.42 | 0.000 | Reference | 0.664 | 1.11 | 4.3 | 0.039 | |
| 76631.67 | 153375.38 | Test | 0.615 | |||||
AICc, corrected Akaike information criterion; k, selection intensity parameter (according to ref. 13); lnL, log likelihood; LR, likelihood ratio from a likelihood ratio test of the RELAX null model assuming k = 1 for all branches and the RELAX alternative model assuming different k for reference and test branches (see ref. 13 for details); ω, mean ω per reference and test branch set.
Table 1.
Selectional strength
| Gene set | ωmean | k | LR | P value |
| PS | R: 0.150, T: 0.225 | 0.72 | 27.9 | <0.001 |
| HK | R: 0.592, T: 0.559 | 6.71 | 75.7 | <0.001 |
| Others | R: 1.020, T: 0.961 | 1.69 | 23.0 | <0.001 |
| UG | R: 0.318, T: 0.678 | 5.63 | 66.5 | <0.001 |
HK, housekeeping; k, selection intensity parameter (13); LR, likelihood ratio; Others, accD, clpP, ycf2; PS, photosynthesis; R, nonparasites + T. versicolor; T, obligate parasites; UG, universal genes.
Fig. S4.
Tests for adaptive evolution in Orobanchaceae. The proportion of sites evolving under different selectional regimes is illustrated as stacked branches, where each stack represents one of the three distinct ω classes: purifying selection (ω < 1) in shades of blue, neutral evolution (ω ∼ 1) in shades of gray, and positive selection (ω > 1) in shades of red. Significant changes in the proportion of sites under positive selection are marked by asterisks behind the taxon names, whose number indicates the significance level as detailed in Inset. See SI Materials and Methods for details of the test method.
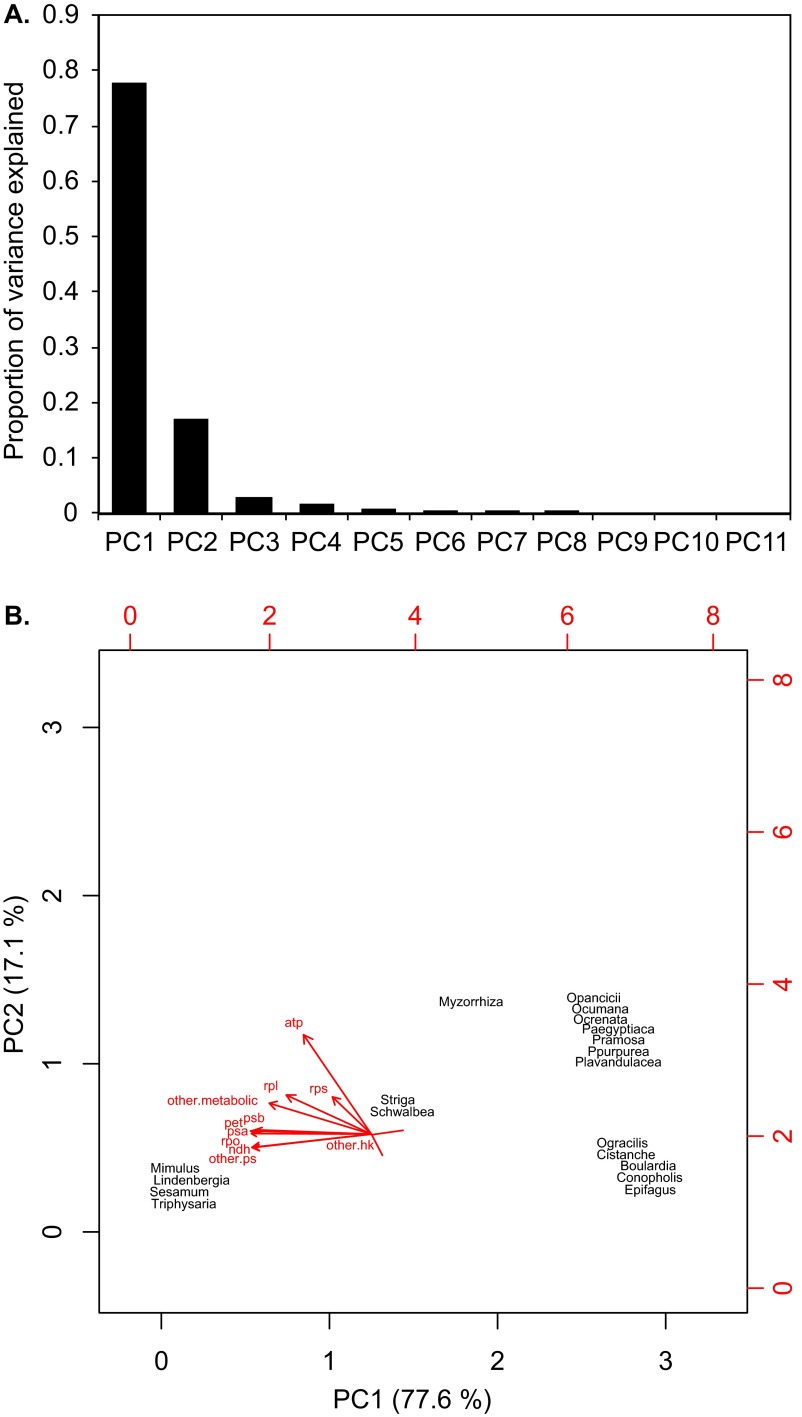
The probabilities of functional complexes to have retained their function along the transition to holoparasitism, which we obtained by averaging over the probabilities of their component genes to be intact as estimated using maximum likelihood reconstructions in BayesTraits 2, have been subjected to phylogenetic principal component analysis (SI Materials and Methods). The first two principal components (PC1, PC2) account for 94.7% of the overall variance (Fig. S5). The plastid photosynthesis gene classes ndh, pet, psa, psb, other photosynthesis-associated genes (ccsA, cemA, ycf3/4), and the rpo genes for the plastid-encoded polymerase contribute strongly (>0.95) to the first component. This result indicates that PC1 is a measure of the putative functioning of complexes associated with light harvesting and electron transport, as well as their transcription and assembly. RbcL contributes to PC1 to a lesser extent (0.84), indicating that this gene covaries with photosynthesis function. Atp genes contribute mainly to PC2 (loading: 0.81), whereas rpl, rps, rbcL, and accD load with less than 0.33. We used the Bayesian information criterion (BIC) and Schwarz weights (SW), the analog to Akaike weights, to evaluate the evidence for an array of alternative phylogenetic regression models. We found that PC1 and PC2 both represent significant factors to explain selection pressures in Orobanchaceae. A model that considers multiway interactions between these two variables (SW 0.72) outperforms a model that incorporates PC1 and PC2 additively (SW 0.26) and models that considered only one component (SW < 0.1). These results suggest that the selection shifts in parasites are shaped mainly by the nonfunctionalization of photosynthesis complexes and factors closely associated with them.
Fig. S5.
Phylogenetic principal component analysis. (A) The screeplot illustrates the variances for each component in descending order. (B) The biplot shows the position of species in ordination space (black) and the different variables (vectors in red). The explained variances (in percent) are provided for PC1 and PC2 in the axis labels.
Microstructural Changes.
The occurrence of length mutations in plastid genes caused by short insertions or deletions (indels) varies strongly between the different gene classes, with intact photosynthesis genes usually accumulating fewer indels (<1 per gene) than housekeeping genes (Fig. S6). Indel rates in genes that are universally present in Orobanchaceae do not differ among nonparasites, photosynthetic parasites, and the nonphotosynthetic parasites M. californica, Orobanche spp., and Phelipanche spp., whereas the remaining holoparasites (Boulardia latisquama, C. phelypaea, C. americana, and E. virginiana) show more length mutations. Phelipanche spp. (1.5–1.67 indels per gene) show slightly more length mutations than Orobanche spp. (0.83–1 indel per gene) and M. californica (0.83 indel per gene), which has indel rates similar to those of photosynthetic plants (0.83 indel per gene). Retained photosynthesis genes of holoparasites often show unique indels. For example, psbZ in O. gracilis, psaJ and psbJ in C. phelypaea, and ndhB, petA, psaC/I, and psbA/D/K in M. californica show length mutations, but none of the photosynthetic taxa have indels in any of these genes. Indels are rare in atp genes of nonphotosynthetic parasites. Mapping substitution rates and microstructural changes onto the dated Orobanchaceae phylogeny suggests that indel accumulation precedes substitution rate changes in some holoparasites in both housekeeping genes and atp genes (Fig. 2A), whereas in other photosynthesis genes (lacking in holoparasites), this effect is less obvious (Fig. S3).
Fig. S6.
Indels in plastid protein-coding genes of Orobanchaceae. The heatmap shows differences in number and distribution of indels for each plastid protein gene. Green to red colors indicate increasing rates of insertions, whereas green to purple colors indicate increasing rates of deletions.
Lifestyle-Dependent Changes of Evolutionary Rates.
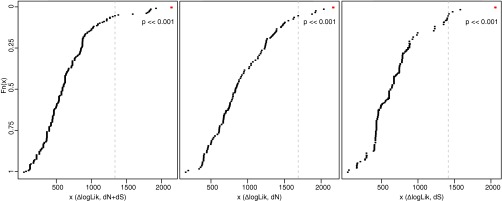
The dependency of evolutionary rates (dN and dS separately and jointly) on lifestyle was tested by using models that fuse sequence and trait evolution (15), thus evaluating whether the transition to obligate parasitism contributes significantly to evolutionary rate changes. The overall best models for the total substitution rate, dN, and dS distinguish between nonparasites plus the facultative hemiparasite T. versicolor versus obligate parasites irrespective of photosynthetic capacity (LRTs against the respective null models that assumed no influence of lifestyle changes, all P < 0.001). Parametric trait bootstrapping (15), which tests whether the observed rate variation is associated with an analyzed trait significantly more often than with uncorrelated traits that evolve in a similar manner, supports that shifts in dN and dS, alone and jointly, are significantly associated with changes in lifestyle (all P < 0.001) (Fig. S7). These results indicate that changes of nucleotide substitution rates coincide with the establishment of obligate parasitism rather than with the loss of photosynthesis.
Fig. S7.
Parametric bootstrap analysis of trait-dependent nucleotide substitution rate shifts. To analyze the effect of lifestyle changes on the total substitution rate (dN+dS) (Left) and on dN (Center) and dS (Right) separately, we simulated 200 datasets and plotted the log-likelihood difference (∆logLik) between the null and the alternative model estimated from these simulated datasets (black dots) against the log-likelihood difference from the real data (red dot). The parametric bootstrap (41) tests whether the observed rate variation correlates with the investigated trait (here lifestyle) significantly more often than with uncorrelated traits that may evolve in a similar manner. We tested whether the observed likelihood ratio (i.e., the difference in logarithmic space) lies within the distribution of likelihood ratios obtained from the simulated datasets (H0, no difference between the observed and simulated likelihood ratio; HA, observed likelihood ratio differs from the simulated data). A dashed line indicates the critical value of the 0.05 significance level for a one-sided test.
Genetic Factors Underlying the Substitution Process.
We studied the association of evolutionary rates (dN, dS, and the total rate µ) and ω with genetic traits (genome rearrangements, plastome size, gene content, GC content, indels), lifestyle, and life history by using uniresponse and multiresponse generalized linear mixed models using Markov chain Monte Carlo methods (MCMC-GLMM) (16). Substitution rates (dN, dS) and ω are each highly correlated with genetic traits. Uniresponse MCMC-GLMM indicates that dN relates more strongly to genetic traits than dS. The best models (according to SW) suggest additive effects of indels, the number of gene losses, the plastome size, and the lifestyle to predict both dN and µ, whereas dS is affected by the number of rearrangements (rather than indels per se), gene losses, plastome size, and the lifestyle (Table 2). Life history is present in none of the top-ranked models. Unlike in the evolutionary rates models, the best ω model requires only the indel rate as factor (SW 0.62), suggesting that indels tend to accumulate in regions with high ω in Orobanchaceae. This univariate model outperforms a bivariate model with additive effects of indels and lifestyle (SW 0.25).
Table 2.
Phylogenetic MCMC-GLMMs
| Model | SWs* |
| µ ∼ indels + gene loss + pt size + lifestyle | 0.334 |
| µ ∼ GR + gene loss + pt size + lifestyle | 0.270 |
| µ ∼ indels + gene loss + pt size | 0.157 |
| dS ∼ GR + gene loss + pt size + lifestyle | 0.255 |
| dS ∼ indels + gene loss + pt size + lifestyle | 0.225 |
| dS ∼ indels + gene loss + pt size | 0.142 |
| dS ∼ GR + pt size + lifestyle | 0.092 |
| dN ∼ indels + gene loss + pt size + lifestyle | 0.388 |
| dN ∼ GR + gene loss + pt size + lifestyle | 0.276 |
| dN ∼ indels + gene loss + pt size | 0.147 |
| dN, dS ∼ indels + gene loss + pt size | 0.346 |
| dN, dS ∼ GR + pt size + lifestyle | 0.213 |
| dN, dS ∼ indels + pt size + lifestyle | 0.145 |
| ω ∼ indels | 0.617 |
| ω ∼ indels + lifestyle | 0.247 |
GR, genome rearrangements based on locally collinear blocks; µ, total rate, pt, plastome.
Calculated separately over the set of models per response variable(s), and only models with a cumulative weight of 0.7 are shown.
SI Materials and Methods
Annotation.
We annotated the newly reconstructed plastomes by using the procedures and settings described earlier (5, 11). Additionally, we queried combined builds of transcriptome data from Phelipanche aegyptiaca (build BC4 with 1,221,257 unigenes) and Striga hermonthica (build BC2 with 726,534 unigenes) (39) to assist the annotation and classification of genes as pseudogenes. We classified genes as functionally lost (pseudogenes) if they were truncated, showed frame shifts, or had a high sequence drift compared with intact genes of the nonparasite Lindenbergia philippensis. The following genes may be pseudogenes (due to either one premature stop codon, an uncertain start codon, or high sequence divergence), but were treated as intact in all analyses: accD in S. hermonthica, Schwalbea americana, and all Phelipanche species; psaJ and psbJ in Cistanche phelypaea; rps16 in Boulardia latisquama; clpP in all Orobanche species; atpE and atpF in O. crenata; psbM and psbZ in O. gracilis; atpA, psbA, and psbJ in Myzorrhiza californica; and rps3, rps11, rps15, and ycf1 in all Phelipanche species. Annotations of some of the earlier published Orobanchaceae plastomes (5) were revised with respect to gene delimitations and their classification as intact or pseudogene after Illumina or Sanger sequencing-based error correction as follows: psbM and psbZ in O. gracilis, rpl22 in Conopholis americana and B. latisquama, rps15 in Phelipanche purpurea, and rps3 in Phelipanche ramosa, ycf1 in all Phelipanche species, as well as trnRUCU in B. latisquama, trnGUCC in Phelipanche lavandulacea, and trnSUGA in Co. americana; Fig. S1 provides a graphical summary of the gene content in all study taxa.
AccD of S. americana contains a premature stop codon, verified by Illumina resequencing; expression data are not available. The accD gene in Phelipanche is 5′ truncated, lacking more than 700 bp compared with other Orobanchaceae. Although the latter half of accD in Phelipanche shows significant similarity to known plastid accD, we could identify only two unigenes with partial similarity (e value > 1e−100) to the annotated plastid accD in transcriptome data of P. aegyptiaca (39). We excluded accD of Phelipanche from all rate and selection tests. In S. hermonthica, the 5′-end of accD is highly diverged, precluding unambiguous identification of the gene start. However, several transcripts cover 85% of the annotated gene region, but transcriptome data do not allow an unambiguous identification of the start codon. We therefore included only the reading frame covered by transcriptome data in the rate analyses, thus lacking 39 bp (13 aa) compared with nonparasites. The matK gene of Epifagus virginiana, C. americana, and C. phelypaea was included in rate and selection tests as described earlier (5). Rps16 of Orobanche cumana lacks its typical intron, and in B. latisquama the gene is 3′-truncated lacking 13 aa due to a premature stop codon. The petA gene of M. californica is 3′-truncated by approximately 10 aa compared with hemiparasites and nonparasites. The gene start of rpl33 is unclear in all Phelipanche species, but transcripts cover 96% of the annotated gene region, and we therefore treated the gene as intact and included the validated coding sequences in rate and selection tests. P. purpurea has two copies of rps15, which differ in their length because of a long indel in frame, and we used the copy more similar to other parasite rps15 genes for all rate and selection tests. trnGUCC in P. lavandulacea has an abnormal D-loop secondary structure due to noncompensatory base-pair changes, and it may therefore not be functional. The gene ycf1, although present in all species, was excluded from all analyses of substitution rates, selection pressures, and indels because of unresolvable uncertainties regarding the correct homology assignment.
Analysis of Gene Loss and Probabilities of Functional Complexes to Have Retained Their Function.
To assess the history of gene losses and the probabilities of functional complexes to have retained their function over time, we reconstructed the ancestral states for unique plastid genes (gene duplicates due to a localization in the IR were ignored); that is, 79 protein-coding genes + 4 rRNAs + 30 tRNAs, using the maximum likelihood approach (with 500 estimation attempts) implemented in BayesTraits 2 (37). The input tree topology of the study taxa was imposed according to the established phylogenetic relationships in Orobanchaceae (10, 11), with branch lengths scaled according to the total rate across all universal plastid genes (branch length optimization is described below in Analysis of Molecular Evolutionary Rates; see Fig. 1 and Fig. S1 for the set of universal plastid genes in Orobanchaceae). We coded all plastid genes as binary traits with the states being either intact or absent/pseudogene [matrix available from Dryad (10.5061/dryad.t2m75) upon publication]. The probabilities of genes being intact or absent/pseudogene at the tree’s internodes were computed by using an unconstrained two-parameter model, which showed a significantly better fit (LRT: P < 0.001) than a model, where a reversal from absent to intact was not permitted by enforcing the respective rate to be zero. The results (graphically summarized in Fig. S1B) are available from Dryad (10.5061/dryad.t2m75) upon publication. The probability of the plastid-encoded fraction of a functional complex to have retained its function was approximated by using the ML-estimated probabilities of being intact obtained for each gene per functional complex. Specifically, we calculated the probability of a functional complex to have retained its function as the average of the probabilities of being intact over all genes coding for components of a given functional complex [available from Dryad (10.5061/dryad.t2m75) upon publication]. This measure (i.e., the probability of a functional complex to have retained its function) provided a continuous (instead of discrete) quantification of the putative degree of functioning, thus avoiding the use of arbitrary probability cutoffs as the decision criterion. Additionally, it indirectly allowed, at least to some extent, for the possibility of continued functioning of a complex after gene loss from the plastome due to functional replacement by a cytosolic gene copy (de novo or after intracellular gene transfer) (1, 2, 11). These data of the potential functioning of the different complexes were further reduced by phylogenetic principal component analysis (47), using a dataset with the probabilities of each functional complex to have retained its function per node and our Orobanchaceae tree (10, 11) as input data. The resulting principal components (PCs), which together explained at least 90% of the overall variance were extracted (i.e., PC 1 and 2) and subjected to further analysis with MCMC-GLMM (16) (described in detail in Phylogenetic MCMC-GLMMs) to evaluate associations of selection pressures in retained genes with gene losses.
Analysis of Molecular Evolutionary Rates.
Nucleotide substitution rates and the ratio of nonsynomymous to synonymous substitution rates (ω) were analyzed with HyPhy 2.1–2.3 (39), running tests both gene-wise and by functional classes. We aligned all datasets codon-wise by using prank 0.14 (41) with a guide tree reflecting the accepted phylogenetic relationships of the study taxa (10, 11), the empirical codon substitution model, and the standard genetic code; no sites were excluded for any of the subsequent analyses. Tests of relative nonsynonymous and synonymous substitution rates (dN and dS, respectively) for all plastid protein genes (single and concatenated according to their functional gene class or combined as dataset of universally retained genes) were carried out by using custom batch scripts and the MG94×GTR hybrid model with a corrected 3 × 4 codon frequency estimator as described recently (11). In brief, to evaluate the significance of substitution rate differences between two species, the log likelihoods of an unconstrained (i.e., allowing individual rates per taxon) and a constrained model (i.e., the substitution rates on the branch of interest are forced to be identical to that of the reference) were compared by building and optimizing individual likelihood functions for each gene and the constrained and unconstrained models. Pairwise rate tests were conducted between each of the parasitic taxa, L. philippensis, and Erythranthe guttata (syn. Mimulus guttatus), using Sesamum indicum as outgroup. Results were visualized as heatmaps by using the heatmap function of the R package stats. All input files and the R script are available from Dryad (10.5061/dryad.t2m75) after publication.
To test for correlations between dN and dS, we optimized the branch lengths of the established Orobanchaceae phylogeny (10, 11) for each concatenated gene dataset (corresponding to the different functional complexes and the set of universal genes) separately by using the MG94×GTR-3×4 model in HyPhy 2.1–2.3 (42), and extracted the resulting dN- and dS-scaled trees. Using the R package ape (45), these trees were transformed into patristic distance matrices that were subsequently used as input for Mantel tests with phylogenetic permutation (12) (run with 1,000 permutations each; i.e., taking nonindependence among species into account). This method allowed the necessary analysis of pairwise distances among taxa and avoided treating evolutionary rates as ordinary (phenotypic or genotypic) traits.
Lifestyle and Substitution Rate Analysis.
We analyzed the effect of lifestyle changes on the evolution of nucleotide substitution rates using traitRate 1.1 (15), which allows trait-dependent changes in evolutionary rates to be detected in a computationally unified framework under the maximum likelihood paradigm. Evolutionary rates were provided as phylogenetic trees with branch lengths rescaled by using the set of universal genes (described in Analysis of Molecular Evolutionary Rates). To evaluate lifestyle-dependent changes of evolutionary rates, we compared, per evolutionary rate (total rate, dS, dN), a model that assumes that the substitution rate evolution correlates with lifestyle changes (M1) with a model that assumes no such correlation (M0) and tested for statistical significance using LRTs (15). Additionally, we used parametric bootstrapping to test whether the observed value of the LRT test statistic D [i.e., 2 × (log-likelihood (M1) − log-likelihood (M0)], was significantly greater than expected for traits that evolve in a similar manner as our trait of interest, but are uncorrelated with the molecular evolutionary rates (41). Thus, this procedure accommodates that existing rate variation may not be due to the trait of interest (i.e., lifestyle) alone. For each evolutionary rate, 200 lifestyle data matrices (i.e., randomly generated trait data for the tips of the tree) were simulated along the original Orobanchaceae phylogeny in traitRate by using the parameters inferred by ML for the original dataset under the M1 model. For each of the 200 simulated trait datasets, we optimized the likelihood function under model M0 and model M1 and calculated the LRT test statistic D, thus obtaining a distribution of D expected when existing substitution rate variation is not associated with lifestyle. The empirical P value (i.e., the proportion of D values from the simulated datasets that are at least as high as the D value from the original data) was estimated by using the R package stats and used for rejecting the null hypothesis of no association between substitution rates and lifestyle.
Analysis of Microstructural Changes.
To analyze the evolution of microstructural changes, we coded insertions and deletions (indels) in all concatenated datasets representing the different functional complexes and the dataset of universal genes with the command-line version of SeqState 1.4 (43) using the simple gap coding procedure (SIC) (44); indels at the alignment borders were not considered (SeqState option: “noborder”). We extracted the SIC-coded indel matrix from the results files and formatted each of these as nexus file. Given the accepted species tree (10, 11) (Figs. 1 and 2), we reconstructed the indel history by maximum parsimony over the tree by using the R packages ape (45) and phangorn (46) [R code available from Dryad (10.5061/dryad.t2m75) upon publication]. The lengths of the tree branches thus were scaled according to the number of indel events per branch. Additionally, we assessed the frequency of indel events in all protein-coding genes and taxa by counting the occurrence of species-specific gaps for all aligned datasets and visualized the results as heatmaps by using the heatmap function of the R package stats.
For the visual inspections of the time series of indel and substitution rate evolution per functional complex, we used penalized likelihood (PL) (48) (implemented in ape) given our phylogenetic tree with branches scaled to reflect the total rate over all universal plastid genes (branch length optimization performed by using Hyphy; described in Analysis of Molecular Evolutionary Rates). We constrained the root age to 51–71 million years ago (11). Because the results of PL were in line with an earlier study that used various Bayesian methods for the molecular dating of Orobanchaceae (11), we refrained from using additional and more sophisticated divergence dating methods here. We then used the reconstructed history of dN, dS (as inferred with Hyphy, described in Analysis of Molecular Evolutionary Rates), and indels (coded with SeqState using SIC and parsimony-optimized) per functional complex to plot and paint three trees per complex according to the number of dN, dS, or indel events per branch using the plot.phylo-function implemented in the R package ape (44).
Analysis of Selectional Changes.
Changes of ω across all functional complexes were tested with RELAX (13). Different test branch sets were evaluated by using Akaike weights to identify the best lifestyle model per gene. Based on a branch-site random effects likelihood method to test for episodic diversifying selection (bS-REL) (14), the RELAX framework uses a selection intensity parameter, k, to test whether and how ω deviates from neutrality (i.e., ω = 1). As relaxation of selection distinctly affects sites under purifying selection (ω < 1) and sites under positive selection (ω > 1), it will move ω toward 1 if selection is relaxed (i.e., ω < 1 increases and ω > 1 decreases). Using partitioned reference and test branches in a given tree, the null model assumes k = 1 for all branches (i.e., test and reference branches have the same ω distribution), whereas in the alternative model, k is allowed to differ for the reference and test branch set. In addition to RELAX, we used bsREL to analyze per branch estimates of the proportions of sites under different selectional regimes, and to specifically test for sites under positive selection. We preferred bsREL over similar approaches because this method controls more efficiently for the rate of false positives and the loss of power by making no assumptions about foreground and background lineages (14). bsREL performs a series of LRTs with subsequent sequential alpha error correction (Bonferroni–Holm correction for multiple testing) to identify all lineages where a proportion of sites, whose extent is estimated by the model, evolves with ω > 1 across the tree and sites. This method therefore allows assessing the extent of adaptive evolution in plastid genes along the transition to holoparasites. R in combination with the ape package (45) was used to combine and visualize the results of bsREL.
Phylogenetic MCMC-GLMMs.
Phylogenetic MCMC-GLMMs were computed with an inverse G-matrix accounting for phylogenetic relationships (16), whereby random effects were assigned to the taxa. We used unfixed and least-informative priors for both the variance-covariance matrix of the random effects and the residuals. For the inverse Wishart distributions of the uniresponse (total rate, dN, dS, and selection pressure assessed by ω) and biresponse (dN and dS) models, we used hyperpriors corresponding to one-half and one-quarter of the dataset’s variance, respectively. As fixed effects, we used genome rearrangements (GR), which we inferred from locally collinear blocks (described in refs. 5 and 11), plastome size, gene content (Fig. S1), total GC content [obtained by using SeqState 1.4 (43)], indels (extracted as branch data from indel-scaled trees; described in Analysis of Microstructural Changes), lifestyle (Table S2), and life history (Table S2). We derived evidence for the best phylogenetic regression model from the array of alternative models from the BIC and SW. Starting from full models with all factors included, we reduced the models stepwise by one factor (according to its significance in the model) until the reduction yielded no better fit according to BIC. Models that allowed multiway interactions between factors were omitted as these converged significantly worse. Per fit, we collected 10,000 samples, sampled every 10th generation, allowing for an initial burn-in of 20%. Using BIC weights, we considered only models in the final set until the cumulative SW reached or exceeded 0.7. We used the same strategy to evaluate whether gene losses (measured as PCs from the probabilities of the plastid-encoded fraction of each of the different functional complexes to have retained their function; see Analysis of Gene Loss and Probabilities of Functional Complexes to Have Retained Their Function) affect selection pressures (measured as ω) in retained genes across the Orobanchaceae tree. Phylogenetic relationships were accounted for as described above. In addition to additive models, we also tested for interactions between the fixed effects in MCMC-GLMM. As above, all PC regression models were ranked by BIC and SWs, including only those whose SW reached or exceeded 0.9.
Table S2.
Studied species
| Taxon name | GenBank accession no. | Lifestyle | Life history |
| Boulardia latisquama | HG514460 | SH | Perennial |
| Cistanche phelypaea | HG515538 | SH | Perennial |
| Conopholis americana | HG514459 | SH | Perennial |
| Epifagus virginiana | M81884 | SH | Annual |
| Erythranthe guttata | PRJNA253667 | NP | Annual |
| Lindenbergia philippensis | HG530133 | NP | Perennial |
| Myzorrhiza californica | HG515539 | SH | Perennial |
| Orobanche crenata | HG515537 | GH | Annual |
| Orobanche cumana | KT387722 | SH | Annual |
| Orobanche gracilis | HG803179 | GH | Biennial |
| Orobanche pancicii | KT387724 | SH | Annual |
| Phelipanche aegyptiaeca | KU212370 | GH | Annual |
| Phelipanche lavandulacea | KU212371 | SH | Annual |
| Phelipanche purpurea | HG515536 | SH | Biennial |
| Phelipanche ramosa | HG803180 | GH | Annual |
| Schwalbea americana | HG738866 | OH | Biennial |
| Sesamum indicum | NC016433 | NP | Annual |
| Striga hermonthica | KU212372 | OH | Annual |
| Triphysaria versicolor | KU212369 | FH | Annual |
FH, facultative hemiparasite; GH, generalist holoparasite; NP, nonparasite; OH, obligate hemiparasite; SH, specialist holoparasite.
Discussion
Changes in gene content and shifts of nucleotide substitution rates have been commonly thought to relate to the relaxation of selective constraints and the loss of photosynthesis in plastid-bearing lineages that have secondarily acquired a parasitic lifestyle. Here, we showed that obligate parasitism, characterized by a parasite’s need for a host plant, and the loss of photosynthesis strongly affects the functional reduction and rate accelerations in plastid genomes of Orobanchaceae, whereas parasitism per se, that is the ability to tap into the vascular tissue of another plant, are of subordinate importance (Fig. 1 and Tables 1 and 2). Both plastid photosynthesis and housekeeping genes evolve at significantly elevated evolutionary rates (including elevated indel rates) not only in holoparasites but already in the obligate hemiparasitic S. americana and S. hermonthica. Their plastomes also show more genomic rearrangements than autotrophs and the facultative hemiparasites T. versicolor and Bartsia inaequalis (17). These results are further corroborated by gene expression data from aboveground tissue of S. hermonthica, which expresses nuclear genes for light harvesting and photosystems with lower abundance than T. versicolor (18). The relaxation of purifying selection in both photosynthesis and housekeeping genes (e.g., rpl, rps) (Fig. 2) accompanying the transition to an obligate parasitic lifestyle also occurs in other parasitic lineages such as the sandalwood order (Santalales), a large group of flowering plants that has evolved root and stem parasites. Here, the plastomes of facultative hemiparasitic Ximenia americana and Osyris alba are highly similar to the nonparasite Heisteria concinna regarding their gene contents and evolutionary rates, whereas the obligate parasites Phoradendron leucarpum and several Viscum species show gene losses and relaxed selection in photosynthesis genes (19).
Accelerated evolutionary rates in plastomes have often been associated with changes in life history, which is the shift from long to short generation times (20). We found no consistent effect of generation time on rate variation (Table 2), which might be due to a high variability in life span, because in parasitic plants, generation time may be determined by host quality (only on annual hosts the parasite must be annual itself) rather than by intrinsic features. The strong coevolution of dN and dS (Table 2) suggests a lineage effect via the actual process of mutation (neutral mutation rate hypothesis), which, among others, depends on species-specific differences in DNA replication and repair efficiencies (21). As the selective pressures on plastid function gradually decrease in parasites, the proteins for DNA processing and DNA maintenance may experience relaxations of selection, just like the plastid genome they replicate or repair.
Changes in selection do not occur in a monotonic fashion. Instead, phases of rate acceleration and relaxed selection that coincide with inferred bursts of gene loss (Fig. 2 A and B) are followed by phases of rate deceleration and intensified selection in the retained functional complexes (Fig. 2B), suggesting that the plastomes of parasites have evolved toward a new rate equilibrium; we propose that this is due to transcript and protein turnover rate-dependent substitution rate shifts. In photosynthetic lineages, the high demand for the photosynthesis-related machinery selects for low nucleotide and amino acid substitution rates to maximize rapid translation through optimized codon use maintained by low dN and dS, and to minimize the risk of unfavorable protein misfolding (20). Therefore, codon use and substitution rates differ notably between the different gene classes in plastomes of flowering plants (22), whereas in parasites, this distinctness diminishes (5). Here, selective pressure on high turnover in the plastid is reduced, because the parasite obtains at least parts of the required organic compounds from its host rather than synthesizing them. Because of the reduced need for a rapid assembly of the thylakoid photosynthesis machinery, purifying selection is relaxed not only in photosynthesis genes, but also in genes encoding the transcription and translation machinery, allowing for indels to accumulate and dN and dS to increase. This finding is in line with the relaxation of selection in the genes for the plastid-encoded polymerase (rpo) seen already in the photosynthetic but obligate parasites S. americana, S. hermonthica (Table S1), mistletoes (19), and the repeatedly observed overall higher evolutionary rates in parasites (Fig. 1). These changes may also involve adaptations in the plastid housekeeping apparatus (Fig. S4). The hypothesized turnover rate-dependent rate shifts will be attenuated in genes that are required over longer periods, such as the ATP synthase (atp) genes or RuBisCO (rbcL), possibly because they take over or continue to carry out alternative functions (23, 24). Therefore, nonphotosynthetic parasites such as M. californica, Orobanche (11), or some Cuscuta species (7, 25), which all retain intact genes for the ATP synthase despite the loss of other photosynthesis genes, also have lower base-level evolutionary rates. The eventual deletion of all dispensable regions may reconstitute the compactness of the plastid chromosome with its typically low amounts of nongenic and low-complexity DNA regions (1).
A Model of Plastome Evolution in Parasites.
Based on our data and previous research (4–7, 9, 11, 19, 25–35), we here propose a model of plastid genome evolution under relaxed selective constraints (Fig. 3). This model is applicable to many other secondarily heterotrophic lineages within primarily phototrophic clades other than Orobanchaceae such as algae and mycoheterotrophic plants.
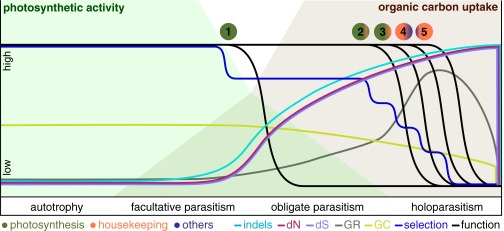
Fig. 3.
General model of plastome evolution under relaxed functional constraints. The model illustrates the evolution of genomic traits (GC, GC content; GR, genomic changes including gene deletions), functionality of different plastid-encoded complexes (blue, other metabolic genes; green, photosynthesis genes; orange, housekeeping genes), evolutionary rates (dN, dS) as well as selection intensity and their associations during the transition from autotrophy with high photosynthetic activity (green background) and no uptake of external organic carbon to holoparasitism with maximal uptake (brown background). As the specialization on the heterotrophic lifestyle increases, more plastid-encoded complexes are lost, altering evolutionary rates and the genome structure. Numbers indicate the predicted functional reductions starting with the loss of nonessential or stress-relevant genes (ndh genes) (1) followed by primary photosynthesis genes (pet and psa/psb genes) and the plastid-encoded polymerase (PEP) (2), genes that have a prolonged function (e.g., atp genes, rbcL) and nonessential housekeeping genes (3), other metabolic genes (e.g., accD, clpP, ycf1/2) (4), and, finally, the remaining housekeeping genes, including trnE (5). Unlike most factors that are likely to show a rather monotonic trend as parasitism evolves, selectional strength may experience several periods of relaxation and intensification. The associations of evolutionary rates, genomic traits, and functional losses between 1 and 3 have been observed, whereas those after 3 are hypothetical. For simplicity, we here depict the functional reduction of a plastome with a coding capacity of flowering plants; more or fewer lineage-specific reconfigurations may occur in other lineages.
Parasitism relaxes constraints on the NADH complex that is essential for electron cycling around photosystem I under stress, rendering ndh genes the first, to our knowledge, to be functionally lost from the plastome (5, 9). More dramatic changes concur with the transition to obligate parasitism, which relieves photosynthesis and, concomitantly, on plastid housekeeping functions of functional constraints (losses 1 and 2 in Fig. 3). This first phase of selection relaxation is characterized by a steady increase of microstructural changes and the acceleration of dN and dS. Following this episode of selectional shift, evolutionary rates in the plastome evolve at a new equilibrium, perhaps matching the modified transcript and protein requirements (11). This molecular evolutionary regime shift is repeated once selective constraints on plastid proteins (e.g., ATP synthase) that continue to function for a longer period are lost or functionally replaced (functional loss 3 in Fig. 3). The relaxation of selective pressure on photosynthesis, alternative photosynthesis-unassociated functions, and on the plastid housekeeping machinery may be linked to the increasing specialization on external carbon (e.g., via distinct host systems with improved efficiency of nutrient acquisition), but the precise coevolutionary mechanisms remain unclear. The lifestyle-specific shifts of evolutionary rate regimes are accompanied by a reduced plastome GC content that may in part be due to relaxed constraints on codon use or nutrient economy (5, 36), although the changing GC content apparently has no direct influence on dN and dS (Table 2). However, low GC content correlates with increases in the amount of structural rearrangements (37) including the deletion of dispensable DNA (5), all factors that directly influence the substitution rates of plastid genes (Table 2) (11).
Reasons for the retention of minimal plastomes in most nonphotosynthetic plants investigated so far are varied. The proportion and nature of lineage-specifically retained genes and nonessential DNA potentially relate to inefficient or impossible protein import, highly reduced translational apparatus, regulatory coupling of genes for biological processes, coordinate assembly and cotranslation of partnered proteins, divergences in the genetic code (including modified start codons), or posttranscriptional editing that all represent barriers for functional gene transfer (38). Even if all protein genes are lost, the plastid-encoded l-glutamyl-tRNA (trnE) may still be required for tetrapyrrole biosynthesis in plants, which requires its functional transfer to the nuclear genome because the nuclear equivalent cannot replace the initiator function (38; but see ref. 35). However, a minimal plastome with only one tRNA that is nearly indifferent from the cytosolic tRNA species would be difficult to detect, even with high-throughput methods and in situ visualization techniques. Functional recompartmentalization of molecular biological processes and functional replacement of plastid-encoded genes through, for instance, functional intracellular gene transfer may, however, eventually relieve the organelle of the pressure to retain own genetic material (losses 4 and 5 in Fig. 3), potentially leading to plastids without plastomes [as in Rafflesia (32); Polytomella (33)].
Materials and Methods
Taxon Sampling and Plastome Sequencing.
Using whole-genome shotgun sequencing (454 FLX and Illumina, paired-end), we reconstructed the plastomes of four holoparasites and two hemiparasites (Table S2) in addition to the Orobanchaceae already sequenced (5, 6, 11), using the same experimental and bioinformatic procedures. We queried combined builds of transcriptome data from Phelipanche aegyptiaca and S. hermonthica (39) to assist the annotation of plastid genes. Details are provided in SI Materials and Methods.
Analysis of Gene Content and Evolutionary Rates.
Based on a matrix of all unique plastid genes and the established phylogenetic relationships (10, 11), we reconstructed gene losses at ancestral nodes by using BayesTraits 2 (40) under the multistate option and 500 maximum likelihood (ML) attempts. We calculated the probability of a functional complex to have retained its function at each node by averaging over the ML estimates of probabilities of its contributing genes to be intact (Fig. S1). Following automated, codon-wise alignments by using prank 0.14 (41), analyses of relative dN and dS, and of the total rate in all plastid protein genes were carried out in HyPhy 2.1–2.3 (42) using custom batch scripts and the MG94×GTR_3×4 codon model; ycf1 was excluded because of uncertain homology assessment. Changes of ω and of the strength of selection measured by k were tested with a series of branch-site random effects likelihood methods (13, 14). For identifying the best lifestyle model per gene, different test branch sets were defined and evaluated by using Akaike weights. Details of all procedures are provided in SI Materials and Methods.
Analysis of Evolutionary Rates, Selection Pressures, and Genetic Factors.
We used traitRate 1.1 (15) with 100 stochastic mappings and LRTs to test for lifestyle-dependent rate changes. To this end, we compared the log likelihoods of a null model that assumes no trait dependency versus a model that includes a trait parameter for each tested rate (total rate, dN, or dS) on the set of universally retained genes (Fig. 1 and Fig. S1) and performed parametric bootstrap analyses (as in ref. 15) of the best trait-rate models using 200 replicates. We measured rearrangements by locally collinear blocks (Fig. S2) (as in refs. 5 and 11). We distinguished lifestyle as nonparasite, facultative hemiparasite, obligate hemiparasite, generalist holoparasite, or specialized holoparasite, and life history as annual, biennial, and perennial (Table S2). Indels were coded with SeqState 1.4 (43) using SIC (44), and their occurrence was reconstructed over the Orobanchaceae tree by using the R packages ape (45) and phangorn (46). Gene losses per branch were calculated as the percentage of nonessential unique genes lost, where nonessential refers to a gene that has been lost in one or more of the study taxa. Phylogenetic MCMC-GLMMs (16) were computed by using unfixed and least-informative priors for both the variance-covariance matrices of the random effects and the residuals, and variance-corresponding hyperpriors. Factors were hierarchically reduced by significance until no better model fit was obtained according to BIC. We considered only models in the final set until the cumulative Schwarz weight per response exceeded 0.7. We used phylogenetic principal component (47) regression to model associations between selection pressure and the probability of the plastid-encoded fraction of a functional complex to have retained its function (from BayesTraits ML probabilities per complex). All components that together explained at least 90% of the variance were used as fixed effects in phylogenetic MCMC-GLMM analysis (as above). To evaluate the time series of genetic changes, we traced dN, dS, and indels on dated phylogenies, which we obtained by using penalized likelihood (48) implemented in ape (45), setting the root age boundary to 51–71 million years ago (11). Details of all coevolutionary analyses are provided in SI Materials and Methods.
Acknowledgments
We thank S. Renner (Munich) and T. Rattei (Vienna) for access to genome data of some holoparasites; and J. Naumann (Pennsylvania State University) and two anonymous reviewers for valuable comments on an earlier version of this manuscript. This work was supported by Austrian Science Fund FWF Grant 19404 (to G.M.S.); National Science Foundation Grants DBI-0701748 and DBI-1238057 (to C.W.d.); and the German Academic Exchange Service (S.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HG514460, HG515538, HG514459, M81884, HG530133, HG515539, HG515537, KT387722, HG803179, KT387724, KU212370, KU212371, HG515536, HG803180, HG738866, NC016433, KU212372, and KU212369) and in Dryad Digital Repository, datadryad.org (10.5061/dryad.t2m75).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607576113/-/DCSupplemental.
References
- 1.Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol Biol. 2011;76(3-5):273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen RK, Ruhlman TA. Plastid genomes of seed plants. In: Bock R, Knoop V, editors. Genomics of Chloroplasts and Mitochondria. Springer; Dordrecht, The Netherlands: 2012. pp. 103–126. [Google Scholar]
- 3.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young ND, dePamphilis CW. Rate variation in parasitic plants: Correlated and uncorrelated patterns among plastid genes of different function. BMC Evol Biol. 2005;5(1):16. doi: 10.1186/1471-2148-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wicke S, et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25(10):3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89(22):10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7(1):45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohan AJ, Wolfe KH. A subset of conserved tRNA genes in plastid DNA of nongreen plants. Genetics. 1998;150(1):425–433. doi: 10.1093/genetics/150.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett CF, et al. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 2014;31(12):3095–3112. doi: 10.1093/molbev/msu252. [DOI] [PubMed] [Google Scholar]
- 10.McNeal JR, Bennett JR, Wolfe AD, Mathews S. Phylogeny and origins of holoparasitism in Orobanchaceae. Am J Bot. 2013;100(5):971–983. doi: 10.3732/ajb.1200448. [DOI] [PubMed] [Google Scholar]
- 11.Cusimano N, Wicke S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2016;210(2):680–693. doi: 10.1111/nph.13784. [DOI] [PubMed] [Google Scholar]
- 12.Lapointe F-J, Garland T., Jr A generalized permutation model for the analysis of cross-species data. J Classif. 2001;18(1):109–127. [Google Scholar]
- 13.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 2015;32(3):820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosakovsky Pond SL, et al. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 2011;28(11):3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayrose I, Otto SP. A likelihood method for detecting trait-dependent shifts in the rate of molecular evolution. Mol Biol Evol. 2011;28(1):759–770. doi: 10.1093/molbev/msq263. [DOI] [PubMed] [Google Scholar]
- 16.Hadfield JD. MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J Stat Softw. 2010;33(2):1–22. [Google Scholar]
- 17.Uribe-Convers S, Duke JR, Moore MJ, Tank DC. A long PCR-based approach for DNA enrichment prior to next-generation sequencing for systematic studies. Appl Plant Sci. 2014;2(1):1300063. doi: 10.3732/apps.1300063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wickett NJ, et al. Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr Biol. 2011;21(24):2098–2104. doi: 10.1016/j.cub.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Petersen G, Cuenca A, Seberg O. Plastome evolution in hemiparasitic mistletoes. Genome Biol Evol. 2015;7(9):2520–2532. doi: 10.1093/gbe/evv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaut B, Yang L, Takuno S, Eguiarte LE. The patterns and causes of variation in plant nucleotide substitution rates. Annu Rev Ecol Evol Syst. 2011;42(1):245–266. [Google Scholar]
- 21.Moriyama T, Sato N. Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front Plant Sci. 2014;5(5):480. doi: 10.3389/fpls.2014.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wicke S, Schneeweiss GM. Next generation organellar genomics: potentials and pitfalls of high-throughput technologies for molecular evolutionary studies and plant systematics. In: Hörandl E, Appelhans M, editors. Next Generation Sequencing in Plant Systematics, Regnum Vegetabile. Koeltz Scientific; Koenigstein, Germany: 2015. pp. 9–50. [Google Scholar]
- 23.Kamikawa R, et al. Proposal of a twin arginine translocator system-mediated constraint against loss of ATP synthase genes from nonphotosynthetic plastid genomes. Mol Biol Evol. 2015;32(10):2598–2604. doi: 10.1093/molbev/msv134. [DOI] [PubMed] [Google Scholar]
- 24.Leebens-Mack J, dePamphilis C. Power analysis of tests for loss of selective constraint in cave crayfish and nonphotosynthetic plant lineages. Mol Biol Evol. 2002;19(8):1292–1302. doi: 10.1093/oxfordjournals.molbev.a004190. [DOI] [PubMed] [Google Scholar]
- 25.McNeal JR, Kuehl JV, Boore JL, dePamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7(1):57. doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 2011;28(7):2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol Evol. 2014;6(1):238–246. doi: 10.1093/gbe/evu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logacheva MD, Schelkunov MI, Penin AA. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol. 2011;3:1296–1303. doi: 10.1093/gbe/evr102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schelkunov MI, et al. Exploring the limits for reduction of plastid genomes: A case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol. 2015;7(4):1179–1191. doi: 10.1093/gbe/evv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam VKY, Soto Gomez M, Graham SW. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol Evol. 2015;7(8):2220–2236. doi: 10.1093/gbe/evv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett CF, Davis JI. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 2012;99(9):1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- 32.Molina J, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol Biol Evol. 2014;31(4):793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith DR, Lee RW. A plastid without a genome: Evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164(4):1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naumann J, et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae) Genome Biol Evol. 2016;8(2):345–363. doi: 10.1093/gbe/evv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellot S, Renner SS. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol Evol. 2015;8(1):189–201. doi: 10.1093/gbe/evv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfe KH, Morden CW, Ems SC, Palmer JD. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: Loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol. 1992;35(4):304–317. doi: 10.1007/BF00161168. [DOI] [PubMed] [Google Scholar]
- 37.Müller AE, et al. Palindromic sequences and A+T-rich DNA elements promote illegitimate recombination in Nicotiana tabacum. J Mol Biol. 1999;291(1):29–46. doi: 10.1006/jmbi.1999.2957. [DOI] [PubMed] [Google Scholar]
- 38.Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11(2):101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, et al. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol Biol Evol. 2015;32(3):767–790. doi: 10.1093/molbev/msu343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53(5):673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 41.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102(30):10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pond SL, Frost SDW, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 43.Müller K. SeqState: Primer design and sequence statistics for phylogenetic DNA datasets. Appl Bioinformatics. 2005;4(1):65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- 44.Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst Biol. 2000;49(2):369–381. [PubMed] [Google Scholar]
- 45.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 46.Schliep KP. phangorn: Phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revell LJ. Size-correction and principal components for interspecific comparative studies. Evolution. 2009;63(12):3258–3268. doi: 10.1111/j.1558-5646.2009.00804.x. [DOI] [PubMed] [Google Scholar]
- 48.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19(1):101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]