Abstract
Genetic factors impact behavioral traits relevant to numerous psychiatric disorders and risk-taking behaviors, and different lines of evidence have indicated that discrete neurobiological systems contribute to such individual differences. In this study, we explored the relationship of genetic variants of the prodynorphin (PDYN) gene, which is enriched in the striatonigral/striatomesencephalic pathway, a key neuronal circuit implicated in positive ‘Go' behavioral choice and action. Our multidisciplinary approach revealed that the single nucleotide polymorphism (SNP) rs2235749 (in high linkage disequilibrium with rs910080) modifies striatal PDYN expression via impaired binding of miR-365, a microRNA that targets the PDYN 3′-untranslated region (3′UTR), and is significantly associated to novelty- and reward-related behavioral traits in humans and translational animal models. Carriers of the rs2235749G allele exhibited increased levels of PDYN 3′UTR in vitro and had elevated mRNA expression in the medial nucleus accumbens shell (NAcSh) and caudate nucleus in postmortem human brains. There was an association of rs2235749 with novelty-seeking trait and a strong genotype–dose association with positive reinforcement behavior in control subjects, which differed in cannabis-dependent individuals. Using lentiviral miRZip-365 constructs selectively expressed in Pdyn-neurons of the NAcSh, we demonstrated that the Pdyn-miR365 interaction in the NAcSh directly influences novelty-seeking exploratory behavior and facilitates self-administration of natural reward. Overall, this translational study suggests that genetically determined miR-365-mediated epigenetic regulation of PDYN expression in mesolimbic striatonigral/striatomesencephalic circuits possibly contributes to novelty seeking and positive reinforcement traits.
INTRODUCTION
Vulnerability to psychiatric disorders is influenced by a complex relationship between hereditary behavioral traits, genetics, as well as environment. Of the heritable traits, reward sensitivity and novelty/sensation seeking (Cloninger, 1987; Zuckerman, 1993) have been strongly linked to goal-directed and risk-taking behaviors. These traits are considered significant contributors to the development of drug abuse (Piazza et al, 1989; Belin et al, 2008; Blanchard et al, 2009; Jupp and Dalley, 2014), are found at high levels in gamblers (Janiri et al, 2007) and are positively associated with depressive symptoms and suicidal behavior in adolescents (Ortin et al, 2012). A key component of the neural circuitry shown to be critical for reinforcement learning and goal-directed behavior is the striatonigral/ventral striatomesencephalic (SN/VSM) pathway, which has also been shown to innervate the pallidum (Kupchik et al, 2015). Medium spiny SN and VSM neurons in the dorsal and ventral striatum, respectively, are well established to constitute the 'Go' (positive reward choice) pathway facilitating behavior (Lobo et al, 2010; Hikida et al, 2010; Kravitz et al, 2012). Human studies (Frank et al, 2007) have documented that genetic polymorphisms of the DARPP-32 gene, used as a non-selective proxy of SN neurons, are associated with impaired positive reinforcement learning. The SN/VSM neurons are, however, more specifically characterized by the predominant expression of the opioid neuropeptide prodynorphin (PDYN) (Heiman et al, 2008). Individual differences within the 3′ untranslated region (3′UTR) of the PDYN gene have been shown to be associated with the abuse of various drugs, specifically cocaine, alcohol, and heroin (Clarke et al, 2009; Yuferov et al, 2009; Taqi et al, 2011). However, no study has yet addressed whether such differences in PDYN are associated with behavioral endophenotypes relevant to psychiatric disorders and risk-taking behaviors and no information exists regarding the association with cannabis dependence, a drug which is now widely used by teens and young adults.
Here, we present converging molecular, in vitro, and in vivo evidence suggesting a central role of PDYN-expressing SN/VSM neurons in behavioral traits related to psychiatric vulnerability. We demonstrate that a functional polymorphism in the 3′UTR of PDYN, rs2235749, impairs the binding of miR-365 as well as PDYN expression in the human striatum. In addition, we show that the miR-365-PDYN interaction, regulated by rs2235749, is specifically associated with positive reinforcement and novelty-seeking traits in rats and humans.
MATERIALS AND METHODS
Participants and Personality Traits Assessment
Healthy subjects were recruited from a population (18–27 years old) participating in neurocognitive and genetic studies (Jutras-Aswad et al, 2012). They had no history of major psychiatric or medical disorders and did not report current or sustained (more than 1 month) past psychotropic medication intake. Subjects were evaluated using an array of questionnaires to obtain information on health and sociodemographic characteristics. Subjects were excluded for major psychiatric disorder based on the Mini International Neuropsychiatric Interview (Sheehan et al, 1998). Fifty normal subjects were enrolled as well as 50 subjects with a lifetime cannabis dependence diagnosis determined using the Structured Clinical Interview for DSM-IV (defined as a maladaptive pattern of substance use, leading to clinically significant impairment of distress, as manifested by three or more criteria of substance dependence occurring in the same 12-month period at any time in the past). Participants had a mean age of 22.7 years. Two-thirds of the sample population were males from a racially diverse but mainly Hispanic background. A vast majority of subjects (93%) had a high school diploma or higher level of education and were single (91%) at the time of the study. Cannabis-dependent subjects and controls had similar racial breakdowns and only differed in the percentage of Hispanic origin (52 vs 30%, p<0.05) and daily cigarette use (54 vs 4%, p<0.001). Subjects abstained from alcohol or drugs (except nicotine) on the day of the testing session as assessed by urine toxicology (or by self-report in cannabis-dependent individuals in relation to cannabis use). In addition, participants were assessed for symptoms of acute intoxication and were directly observed for 2 h before testing to ensure that they were not intoxicated during the study.
Personality traits were studied using the revised edition of the Temperament and Character Inventory (TCI-R), a self-report questionnaire designed to assess four temperament and character dimensions (Cloninger, 1999). Our study focused on Novelty Seeking and Reward Dependence subscales, which were used as a measure of temperamental proneness to positive reward sensitivity. We also administered the Zuckerman-Kuhlman Personality Questionnaire (ZKPQ) (Zuckerman, 2002) with a specific focus on the Impulsive Sensation-Seeking subscale to differentiate novelty seeking from the overall, less specific level of impulsivity.
Probabilistic Reward Learning Task
A cognitive probabilistic learning task (Frank et al, 2007) was used to examine positive and negative reinforcement-based decision-making. The task involved a training phase in which participants learned to discriminate between feedback probabilities (positive feedback: green check mark; negative feedback: red cross) associated with different pairs of symbolic stimuli. At the end of the training phase (128 trials), participants should have learned that symbol A is the most favorable choice and the one most likely to get positive feedback, whereas symbol B is the least favorable one and should be avoided. In the course of the testing phase, during which no feedback was given, symbols A and B were paired with the other four symbols to create 8 new pairs for a total of 96 trials. Performance on ‘Choose A' pairs was used as a measure of positive reinforcement learning, whereas performance on ‘Avoid B' pairs reflected negative reinforcement learning. Evaluation of behavior was carried out by investigators blinded to genotype.
Genotyping
Genotyping of DNA (from saliva samples or cerebellar brain tissue) was performed using Taqman predesigned single nucleotide polymorphism (SNP) genotyping assays (Applied Biosystems, Foster City, CA) and ABI 7900HT. PDYN SNPs rs2235749 and rs910080 (assay IDs 2507439 and 2507541, respectively) were chosen based on published data that specifically implicated the 3′UTR SNPs in substance dependence (Xuei et al, 2006; Yuferov et al, 2009). Linkage disequilibrium (LD) analysis was also carried out for SNPs in the PDYN gene +/−10 kb using the 1000 Genomes data based on racially mixed reference populations and with a minor allele frequency of at least 0.05. Samples were also analyzed for 80 ancestry indicative markers using an enhanced 34-plex assay (Fondevila et al, 2013) in order to confirm genetic ancestry.
mRNA Expression in the Postmortem Brain
Fresh-frozen striatal specimens were obtained from normal adult Caucasian subjects without head trauma from our brain bank collection (specimens collected at the Department of Forensic Medicine, Semmelweis University, Hungary) (Drakenberg et al, 2006). The specimens were collected under the guidelines approved by the local Human Ethical Committee within 24 h after death. The demographic characteristics of the subjects (N=16) were: 35.9±3.2 years old; 14 males/2 females, 6.71±0.05 brain pH; postmortem interval within 22.7±1.3 h; cause of death being cardiac failure (N=12), electric shock (N=2), viral infection (N=1), pulmonary embolus (N=1).
In situ hybridization histochemistry was performed on 20-μm-thick cryosections to measure PDYN mRNA expression levels with discrete anatomical specificity, as described previously (Fagergren et al, 2003; Drakenberg et al, 2006). For microRNA assessment, small RNAs were isolated using miRVana miRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA; cat. #AM1560), cDNA was synthesized using the qScript microRNA cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD; cat. #95107) and measured by qPCR using pre-designed assays from Quanta Biosciences (HSMIR-0365A-3P, HSMIR-0135A, and HSMIR-022-3P).
Stereotaxic Surgeries and Rat Behavioral Model
Adult (70–85 days) male Long-Evans rats, maintained on reversed 12-h dark/light cycle with food/water ad libitum, were stereotaxically infused (0.5 μl) with lentiviral vectors expressing GFP, as well as miRZip-365 under the control of the Pdyn promoter (n=9) or empty control vectors (n=9; Supplementary Figure S1A; custom cloned by Systems Biosciences, Mountain View, CA). Vectors were bilaterally infused at 0.1 μl/min into the NAcSh (10° angle from midline relative to bregma: AP+1.7 mm; ML+2.3 mm; DV-6.8 mm from dura). Behavioral paradigms began 12 days later to allow for optimal expression of the vector as determined by pilot studies (Supplementary Figure S2). Anatomical validation of the vector location was conducted after the behavioral procedures examining GFP fluorescence (control) and Nissl staining (general needle track and morphology). Increased Pdyn mRNA expression in the target brain region was validated using in situ hybridization histochemistry.
To determine novelty-seeking behavior (locomotor activity in a novel environment), animals were placed in an Open Field apparatus (16” × 16” Accuscan, Columbus, OH) and movements automatically tracked by beam breaks during 60 min.
To assess food self-administration behavior, animals were food-restricted overnight prior to testing. Animals were placed in an operant chamber (29.5 cm × 32.5 cm × 23.5 cm) housed in sound-attenuating boxes (MED Associates, St Albans, VT) with two levers; depression of one (designated the active lever) resulted in the delivery of one standard food chow pellet under a fixed-ratio schedule of reinforcement, whereas depression of the other (designated the inactive lever) had no programmed consequences. To test palatable food self-administration assessment, animals had ad lib to normal chow and depression of reinforcement lever resulted in the delivery of a chocolate pellet. All animal testing was carried out using a counter-balanced experimental design at a similar time of the day.
Cell Culture and Luciferase Assays
Trizol was used to extract mRNA from putamen punches of two postmortem human subjects that differed in their rs2235749/rs910080 genotype. Subsequently, we synthesized cDNA (qScript cDNA SuperMix, Quanta Biosciences; cat. #95048), amplified the region containing rs2235749 (forward primer: 5′-CAGTTCCCTCAGCCCTCATT-3′, reverse primer: 5′-ACTCCAGAAGCAGCAGCTAG-3′) and confirmed the genotype by Sanger sequencing. 3′UTR segments corresponding to the A and G alleles were then ligated into pFRL2 vector (Malo et al, 2003) attached downstream of firefly luciferase, driven by a CMV promoter (Supplementary Figure S1B). The constructs were used to transform HEK 293 cells that were subsequently incubated at 37 °C for 3 days. Protein levels were subsequently measured using the Dual-Glo luciferase assay system (Promega, Madison, WI, cat. #E2920). Firefly luciferase(-PDYN) signals were normalized to renilla luciferase, expressed from a separate promoter within pFRL2, to account for potential differences in transfection efficiency. Co-transfection experiments were carried out using hsa-miR-365a-3p mirVana miRNA mimic (Life Technologies, Carlsbad, CA; cat. #4464066) and baseline levels were assessed using dsRNA with no known endogenous targets from SwitchGear Genomics (Active Motif, Carlsbad, CA; cat. #MIM9001).
Statistical Analysis
mRNA expression (DPM/mg) from in situ hybridization histochemistry data was normalized using natural logarithm. General linear stepwise regression analysis was used to evaluate genotype group differences with covariates: eg, age, postmortem interval, brain pH, and sex. Statistical evaluations were performed using the JMP 12 (SAS Institute, Cary, NC) software package. The relationship between genotype, behavioral traits and cannabis use outcomes were analyzed using ANOVA and Pearson correlation, while group (based on a cannabis-dependence diagnosis) and gene × group interaction effects were calculated using general linear models. Genotyping patterns with phenotype were assessed with Fisher's exact test. For the rat analyses, repeated measures ANOVA with Holm-Sidak post hoc multiple comparisons were conducted.
RESULTS
3′UTR Polymorphisms of the PDYN Gene are Associated with Behavioral Traits Related to Novelty Seeking and Reward Learning
To assess the role of PDYN variants with behavioral endophenotypes related to novelty seeking and reward learning, SNPs within the 3′UTR of PDYN were genotyped. As novelty seeking and reward learning are important risk conferring factors with respect to neuropsychiatric disorders, especially addiction, we focused on two SNPs (rs2235749, rs910080) implicated in substance use disorders (Xuei et al, 2006; Yuferov et al, 2009). All genotypes conformed to Hardy–Weinberg equilibrium. On basis on the 1000 Genome data analysis of the PDYN gene (+/− 10 kb), rs2235749 was in strong LD with several 3′UTR SNPs including rs910080 and was thus a good proxy for the 3′UTR (Supplementary Figure S3). As there was a near complete overlap of the rs2235749 and rs910080 polymorphisms in each subject consistent with the strong LD (D'>0.9) between these SNPs (Wei et al, 2011), only the genotype data of the rs2235749 variant are presented below for the postmortem and clinical cohorts. Racial composition did not differ among the different genotypes and self-reported race showed a complete overlap with ancestry informative markers.
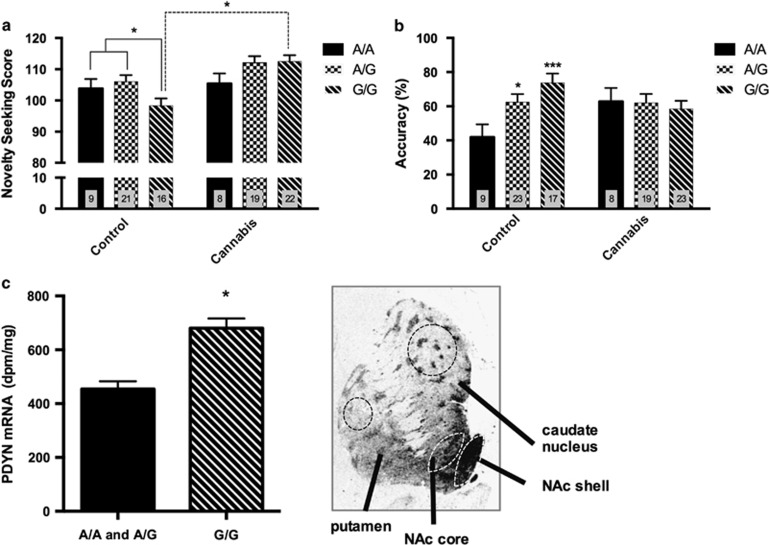
There was a significant overall association between novelty seeking and the rs2235749 variant such that G/G homozygous individuals had decreased novelty seeking compared with other groups (Figure 1a, p<0.05). In addition, there was a significant overall difference for novelty seeking (F5,94=5.76, p=0.001) with group (p=0.0001) and a group × PDYN rs2235749 interaction (p=0.042). Cannabis subjects had higher novelty-seeking trait as compared with controls that was driven by carriers of the rs2235749G allele who had higher novelty-seeking scores in the cannabis dependence group as compared with controls (p<0.01). No association was evident based on SNPs for the age at first use of cannabis or age at onset of dependence. Impulsive risk-taking and reward-dependence traits that were more prevalent in cannabis subjects (p=0.013) were not predicted by the SNPs studied although there was a significant correlation between novelty seeking and impulsive risk taking (r=0.6355, p<0.0001).
Figure 1.
PDYN 3′UTR SNP rs2235749 is associated to novelty seeking, positive reward learning, and striatal PDYN mRNA levels in control human subjects. (a) Novelty-seeking scores in relation to PDYN rs2235749 genotype in control and cannabis-dependence groups. (b) PDYN SNPs in relation to the accuracy of Choose A (positive reinforcement) performance in controls and cannabis users. (c) PDYN mRNA expression levels (DPM/mg) in the nucleus accumbens shell (NAcSh) of normal human subjects. PDYN in situ hybridization histochemistry micrograph with dashed lines representing sampled striatal regions. Error bars indicate SEM. *p<0.05, ***p<0.001.
Performance on the cognitive probabilistic learning task examined in regard to positive (Choose A) and negative (Avoid B) reward behaviors revealed a significant group × PDYN rs2235749 interaction for Choose A behavior (p=0.0242; Figure 1b). A genotype dose effect was apparent such that control homozygous G/G subjects had higher Choose A behavior than heterozygous A/G (p=0.037) and homozygous A/A individuals (p=0.0002) (Figure 1b). However, there was no association of genotype in cannabis subjects. There was no significant genotype relationship with Avoid B avoidance behavior in any group (not shown).
3′UTR PDYN Polymorphisms Correlate to PDYN mRNA Levels In Vivo and In Vitro
In order to investigate the functional relevance of rs2235749 and to assess whether the polymorphism relates to gene expression, we measured PDYN mRNA levels in the human striatum of postmortem specimens from normal subjects. Only three subjects for the SNP rs2235749 were A/A genotype and thus were combined with A/G subjects because their mRNA levels completely overlapped the A/G group. PDYN mRNA expression levels were increased 1.5-fold in the medial NAcSh of homozygous G allele subjects compared with rs2235749A carriers (F1,14=23.24, p=0.0003; Figure 1c). No significant association was detected for this polymorphism in the NAc core (p=0.81) or putamen subregions (p=0.12). However, there was a significant association with PDYN mRNA expression in the associative caudate nucleus region (F1,14=6.37, p=0.025).
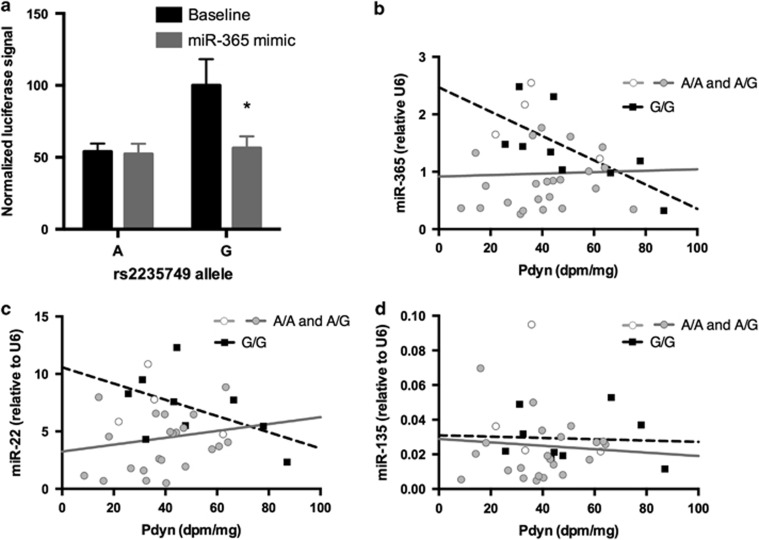
In order to elucidate the mechanism by which PDYN 3′UTR polymorphisms contribute to differential expression, we studied the causal relationship of the allelic differences on PDYN levels in vitro. 3′UTR sequences corresponding to either the A or the G allele of rs2235749 were inserted into a pFRL2 vector downstream of a CMV promoter driven firefly luciferase, and a dual luciferase assay was performed to assess the expression of the two constructs in HEK 293 cells. Interestingly, levels of the rs2235749G construct were significantly higher compared with rs2235749A (fold change 1.49, p=0.016; Figure 2a). These data are in line with our postmortem gene expression data and together suggest an important functional role for the rs2235749 SNP.
Figure 2.
rs2235749 affects the binding of miR-365 to PDYN. (a) Double luciferase assay showing PDYN mRNA levels in HEK 293 cells co-transfected with PDYN 3′UTR pFRL2 constructs corresponding to rs2235749G or rs2235749A alleles and miR-365 mimic or scrambled control vector (b–d) correlations between PDYN mRNA and miR-365 (b), miR-22 (c), and miR-135 (d) levels in the postmortem human striatum. Statistical analysis was carried out on pooled (A/A and A/G vs G/G) genotypes. Error bars indicate SEM. *p<0.05.
rs2235749 Affects the Binding of miR-365 to PDYN mRNA
microRNAs are known to target the 3′UTR of transcribed mRNAs and thereby to regulate mRNA levels. One possibility to explain the differential PDYN expression in relation to rs2235749 is that the polymorphism might affect the binding of one or more microRNAs. To identify microRNAs potentially targeting the 3′UTR PDYN region, we performed in silico analyses on predicted gene targets from multiple databases. Intriguingly, rs2235749 is located just 5 base pairs upstream from a predicted target site of miR-365 (TargetScanHuman 6.2). Evidence supporting a potential functional interplay between rs2235749, miR-365, and the PDYN gene expression was apparent in our postmortem cohort. Striatal PDYN mRNA levels were negatively correlated to miR-365 expression in rs2235749G/G subjects (r=−0.69; p=0.039), but not in A allele carriers (r=0.03, p=0.866) (Figure 2b). Two other microRNAs, miR-22 and miR-135, with poorly conserved predicted binding sites in the PDYN 3′UTR (relatively close to rs2235749) showed no correlation to PDYN mRNA, irrespective of rs2235749 (for miR-22, r=−0.51 and p=0.16 in G/G subjects and r=0.17 and p=0.40 in A allele carriers; for miR-135, r=−0.057 and p=0.89 in G/G subjects and r=−0.074 and p=0.72 in A allele carriers; Figure 2c and d). The same was also true for four additional microRNAs (miR-223, miR-335, miR-485, and miR-873) with poorly conserved predicted binding sites located near rs910080 (not shown).
In order to assess whether rs2235749 indeed affected miR-365 binding, HEK 293 cells were co-transfected with pFRL2 constructs corresponding to either A or G allele containing PDYN 3′UTRs, as well as a miR-365 mimic or a non-binding scrambled double-stranded RNA (negative control). Using a dual luciferase assay, we found that the miR-365 mimic significantly decreased luciferase intensity, confirming the binding of this microRNA to the PDYN 3′UTR. Strikingly, this effect fully depended on rs2235749 genotype: the stability of G allele-corresponding constructs was significantly decreased in the presence of the microRNA, while no such effect was observed with the A allele (Figure 2a).
miR-365 Regulates Pdyn Expression, Novelty-Seeking Behavior, and Positive Reinforcement-Based Decision-Making In Vivo
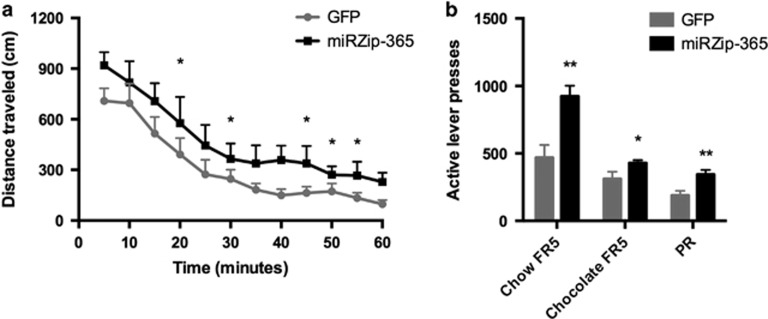
To confirm the functional relevance of the proposed PDYN-miR-365 interaction in vivo, a lentiviral construct expressing miRZip-365 or empty vector was infused into the NAcSh of adult male Long-Evans rats. miRZips are synthetic single-stranded anti-microRNAs designed to suppress microRNA function. By expressing a miRZip construct specifically targeting miR-365, our aim was to inhibit the binding of the microRNA to Pdyn mRNA molecules and to assess how disrupting this system affects Pdyn expression as well as novelty-seeking and positive reinforcement learning behaviors. Importantly, the expression of our lentiviral construct was driven by the Pdyn promoter, thus ensuring specificity to the striatonigral/striatomesencephalic pathway. We validated the constructs using immunohistochemistry and in situ hybridization histochemistry: viral expression was determined to be localized to the NAcSh, and miRZip-365 animals had increased Pdyn mRNA beginning 12 days after stereotaxic surgery (p=0.026).
Open Field locomotor testing demonstrated that NAcSh miRZip-365 infusion increased novelty-seeking behavior apparent in both time spent moving (p=0.05) and total distance traveled (p=0.026) as compared with controls (Figure 3a). Positive reinforcement behavior was subsequently assessed using a food self-administration paradigm. In line with our hypothesis, miRZip-365 expression significantly increased active lever pressing for normal food reward (p=0.006), palatable chocolate pellets (p=0.01), and during progressive ratio to determine motivated behavior (p=0.003), confirming enhanced positive reinforcement upon downregulation of miR-365 (Figure 3b). Importantly, animals did not differ in inactive lever pressing during the food self-administration sessions emphasizing the specificity of the reward-directed behavior, and did not show differences in general locomotor activity.
Figure 3.
miR-365 inhibition in the nucleus accumbens shell impairs novelty-seeking and positive reward learning behavior in rats. (a) Novelty-seeking behavior in adult male Long-Evans rats bilaterally injected with a lentiviral construct expressing miRZip-365 driven by the Pdyn promoter. (b) Food self-administration behavior in adult male Long-Evans rats bilaterally injected with a lentiviral construct expressing miRZip-365 driven by the Pdyn promoter. Error bars indicate SEM. *p<0.05, **p<0.01.
DISCUSSION
We propose a novel, genetically influenced microRNA-mRNA regulatory relationship between PDYN and miR-365 that contributes to behavioral endophenotypes relevant to addiction. Our in vitro and in vivo data, including direct study of human subjects, revealed that SNPs in the 3′UTR of the PDYN gene strongly affect the binding of miR-365 to this region, relate to PDYN mRNA expression levels in the human NAcSh and are associated with novelty seeking and positive reinforcement-based decision-making. These findings suggest that a genetically determined PDYN tone within the ventromedial striatal circuitry, established through impaired targeting of miR-365 to the PDYN 3′UTR, may contribute to individual differences linked to behavioral traits relevant to reward sensitivity.
PDYN in the striatum is expressed in a ventromedial-to-dorsolateral gradient with most abundant levels in the NAcSh (Hurd and Herkenham, 1995). While a significant relationship was apparent between PDYN mRNA expression in the associative subdivision of the human caudate nucleus and SNP genotype, the genetic association with mRNA expression levels was most pronounced in the NAcSh. In addition to the well-documented involvement of the SN ‘direct' pathway of the dorsal striatum in ‘Go' behavior, a similar organization exists in the ventral striatum for reinforcement behavior (Lobo et al, 2010; Hikida et al, 2010; Kravitz et al, 2012). The ventral striatal ‘direct' output VSM pathway is organized such that mesencephalic projections of Pdyn cells in the NAc core primarily target the substantia nigra pars compacta, whereas those in the shell subdivision innervate the ventral tegmental area (Zhou et al, 2003). The striato-ventral tegmental area output of the NAcSh would be predicted to directly modulate mesolimbic neurons relevant to reward and goal-motivated behavior. Several human and primate studies have documented that in vivo NAc activity is correlated with individual differences in sensation and novelty seeking (McClure et al, 2003; Montague et al, 2004; Abler et al, 2006). Interestingly, rats with increased vulnerability for alcohol intake and novelty-seeking behavioral traits have enhanced Pdyn in the NAcSh, but not NAc core or dorsal striatum (Guitart-Masip et al, 2006). Moreover, high responder rats to novelty stressor show enhanced drug self-administration and are characterized by elevated NAc Pdyn mRNA expression (Lucas et al, 1998). Consistently, animals in our study with increased Pdyn in the NAcSh, induced by the lentiviral miRZip-365 construct, exhibited enhanced novelty-induced locomotor behavior. An association between the PDYN rs2235749 SNP and novelty-seeking trait was also observed in our human study. Homozygous carriers of the rs2235749G allele, which related to enhanced NAcSh PDYN mRNA expression, had reduced novelty-seeking scores, but interestingly, novelty seeking was enhanced in G-allele cannabis-dependent subjects. Novelty seeking is a multifaceted behavioral construct; thus further studies are needed to examine its different dimensions in relation to PDYN and the interaction with cannabis use and dependence.
The relationship of the PDYN rs2235749 variant in relation to positive reinforcement behavior was well matched in both human subjects and our animal model. Human subjects showed a significant genotype–dose relationship to Choose A behavior, in line with the expected increase in positive decision-making choice among carriers of the genotypes associated with higher NAcSh PDYN mRNA expression. Using the same neurocognitive task known to dissociate vulnerability in probabilistic reward ‘Go' from avoidance learning ‘No/Go' based on discrete cortico-striatal circuits (Frank et al, 2007), our results did, however, demonstrate that PDYN is specifically associated with decisions related to positive, but not negative, reinforcement. The fact that various lines of evidence from animal models have validated that the ‘direct' and ‘indirect' striatal pathways dichotomize reward ‘Go' and avoidance ‘No/Go' behaviors (Hikida et al, 2010; Lobo et al, 2010) strengthens the human findings. Indeed, rats injected with miRZip-365 demonstrated increased self-administration of natural and palatable food and were more motivated to obtain the reward.
Interestingly, in contrast to controls, the association between PDYN genotype with performance on the cognitive probabilistic learning task was not present in cannabis subjects. Whether these differences predated the onset of cannabis dependence in at-risk individuals or are due to a possible residual effect of recent cannabis use on testing measures is difficult to determine in the current study. Cannabis-dependent participants were directly observed for a few hours before performing cognitive tasks and did not show symptoms of acute intoxication. Nevertheless, residual effects may have masked the effect of genotype variation on learning task performance and novelty-seeking behavior in the cannabis-dependent group. A number of studies have shown acute and short-term effects of cannabis on various domains of cognition (reviewed in Broyd et al, (2015)), which further underscores the need for future research that could disentangle pre-existing, acute and residual cognitive deficit in cannabis-dependent populations.
An important aspect of the current study was the unique mechanistic insights provided about the functional relationship between 3′UTR SNPs and microRNA regulation of PDYN that contribute to behavior. microRNAs, small non-coding RNAs that regulate mRNA levels by binding to the 3′UTR, have been shown to have critical roles in neurodevelopment and survival, as well as adult brain function including synaptic plasticity and memory formation (McNeill and Van Vactor, 2012; Kenny, 2014). microRNAs have also been implicated in reward and addiction such that striatal deficiency of Argonaute-2, a master regulator of the maturation and function of all microRNAs, directly alters cocaine self-administration behavior (Schaefer et al, 2010). The previous study of Argonaute-2 directly targeted dopamine D2 receptor-expressing cells that are abundant within the striatopallidal pathway. Our study now emphasizes the direct involvement of microRNAs in the VSM circuit in regulating addiction-related phenotypes. Very limited publications currently exist about miR-365 with most related to cancer and none regarding brain function. Here, we propose a novel, genetically influenced regulatory relationship between PDYN and miR-365 that contributes to behavioral endophenotypes associated with positive valence that may be relevant to addiction and related psychiatric disorders.
Whether the association between individual differences in PDYN variants and behavioral traits do contribute to addiction disorders in humans remains to be established. Kreek and colleagues have shown that the 3′UTR PDYN SNPs currently studied are in high LD and together with the rs910079 SNP comprise a haplotype block associated with striatal PDYN mRNA levels as well as cocaine dependence and combined cocaine/alcohol codependence (Yuferov et al, 2009). 3′UTR PDYN SNPs have also been observed to be associated with heroin abuse (Clarke et al, 2009). Moreover, the rs2235749 SNP currently shown to affect miR-365 binding to PDYN mRNA was correlated to impaired DNA methylation and PDYN expression in the dorsolateral prefrontal cortex of alcohol-dependent individuals (Taqi et al, 2011). Although insight is still lacking regarding the functional mechanisms of 3′UTR SNPs, it is clear that the 3′UTR has a significant role in the stability of the mRNA, as well as translation and transcriptional regulation. Importantly, our data demonstrate a causal role of the 3′UTR SNP on mRNA levels and specific behaviors associated with the VSM pathway.
As with all human studies, and considering the limitations of mixed ethnicity and small sample size, replication in another population will be important to validate the results of our association studies. Nevertheless, the human findings provided the foundation for the experimental models that did support the suggestion that the Pdyn striatonigral/VSM circuit and, more specifically, the miR-365–Pdyn relationship is directly relevant to novelty seeking and positive reinforcement decision-making. How cannabis exposure factors in to this equation will need to be addressed in follow-up studies that assess the drug's effect on Pdyn expression and behavior in translational animal models. Another important consideration is the population admixture of the sample population. It has been shown that the PDYN SNPs evaluated in our study are in high LD, but are not related to ethnicity (Yuferov et al, 2009), and there was no evidence in the present study to suggest that the genetic effects observed were due to ethnicity. Nevertheless, future studies involving larger populations will help clarify these issues especially in relation to novelty-seeking traits and allow the possibility to examine other markers specifically enriched in VSM neurons. Moreover, although we focused on the 3′UTR based on our initial hypothesis, other polymorphisms of PDYN within the promoter region have also been shown to be relevant for gene expression (Zimprich et al, 2000; Nikoshkov et al, 2008) and in relation to substance abuse (Ray et al, 2005; Clarke et al, 2012; Saify et al, 2014). The contribution of different variants within the PDYN gene to discrete behaviors needs to be addressed in larger populations that would provide the statistical power to examine such questions.
Overall, the current translational study reveals a significant contribution of 3′UTR PDYN polymorphisms to mesolimbic NAcSh PDYN expression and positive reward sensitivity traits in humans. Our in vitro data causally demonstrate that this interaction is regulated by impaired binding of miR-365 to the PDYN 3′UTR dependent on rs2235749 allele. Furthermore, direct manipulation of miR-365 activity in Pdyn-expressing neurons in the NAcSh directly relates to novelty-seeking, reward, and motivated behavior. These findings emphasize a novel mechanism involving miR-365–PDYN interaction relevant to individual behavioral traits that might enhance vulnerability for addiction and other psychiatric disorders.
FUNDING AND DISCLOSURE
The authors declare no conflict of interest.
Acknowledgments
We would like to thank James Callens and Nayana Patel for their technical support, Dr Chloe Tessereau for assistance with the LD analysis and Dr Corey Watson for assistance with the analysis of ancestral informative markers. This research was supported by Icahn School of Medicine at Mount Sinai Fund (YLH), NIH DA15446 (YLH), Centre hospitalier de l'Université de Montréal Research Fellowship Award (DJA), NIH T32 5T32GM062754 (SAA), NIH T32 DA007135 (MMJ and MM) and UNCF/Merck Dissertation Fellowship (SAA).
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Abler B, Walter H, Erk S, Kammerer H, Spitzer M (2006). Prediction error as a linear function of reward probability is coded in human nucleus accumbens. NeuroImage 31: 790–795. [DOI] [PubMed] [Google Scholar]
- Belin D, Mar AC, Dalley JW, Robbins TW, Everitt BJ (2008). High impulsivity predicts the switch to compulsive cocaine-taking. Science 320: 1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard MM, Mendelsohn D, Stamp JA (2009). The HR/LR model: Further evidence as an animal model of sensation seeking. Neurosci Biobehav Rev 33: 1145–1154. [DOI] [PubMed] [Google Scholar]
- Broyd SJ, van Hell HH, Beale C, Yücel M, Solowij N (2015). Acute and chronic effects of cannabinoids on human cognition- a systematic review. Biol Psychiatry 79: 557–567. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Ambrose-Lanci L, Ferraro TN, Berrettini WH, Kampman KM, Dackis CA et al (2012). Genetic association analyses of PDYN polymorphisms with heroin and cocaine addiction. Genes Brain Behav 11: 415–423. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Krause K, Li T, Schumann G (2009). An association of prodynorphin polymorphisms and opioid dependence in females in a Chinese population. Addict Biol 14: 366–370. [DOI] [PubMed] [Google Scholar]
- Cloninger CR (1987). A systematic method for clinical description and classification of personality variants. A proposal. Arch Gen Psychiatry 44: 573–588. [DOI] [PubMed] [Google Scholar]
- Cloninger CR (1999). A new conceptual paradigm from genetics and psychobiology for the science of mental health. Aust N Z J Psychiatry 33: 174–186. [DOI] [PubMed] [Google Scholar]
- Drakenberg K, Nikoshkov A, Horvath MC, Fagergren P, Gharibyan A, Saarelainen K et al (2006). Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc Natl Acad Sci USA 103: 7883–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagergren P, Smith HR, Daunais JB, Nader MA, Porrino LJ, Hurd YL (2003). Temporal upregulation of prodynorphin mRNA in the primate striatum after cocaine self-administration. Eur J Neurosci 17: 2212–2218. [DOI] [PubMed] [Google Scholar]
- Fondevila M, Phillips C, Santos C, Freire Aradas A, Vallone PM, Butler JM et al (2013). Revision of the SNPforID 34-plex forensic ancestry test: assay enhancements, standard reference sample genotypes and extended population studies. Forensic Sci Int Genet 7: 63–74. [DOI] [PubMed] [Google Scholar]
- Frank MJ, Moustafa AA, Haughey HM, Curran T, Hutchison KE (2007). Genetic triple dissociation reveals multiple roles for dopamine in reinforcement learning. Proc Natl Acad Sci USA 104: 16311–16316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitart-Masip M, Gimenez-Llort L, Fernandez-Teruel A, Canete T, Tobena A, Ogren SO et al (2006). Reduced ethanol response in the alcohol-preferring RHA rats and neuropeptide mRNAs in relevant structures. Eur J Neurosci 23: 531–540. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE et al (2008). A translational profiling approach for the molecular characterization of CNS cell types. Cell 135: 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida T, Kimura K, Wada N, Funabiki K, Nakanishi S (2010). Distinct roles of synaptic transmission in direct and indirect striatal pathways to reward and aversive behavior. Neuron 66: 896–907. [DOI] [PubMed] [Google Scholar]
- Hurd YL, Herkenham M (1995). The human neostriatum shows compartmentalization of neuropeptide gene expression in dorsal and ventral regions: an in situ hybridization histochemical analysis. Neuroscience 64: 571–586. [DOI] [PubMed] [Google Scholar]
- Janiri L, Martinotti G, Dario T, Schifano F, Bria P (2007). The Gamblers' Temperament and Character Inventory (TCI) personality profile. Subst Use Misuse 42: 975–984. [DOI] [PubMed] [Google Scholar]
- Jupp B, Dalley JW (2014). Behavioral endophenotypes of drug addiction: etiological insights from neuroimaging studies. Neuropharmacology 76: 487–497. [DOI] [PubMed] [Google Scholar]
- Jutras-Aswad D, Jacobs MM, Yiannoulos G, Roussos P, Bitsois P, Nomura Y et al (2012). Cannabis-dependence risk relates to synergism between neuroticism and proenkephalin SNPs associated with amygdala gene expression: a case-control study. PLoS One 7: e39243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ (2014). Epigenetics, microRNA, and addiction. Dialogues Clin Neurosci 16: 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, Tye LD, Kreitzer AC (2012). Distinct roles for direct and indirect pathway striatal neurons in reinforcement. Nat Neurosci 15: 816–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupchik YM, Brown RM, Heinsbroek JA, Lobo MK, Schwartz DJ, Kalivas PW (2015). Coding the direct/indirect pathways by D1 and D2 receptors is not valid for accumbens projections. Nat Neurosci 18: 1230–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Covington HE 3rd, Chaudhury D, Friedman AK, Sun H, Damez-Werno D et al (2010). Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science 330: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas LR, Angulo JA, Le Moal M, McEwen BS, Piazza PV (1998). Neurochemical characterization of individual vulnerability to addictive drugs in rats. Eur J Neurosci 10: 3153–3163. [DOI] [PubMed] [Google Scholar]
- Malo MS, Abedrapo M, Chen A, Mozumder M, Pushpakaran P, Alkhoury F et al (2003). Improved eukaryotic promoter-detection vector carrying two luciferase reporter genes. BioTechniques 35: 1150–1154. [DOI] [PubMed] [Google Scholar]
- McClure SM, Berns GS, Montague PR (2003). Temporal prediction errors in a passive learning task activate human striatum. Neuron 38: 339–346. [DOI] [PubMed] [Google Scholar]
- McNeill E, Van Vactor D (2012). MicroRNAs shape the neuronal landscape. Neuron 75: 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague PR, Hyman SE, Cohen JD (2004). Computational roles for dopamine in behavioural control. Nature 431: 760–767. [DOI] [PubMed] [Google Scholar]
- Nikoshkov A, Drakenberg K, Wang X, Horvath MC, Keller E, Hurd YL (2008). Opioid neuropeptide genotypes in relation to heroin abuse: dopamine tone contributes to reversed mesolimbic proenkephalin expression. Proc Natl Acad Sci USA 105: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortin A, Lake AM, Kleinman M, Gould MS (2012). Sensation seeking as risk factor for suicidal ideation and suicide attempts in adolescence. J Affect Disord 143: 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza PV, Deminiere JM, Le Moal M, Simon H (1989). Factors that predict individual vulnerability to amphetamine self-administration. Science 245: 1511–1513. [DOI] [PubMed] [Google Scholar]
- Ray R, Doyle GA, Crowley JJ, Buono RJ, Oslin DW, Patkar AA et al (2005). A functional prodynorphin promoter polymorphism and opioid dependence. Psychiatr Genet 15: 295–298. [DOI] [PubMed] [Google Scholar]
- Saify K, Saadat I, Saadat M (2014). Association between VNTR polymorphism in promoter region of prodynorphin (PDYN) gene and heroin dependence. Psychiatry Res 219: 690–692. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Venø MT, Fowler CD, Min A, Intrator A et al (2010). Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med 207: 1843–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E et al (1998). The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry 59(Suppl 20): 22–33; quiz 34-57. [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Sheedy D, Harper C, Alkass K et al (2011). Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol 16: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SG, Zhu YS, Lai JH, Xue HX, Chai ZQ, Li SB (2011). Association between heroin dependence and prodynorphin gene polymorphisms. Brain Res Bull 85: 238–242. [DOI] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L et al (2006). Association of the kappa-opioid system with alcohol dependence. Mol Psychiatry 11: 1016–1024. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Ji F, Nielsen DA, Levran O, Ho A, Morgello S et al (2009). A functional haplotype implicated in vulnerability to develop cocaine dependence is associated with reduced PDYN expression in human brain. Neuropsychopharmacology 34: 1185–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Furuta T, Kaneko T (2003). Chemical organization of projection neurons in the rat accumbens nucleus and olfactory tubercle. Neuroscience 120: 783–198. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Kraus J, Woltje M, Mayer P, Rauch E, Hollt V (2000). An allelic variation in the human prodynorphin gene promoter alters stimulus-induced expression. J Neurochem 74: 472–477. [DOI] [PubMed] [Google Scholar]
- Zuckerman M (1993). P-impulsive sensation seeking and its behavioral, psychophysiological and biochemical correlates. Neuropsychobiology 28: 30–36. [DOI] [PubMed] [Google Scholar]
- Zuckerman M (2002). Zuckerman-Kuhlman Personality Questionnaire (ZKPQ): an alternative five-factorial model. In: Raad BD, Perugini M (eds). Big Five Assessment. Hogrefe & Huber: Seattle, WA, USA. pp 377–396. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.