Abstract
Exposure to serious or traumatic events early in life can lead to persistent alterations in physiological stress response systems, including enhanced cross talk between the neuroendocrine and immune system. These programming effects may be mechanistically involved in mediating the effects of adverse childhood experience on disease risk in adulthood. We investigated hormonal and genome-wide mRNA expression responses in monocytes to acute stress exposure, in a sample of healthy adults (n=30) with a history of early childhood adversity, and a control group (n=30) without trauma experience. The early adversity group showed altered hypothalamus-pituitary-adrenal axis responses to stress, evidenced by lower ACTH and cortisol responses. Analyses of gene expression patterns showed that stress-responsive transcripts were enriched for genes involved in cytokine activity, cytokine–cytokine receptor interaction, chemokine activity, and G-protein coupled receptor binding. Differences between groups in stress-induced regulation of gene transcription were observed for genes involved in steroid binding, hormone activity, and G-protein coupled receptor binding. Transcription factor binding motif analysis showed an increased activity of pro-inflammatory upstream signaling in the early adversity group. We also identified transcripts that were differentially correlated with stress-induced cortisol increases between the groups, enriched for genes involved in cytokine–cytokine receptor interaction and glutamate receptor signaling. We suggest that childhood adversity leads to persistent alterations in transcriptional control of stress-responsive pathways, which—when chronically or repeatedly activated—might predispose individuals to stress-related psychopathology.
INTRODUCTION
Adverse or traumatic events early in life represent well-established risk factors for developing health problems (Repetti et al, 2002; Gilbert et al, 2009). Childhood abuse, neglect, and early loss have been consistently associated with an increased risk of mental health problems in adulthood, including major depression and anxiety disorders (Kendler et al, 1999; Chapman et al, 2004). These observations raised the question of how the long-lasting health consequences of early adverse rearing conditions are sustained. Animal models provide powerful evidence that environmental influences acting early in life can have direct and enduring effects on physiology. Adverse environmental conditions seem to program neurobiological systems that are implicated in regulating emotion and stress responses (Zhang and Meaney, 2010). This ‘defensive programming' of neural, endocrine, and immune responses is thought to serve adaptive purposes, but is at the same time associated with certain costs, including vulnerability to stress-related disorders (Zhang et al, 2006). An important question concerns the biological mechanisms linking childhood trauma, defensive programming of stress response systems, and health risk in adulthood. Stress-related alterations in immune response gene expression programs have been hypothesized to be one of the pathways through which environmental adversity influences disease processes (Irwin and Cole, 2011; Cacioppo et al, 2015). A growing number of studies in adults experiencing a range of current chronic stressors or adversities have shown that immune response genes are highly sensitive to social-environmental conditions. A specific transcriptional profile, characterized by enhanced expression of pro-inflammatory immune response genes and a reciprocal downregulation of antiviral immune response genes, has been observed, eg, in people experiencing loneliness and chronic isolation (Cole et al, 2007), or in individuals diagnosed with breast cancer (Antoni et al, 2012; Cole, 2014). Among the heterogeneous leukocyte population, bioinformatic decomposition identified monocytes and dendritic cells as most responsive to changes in socioenvironmental conditions (Cole et al, 2011). Analysis of isolated leukocyte subpopulations confirmed that monocytes mediate many of the transcriptional effects of social adversity, eg, of traumatic stress (O'Donovan et al, 2011), imminent bereavement, or chronic stress in caregivers of terminally ill patients (Miller et al, 2008, 2014). Furthermore, there is evidence for a link between inflammation and early adversity (Baumeister et al, 2015). Increased pro-inflammatory cytokine levels have been observed in patients (Pace et al, 2006) and healthy individuals reporting a history of childhood maltreatment (Carpenter et al, 2010), and there seems to be a coupling of inflammation and depression risk related to childhood adversity (Miller and Cole, 2012). Low socioeconomic status (SES) during childhood was also associated with increased pro-inflammatory signaling (Miller et al, 2009).
Taken together, monocyte gene expression patterns are influenced by environmental circumstances, and there is evidence for a link between early adverse experience and inflammatory signaling in adulthood. However, previous investigations have all assessed basal gene expression profiles, mostly in adults experiencing current chronic stress or in patients suffering from mental disorders. Thus, it remains unclear whether the propensity towards a certain transcriptional shift in the context of acute stress might be programmed by traumatic experience in childhood. We investigated genome-wide mRNA profiles in monocytes before and after acute exposure to the standardized and well-established laboratory stress paradigm, the Trier Social Stress Test (TSST), in a study group of healthy adults with a history of early adversity compared with a matched control group. The aim was to test whether the responsiveness of the monocyte transcriptome to acute stress might be programmed by childhood trauma. We hypothesized dysregulations in the hypothalamic-pituitary-adrenal axis activity following stress exposure, characterized by a relative hypo-responsiveness in ACTH and cortisol responses in the early adversity group. With regard to transcriptional dynamics, we hypothesized increased pro-inflammatory transcription factor activity as well as altered stress-induced expression of immune response genes in the early adversity group. Taking advantage of the genome-wide approach, we also conducted exploratory analyses to identify genes that were differentially regulated between the early adversity and the control group following stress exposure.
MATERIALS AND METHODS
Participants and Study Procedure
A total of 60 healthy adults aged between 45 and 60 years of age were recruited via articles in local newspapers and community-posted flyers. The German 28-item version of the Childhood Trauma Questionnaire (CTQ, Bernstein et al, 2003; Wingenfeld et al, 2010) was used to assess five categories of childhood trauma (sexual, physical and emotional abuse, and physical and emotional neglect) in all individuals. Cutoff scores for moderate-severe exposure were used to classify subjects as positive for a history of childhood adversity (n=30), which was validated in a structured interview with the Early Trauma Inventory (ETI, Bremner et al, 2000; Wingenfeld et al, 2011). Participants who met the criteria for mental disorders at the time of assessment or during the preceding 12 months (screened for with Structured Clinical Interview for DSM Disorders (SKID I & II, Wittchen et al, 1996) were excluded from study participation. The control group consisted of 30 participants who scored below cutoff on all CTQ subscales, and who were matched for gender, age, and current SES as well as childhood SES. All participants filled in the Resilience Scale (RS-25, Schumacher et al, 2005), and current psychopathological symptoms were assessed with the Brief Symptom Inventory (Franke, 2000). Intake of psychoactive or cortisol-containing medication was an exclusion criterion, and all female participants had to be free of oral contraceptives and reported to the lab during the luteal phase of their menstrual cycle. Participants were paid 100€ for participation. The study was approved by the Ethics Committee of the Albert-Ludwigs-University Freiburg (183/11).
Psychosocial Stress Protocol and Stress Response Measures
Stress was induced with the TSST, a standardized 15-min laboratory stress protocol, which consists of a mock job interview and an unanticipated mental arithmetic task (see Kirschbaum et al, 1993). All experimental sessions started at 2 p.m. Blood samples for the analyses of ACTH and cortisol were drawn via an indwelling catheter at 45 and 2 min prior, and 1, 10, 20, 30, 45, and 90 min post exposure to the TSST. Total cortisol and ACTH concentrations were measured with an enzyme-linked immunosorbent assay (IBL, Germany) at the University of Trier. Interassay and intrassay coefficients of variation were both under 6.9%.
Subjective stress was assessed at five time points: at −2, +1, +10, +20, and +30 min relative to stress, using the Social Emotional Response Scale (unpublished), which assesses the dimensions arousal (calm, jittery, tense, intense, relaxed, content), self-directed emotions (guilty, ashamed, blameworthy, angry at self, dissatisfied with self), and anxiety (fearful, worried).
General Linear Models were computed to assess the repeated measures effect time, the between-subjects effect group as well as the interaction time × group for endocrine and subjective responses to the TSST exposure. Greenhouse-Geisser corrections were applied where appropriate, and only adjusted results are reported. η2 values are given as an effect size measure.
Gene Expression
For gene expression analyses, 10 ml EDTA blood samples for RNA extraction were collected at 45 min before, as well as 45 and 180 min after the TSST. CD14+ monocytes were isolated via immunomagnetic cell separation (MACS; Miltenyi Biotec, Germany). Fluorescence-activated cell sorting analyses showed high purity of the isolated monocyte population (mean=92.92%, SE=0.59). Isolated cells were resuspended in lysis buffer RA1, shock-frozen in liquid nitrogen, and stored at −80 °C until RNA was extracted (Macherey-Nagel, Germany). RNA integrity number values, assessed with the Agilent 2100 bioanalyzer (Agilent Technologies), ranged from 8.0 to 10.0 (mean=9.7, SE=0.03). A total of 100 ng of RNA was used for genome-wide transcriptional profiling on the Agilent Whole Human Genome Oligo Microarrays 8 × 60 K V2. All samples were randomized within and between arrays to avoid potential batch effects. The assays were performed at the Molecular Service Center (Miltenyi Biotech) following the manufacturer's standard protocol. Quantile-normalized gene expression values were log2-transformed for further analyses with the R (version 3.1.1) package limma (Smyth et al, 2005). For the analyses of group differences in stress-induced gene expression changes, only transcripts that showed a difference in gene expression between the pre-stress and either post-stress time-point with an absolute fold change >1.2 in at least 20% of all samples were further used in the analysis (n=21.414 transcripts). For cross-sectional analyses (Supplementary Table 8), low-expressed transcripts (mean expression below median value per time point) were excluded from further analyses (n=25.341 were retained). In the limma approach, the combined information of all genes is used to adjust the individual variance estimates of genes with an Empirical Bayes approach. For all gene-wise linear models, response variables were created by subtracting the log2-transformed pre-stress expression values from the 45 and 180 min post-stress values, respectively. Effects of time (or equivalently stress), and the interaction between time and group were estimated and adjusted for gender and history of mental disorders. P-values were corrected for multiple testing by the method of Benjamini and Hochberg (Benjamini and Hochberg, 1995). Genes with a fold change of at least 1.2 and a false discovery rate adjusted p-value ⩽0.05 in the pairwise comparison of pre-stress to either post-stress time-point were considered differentially expressed. Both uncorrected and corrected p-values are reported. The ENRICHR toolkit (Chen et al, 2013) was used to identify enriched Gene Ontology Molecular Process categories or canonical pathways among the stress-responsive genes.
Correlation between Neuroendocrine Stress Measures and Gene Expression
Spearman's rank correlation was used to analyze the correlation between TSST-induced cortisol increases (peak levels minus baseline) and changes in gene expression. Analyses were performed separately for groups to identify differential responsiveness to cortisol. To test for significant differences between early adversity group and control group in correlations between stress-induced cortisol increases and gene expression changes, the correlation coefficients were normalized using a standard Fisher's z-transformation, with standard errors (N−3)−1/2, and statistically compared based on the normal approximation of the standardized difference between the normally distributed transformed correlation coefficients.
Transcription Factor-Binding Motifs
Activity of specific transcriptional control pathways was assessed by TELiS bioinformatics analysis of transcription factor-binding motifs (TFBMs) in the promoters of differentially expressed genes (Cole et al, 2005). TFBM frequencies were assessed by using three parametric variations in the size of proximal promoter sequence scanned ((−300 bp, +0 bp) relative to the RefSeq transcription start site, (−600, +0), and (−1,000, +200)) and three parametric variations in the TFBM detection stringency (TRANSFAC MatInspector algorithm, mat_sim=0.80, 0.90, 0.95). Results from each parametric scan were summarized as a ratio of TFBM prevalence in upregulated vs downregulated promoters, and results were averaged over the nine parametric combinations of promoter length and scan stringency for statistical testing with a dependent measures t-test.
Monocyte Subsets
To determine whether differences in activation or prevalence of CD16− and CD16+ monocyte subsets might contribute to observed transcriptional differences within the overall monocyte population, we conducted Transcript Origin Analyses (Cole et al, 2011) and Transcriptome Representation Analyses (Powell et al, 2013), utilizing reference gene expression profiles from isolated CD16− and CD16+ monocytes as previously reported (data from GEO series GSE18565 (Ingersoll et al, 2010). Standard errors for Transcript Origin Analyses and Transcriptome Representation Analyses estimates were derived by bootstrap resampling of residuals from general linear model results for each contrast. In Transcriptome Representation Analyses, cell type-diagnostic transcripts were defined by ⩾6 SD difference between CD16− and CD16+ samples in the reference study.
RESULTS
Sample Characteristics
Thirty participants who experienced adverse events during childhood were compared with a matched control group who reported absence of childhood adversity. The control group was matched for gender, age, current SES, and SES in childhood. Table 1 shows the Childhood Trauma Questionnaire (CTQ) total score and number of participants who met cutoff criteria for moderate to severe exposure of the five CTQ categories. In the early adversity group, 76.6% of the participants experienced three or more and 56.6% experienced four or more types of abuse and neglect.
Table 1. Age, Gender, and Total CTQ Scores in the Sample; Number and Percentage of Participants Meeting Moderate-Severe Cutoff Criteria for CTQ Categories.
| Early adversity (n=30) | Control (n=30) | P | |
|---|---|---|---|
| Age; mean±SD | 52.57±5.52 | 51.47±4.64 | 0.407 |
| Sex; number of females (%) | 20 (66.7) | 20 (66.7) | 1.000 |
| CTQ total score; mean±SD | 67.10±16.04 | 35.50±5.89 | <0.001 |
| CTQ categories; n (%) | |||
| Sexual abuse | 15 (50.0) | — | |
| Physical abuse | 15 (50.0) | — | |
| Emotional abuse | 22 (73.3) | — | |
| Emotional neglect | 26 (86.7) | — | |
| Physical neglect | 21 (70.0) | — | |
| Sexual or physical abuse | 23 (76.7) | — | |
| History of mental disorder, n (%) | 18 (60%) | 8 (26.7%) | 0.20 |
| Depression history, n (%) | 12 (40%) | 3 (10%) | 0.20 |
| BSI global severity index | 0.46±0.41 | 0.34±0.30 | 0.195 |
| RS-25 total | 136.9±26.7 | 135.9±27.5 | 0.888 |
Abbreviations: BSI, Brief Symptom Inventory; RS-25, Resilience Scale, 25 item version.
Participants with history of childhood adversity report a higher number of previous mental disorders and depression. There were no differences in resilience (RS-25) or current psychopathological symptoms between groups. Furthermore, there were no significant differences between the groups regarding childhood or current family income, parental education, own education, and bio-behavioral characteristics (body mass index, waist-to-hip ratio, smoking; see Supplementary Table 1 for details).
Stress Reactivity
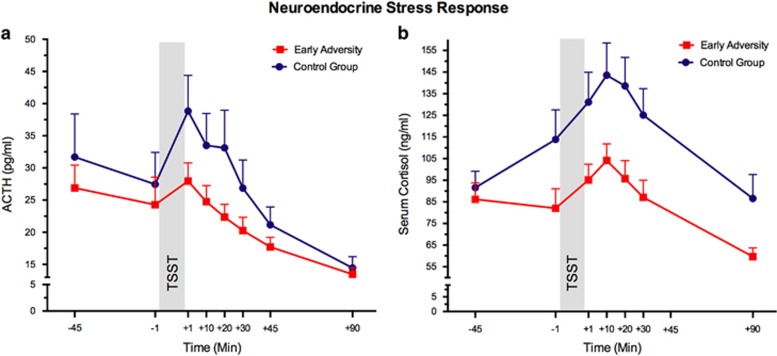
Early adversity was associated with lower ACTH and significantly lower cortisol responses (Figure 1a and b; ACTH: main effect group: F1,56=3.41, p=0.070, η2=0.06; interaction time by group: F2.37,132.52=0.94, p=0.406, η2=0.02. Cortisol: main effect group: F1,56=6.57, p=0.013, η2=0.11; interaction: F3.33,186.63=2.77, p=0.037, η2=0.05). The TSST also led to a significant increase in subjective stress, measured on the dimensions tense arousal, self-directed emotions, and anxiety (main effect time: all F>3.32; all p<0.010). There were no differences in the magnitude or response patterns for subjective stress between the groups (all F<2.16, all p>0.150).
Figure 1.
HPA axis responses to psychosocial stress. (a) Mean plasma adrenocorticotropin (ACTH) levels and (b) serum cortisol levels before and after the TSST (shaded area). Error bars represent SEM.
Gene Expression
Psychosocial stress exposure induced changes in mRNA levels of 570 transcripts (false discovery rate adjusted: 87) at +45 min, and 1044 transcripts (false discovery rate adjusted: 475) at 180 min post-stress exposure, respectively (see Supplementary Tables 2 for lists of upregulated and downregulated genes). In addition to these genes regulated by stress in both groups, we identified further genes that were differentially regulated between the groups following stress exposure. At 45 min post stress, 404 genes showed differential expression patterns, and 3 h after cessation of the TSST, 608 genes were differentially expressed between the two groups (Supplementary Tables 6 and 7). Permutation tests with 10 000 iterations showed that in only 43 out of 10 000 iterations, a greater number of differentially expressed genes compared with the original data were observed.
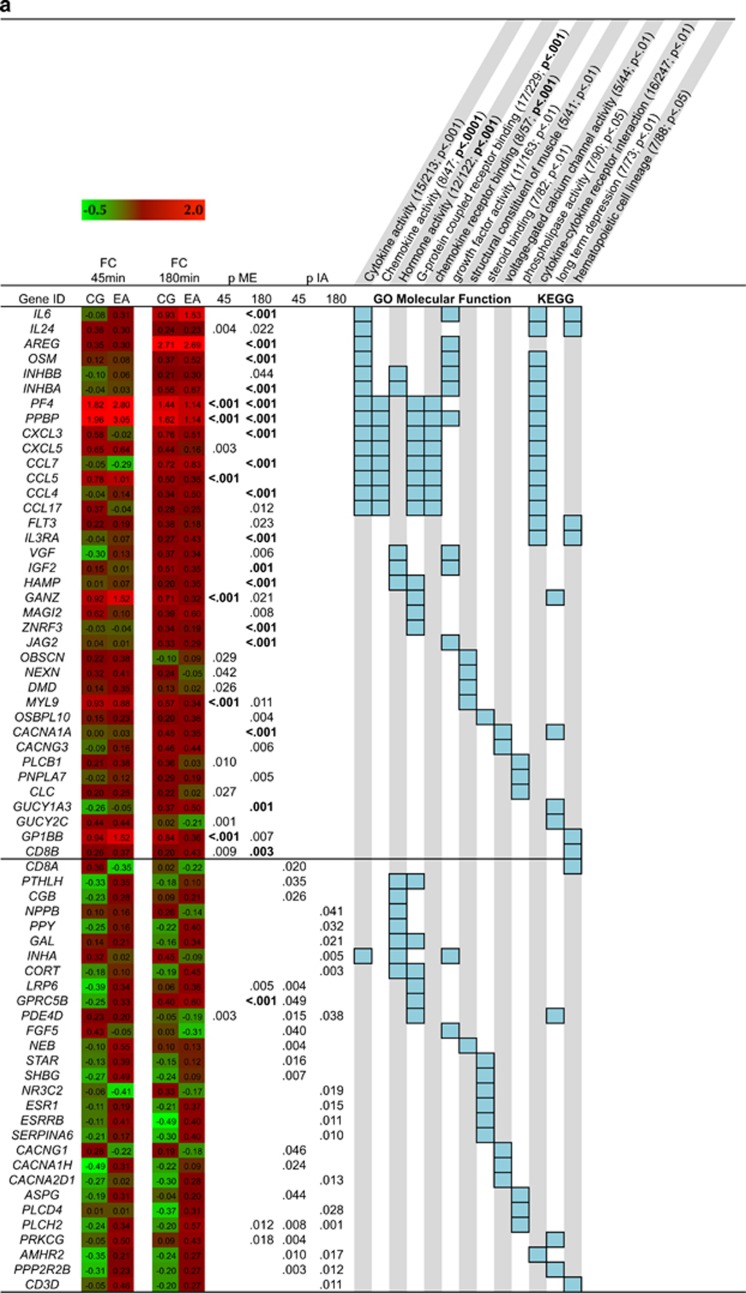
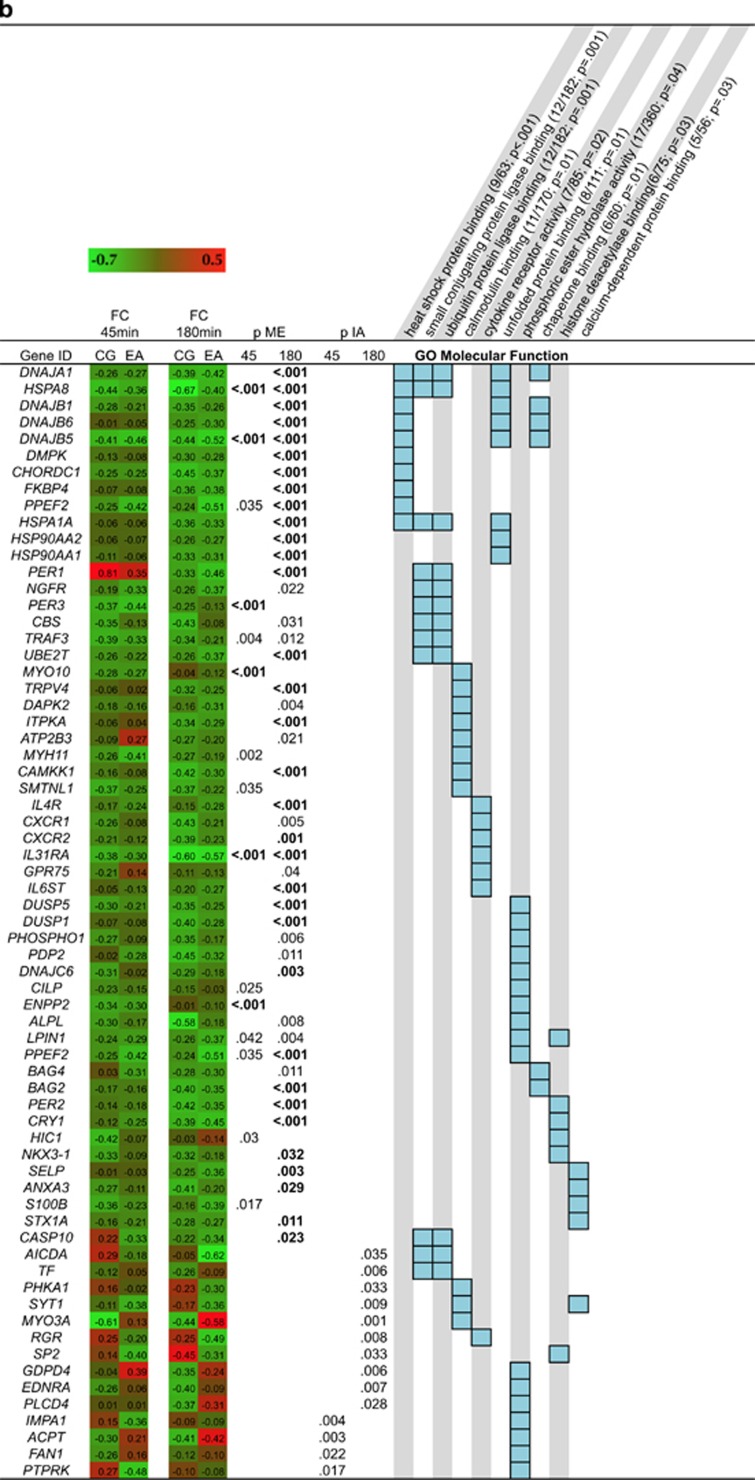
Pathway analyses showed that genes upregulated in response to stress in both groups were enriched with immune system-related (eg cytokine and chemokine activity) and signal transduction-related pathways (eg G-protein-coupled receptor binding; Figure 2a). The downregulated genes were enriched for transcripts involved in protein processing (eg heat shock protein binding, chaperon binding, unfolded protein binding) and cytokine receptor activity (Figure 2b). Differences between groups in stress-induced regulation of gene transcription were observed for genes involved in steroid binding, hormone activity and G-protein coupled receptor binding (Figure 2a), and phosphoric ester hydrolase activity (Figure 2b).
Figure 2.
Heat map showing the log2 fold-change of upregulated genes (a) and downregulated (b) from pre-stress to +45 min and to +180 min post stress, respectively. P-values (bold indicates FDR <0.05) for the main effect (ME) of stress for the two time points and the p-value for the interaction effect (IA) are shown. Light blue boxes indicate involvement in gene ontology (GO) or KEGG pathways. Only pathways with an overlap of at least five transcripts are shown. The overlap and the p-value (bold FDR<0.05) are shown in parentheses next to the respective pathway names.
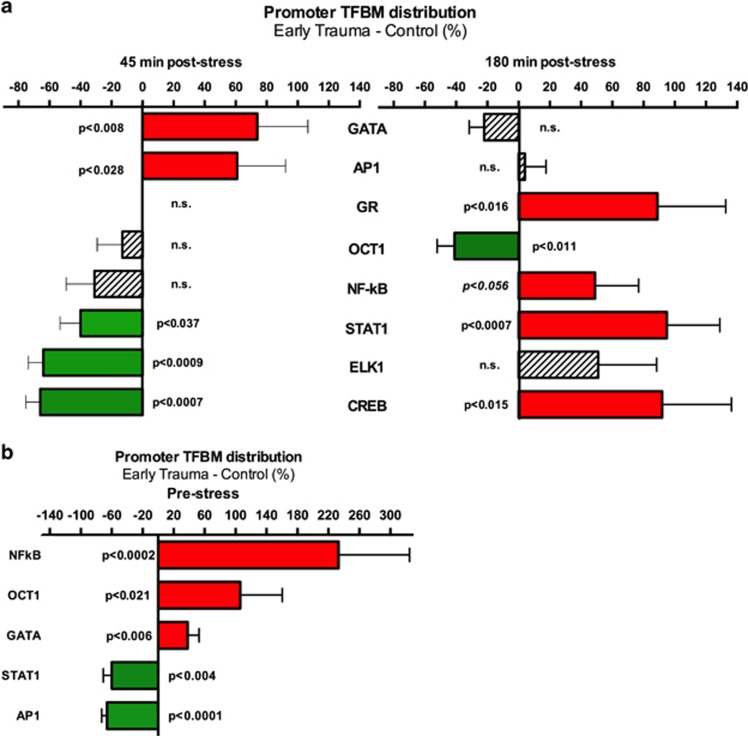
Next, the Transcription Element Listening System (TELiS) promoter-based bioinformatics analysis (Cole et al, 2005) was applied to test whether transcription factors previously associated with different types of social stress or adversity (Cole et al, 2007; Chen et al, 2008, 2010; Miller et al, 2008, 2009; Bower et al, 2010; O'Donovan et al, 2011; Antoni et al, 2012; Fredrickson et al, 2013) might have a role in the observed stress-associated group differences in gene expression. At 45 min post-stress exposure, results indicated greater TSST-induced activation of the pro-inflammatory AP-1 and GATA transcription factors involved in the early adversity group. At the 180 min follow-up, the early adversity group showed a significantly greater increase over time in activation of STAT1, CREB, GR, and a marginally greater increase in NF-kB activity (Figure 3a).
Figure 3.
TELiS bioinformatics analyses quantified response element prevalence in promoters of differentially expressed genes. (a) Transcription factor-binding motif (TFBM) prevalence for the two post-stress time points, (b) for the pre-stress time point. Data represent mean fold difference (±SE) in prevalence of TFBMs, averaged over nine parametric combinations of promoter length and motif detection stringency.
Furthermore, and consistent with previous studies of social adversity, analysis of TFBMs in the promoters of differentially expressed genes at the pre-stress time point identified an overrepresentation of binding sites of NF-κB and GATA factors in the early adversity group. Analyses also indicated overrepresentation of Oct1, and underrepresentation of STAT1 and AP1 in the early adversity group (Figure 3b).
Correlations between Stress-Induced Cortisol Increases and Transcriptional Changes
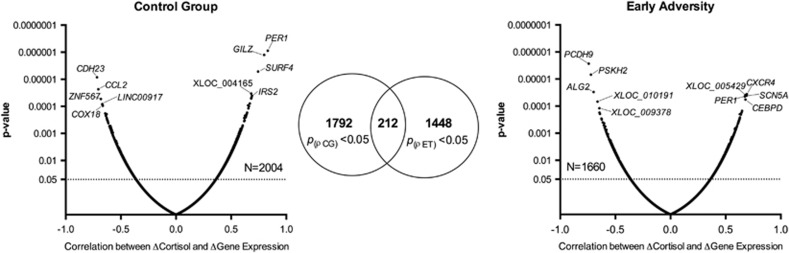
Correlation analyses between stress-induced cortisol increases and gene expression changes 45 min post stress showed substantial differences in cortisol-associated transcriptional changes between groups. As shown in Figure 4, the majority of transcripts were significantly correlated in one group only, with little overlap between groups (see also Supplementary Table 9).
Figure 4.
Spearman's rho-coefficients and p-values of correlations between stress-induced expression change and cortisol increases in the control group (left panel) and the early adversity group (right panel). The top five correlated transcripts (both positively and negatively) are shown. Venn diagrams show little overlap of correlated transcripts between groups.
Pathway analyses (Table 2) showed that the differentially regulated transcripts were enriched for genes involved in cytokine—cytokine receptor interaction (p=0.01 in control group) and neuroactive ligand receptor interaction (p=0.01 in both groups). At +180 min post stress, compared with 45 min post-stress time point, a lower number of transcripts (n=1313 in CG, 1074 in EA, with an overlap of n=62) were significantly correlated with cortisol increases. Pathway analyses showed that correlated genes in the control group clustered in MAPK signaling pathway (p=0.02) and glycerolipid metabolism (p=0.009), whereas an enrichment in focal adhesion (p=0.04) and axon guidance (p=0.01) pathways was observed for the EA group. See Supplementary File 10 for details.
Table 2. Spearman's rho (ρ) Coefficients in Both Groups of Differentially Correlated Transcripts Involved in the Identified Pathways (Table 2A: Cytokine–Cytokine Receptor Interaction, Table 2B: Neuroactive Ligand Receptor Interaction).
| Transcript | ρ CG | ρ EA |
|---|---|---|
| Table 2A: KEGG cytokine–cytokine receptor interactions | ||
| IL6ST | 0.53 | −0.57 |
| CCL18 | 0.52 | −0.35 |
| CXCR4 | 0.46 | 0.69 |
| CCL11 | 0.45 | 0.06 |
| IL21 | 0.43 | −0.15 |
| OSMR | 0.39 | 0.03 |
| CCL7 | 0.38 | −0.23 |
| TNFRSF1A | 0.37 | −0.3 |
| LEP | 0.37 | −0.13 |
| IL1R2 | 0.37 | 0.02 |
| IL18RAP | 0.36 | 0.18 |
| TNFRSF19 | 0.17 | −0.39 |
| TNFRSF6 | 0.11 | 0.42 |
| TNFRSF14 | −0.4 | −0.35 |
| CXCR3 | −0.41 | 0.43 |
| ACVR2B | −0.42 | −0.15 |
| CX3CR1 | −0.43 | −0.25 |
| PRLR | −0.43 | −0.22 |
| TNFRSF25 | −0.43 | 0.18 |
| CXCL10 | −0.43 | 0.24 |
| CCL8 | −0.46 | 0.02 |
| FLT1 | −0.47 | −0.07 |
| CCL2 | −0.7 | 0.17 |
| Table 2B: KEGG neuroactive ligand receptor interaction | ||
| ADORA3 | 0.68 | 0.4 |
| LTB4R | 0.54 | −0.07 |
| P2RY1 | 0.53 | −0.05 |
| GHRHR | 0.47 | 0.08 |
| GRID1 | 0.44 | 0.01 |
| GRIA4 | 0.39 | 0.38 |
| LEP | 0.37 | −0.13 |
| HTR2B | 0.12 | −0.42 |
| P2RX7 | 0.07 | −0.54 |
| ADORA2A | 0.02 | −0.48 |
| TSHB | −0.02 | 0.56 |
| PTGDR | −0.12 | 0.58 |
| GRIK3 | −0.13 | 0.35 |
| SSTR5 | −0.27 | 0.42 |
| MC2R | −0.31 | 0.55 |
| GHSR | −0.31 | 0.59 |
| CALCRL | −0.34 | 0.05 |
| GRM1 | −0.37 | 0.11 |
| P2RY6 | −0.4 | 0.14 |
| GABRE | −0.4 | 0.08 |
| GLP2R | −0.4 | 0.14 |
| HRH1 | −0.42 | 0.01 |
| PRLR | −0.43 | −0.22 |
| PTGFR | −0.45 | −0.05 |
| GRIN2A | −0.45 | −0.09 |
| ADRB3 | −0.47 | 0.11 |
| GRIK2 | −0.49 | 0.06 |
| TRHR | −0.6 | 0.06 |
| AVPR2 | −0.63 | 0.08 |
The mean difference between correlation coefficients was 0.49 in the cytokine–cytokine receptor interaction pathway and 0.52 in the neuroactive ligand receptor interaction pathway
Monocyte Subsets
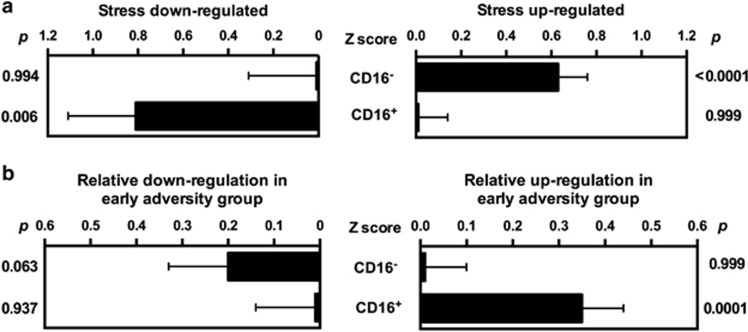
To determine whether the observed differences in the overall monocyte gene expression profile might be driven by differential transcriptional activation or hematological mobilization of subpopulations of immature/pro-inflammatory CD16− ‘classical' monocytes or mature ‘non-classical/ trophic' CD16+ monocytes, Transcript Origin Analyses were performed to assess whether differentially expressed genes were predominately characteristic of individual subpopulations. Results in Figure 5 indicate that genes upregulated by 45 min after TSST derived predominately from the immature CD16− monocyte subpopulation (p<0.0001), whereas downregulated transcripts derived predominately from the more mature CD16+ monocyte subpopulation (p=0.006). The early adversity group did not differ from controls in the magnitude of CD16− monocyte transcript upregulation (p>0.90) but showed significantly greater upregulation of CD16+ monocyte-characteristic genes from baseline to the 45 min post-TSST time point (p<0.001). Groups did not differ in their average baseline expression of CD16− and CD16+ monocyte-derived transcripts (all p>0.05). These monocyte subset-related differences in gene expression persisted through the 180 min time point, with Transcript Origin Analyses results continuing to identify upregulated genes as characteristic of CD16− monocytes (p=0.009) and downregulated genes as characteristic of CD16+ monocytes (p<0.0001). However, by the 180 min time point, the early adversity group did not differ from controls in the expression of genes characteristic of either monocyte subset (all p>0.05).
Figure 5.
Transcript origin analysis assessing (a) the cellular origin of genes downregulated and upregulated within the overall monocyte population 45 min post stress across the whole group and (b) between the control and early adversity group.
DISCUSSION
Exposure to unfavorable circumstances in early life can lead to persistent changes in physiological stress response systems, and these programming effects may be mechanistically involved in mediating the effects of adverse childhood experience on disease risk in adulthood. In the present study, we (i) further corroborate findings of early adversity-related alterations in neuroendocrine responses to acute stress, (ii) provide evidence for altered stress-induced regulation of genes in monocytes in adults with a history of childhood maltreatment, (iii) show differences in cortisol-associated changes in gene expression between the groups, and (iv) identify increased activity of pro-inflammatory upstream signaling in the early adversity group as a potential mechanism of both baseline and stress-associated differences in gene expression.
To the best of our knowledge, this is the first investigation of genome-wide transcriptional profiles following acute stress exposure in adults with a history of severe childhood maltreatment. Results showed that across both studied groups, psychosocial stress led to robust changes in gene expression 45 min and 180 min post stress, with 15% and 46% of the nominally significant transcripts surviving correction for multiple testing at the +45 and +180 time point, respectively. In addition to gene expression changes across both groups, we found a considerable number of genes that were differentially expressed between groups at 45 min (n=404) and 180 min (n=608) post stress, with group differences in stress-induced fold-changes in gene expression ranging from 1.20 to 2.40. Among the differentially expressed genes, several transcripts coding for genes involved in signal transducer activity were found, most notably those involved in G-protein-coupled receptor binding, hormone activity, and steroid binding, including the mineralocorticoid receptor (NR3C2), corticosteroid and sex hormone-binding globulins (SERPINA6, SHBG), estrogen and estrogen-related receptors (ESR1, ESRRB), and steroidogenic acute regulatory protein (STAR). It is unclear whether stress-associated transcriptional changes in monocytes mirror those in brain areas involved in stress processing. The question whether peripheral gene expression signatures are indicative of neuronal processes involved in cellular and behavioral adaptation under stress has to be addressed in animal models (Daskalakis et al, 2014).
One goal of the study was to test the hypothesis of increased pro-inflammatory transcription factor activity in the early adversity group. Bioinformatic analyses interrogating response element prevalence in promoters of differentially expressed genes provided evidence for increased stress-associated pro-inflammatory signaling linked to early adversity. Sequence-based analysis of gene-regulatory regions combined with statistical prevalence analyses to identify transcription-factor binding motifs using TELiS showed a greater stress-induced activity of pro-inflammatory transcription factors AP1 (at +45 min) and NF-kB (at +180 min) in the group with early trauma history. We also observed time-point-specific differences in activity of transcription factors involved in β-adrenergic signaling, and increased GR activity in the early trauma group at the 3 h follow-up time point.
Notwithstanding small group differences, and notwithstanding the fact that transcription factor activation profiles represent indirect bioinformatics inferences, results of the present study are consistent with previous investigations reporting larger pro-inflammatory responses to acute stress in adults with early in life adversity (Pace et al, 2006; Carpenter et al, 2010), and more generally, with the idea that childhood adversity may enhance cross talk between the neuroendocrine and the immune system. Under repeated acute or chronic stress, a stress system programmed towards a pro-inflammatory response bias might contribute to a chronic inflammatory environment (Nusslock and Miller, 2015), which is an established vulnerability factor for stress-related disorders, including major depression (Dantzer et al, 2008; Irwin and Cole, 2011; Miller and Cole, 2012).
Differences were also evident in neuroendocrine stress response profiles. We observed a relative hypothalamus-pituitary-adrenal axis hypo-reactivity in the group of adults with childhood trauma history, evidenced by lower ACTH and cortisol responses to acute stress exposure. This is in line with the majority of studies reporting dampened stress system activity associated with early adversity (Carpenter et al, 2007; Miller et al, 2007; Elzinga et al, 2008; Lovallo et al, 2011; Power et al, 2012), although the opposite has also been reported (Heim et al, 2000).
Analyses of the link between cortisol increases and gene expression changes showed a large number of differentially responsive transcripts. Correlational analyses further support adversity-related differences in stress-induced transcriptional regulation of genes involved in immunoregulation and signal transducer activity.
Finally, our data suggest that within the overall monocyte population, the relatively immature CD16− monocyte subset is the major contributor to transcriptional activation in response to TSST, and these effects persist for at least 180 min. These effects do not appear to be attributable to differential hematological redistribution of the two subsets, however, and thus appear to reflect a selective stress-induced transcriptional activation of CD16− monocytes. The general profile of monocyte subset-specific activation was similar in early adversity and control populations, suggesting that the limited group differences in transcriptome response did not derive predominately from group differences in the magnitude of subset-specific transcriptional activation.
Certain limitations need mention. The early adversity vs control group differences in the magnitude of TSST-induced gene expression did not generally reach statistical significance after correction for multiple testing, so these group differences should be regarded as exploratory and need to be replicated in future studies. Furthermore, the second post-stress measure was taken 3 h after TSST exposure, a time point where greatest new RNA accumulation would be expected. Additional longer follow-up might have been informative with regard to differences in inflammatory gene expression programs, given the long-term effects of cortisol-mediated immune suppression. One explaining factor for the small differences might be the investigated population, as participants were currently healthy, ie, diagnosis-free for at least a year. It is possible that at least part of the investigated adults with a history of childhood maltreatment might be resilient against the pathogenic effects of early adverse experience. The significantly higher percentage of previous mental disorders (see Table 1) speaks, however, against the notion of a generally resilient sample. Furthermore, the present analyses all control for any confounding of history of mental disorders and sex differences with early adversity status and continue to find notable early adversity sensitization of stress effects. Owing to the limited sample size for separate analyses of females and males, the present study is not able to address whether sex differences might exist in the magnitude of these effects, and that remains an important topic for future research.
To conclude, the findings show that early trauma alters stress-induced gene expression in CD14+ monocytes, particularly of genes involved in steroid hormone activity and signal transduction, and that these differences are associated with increased activity of pro-inflammatory upstream signaling. The investigation of the downstream effects of stress mediators, such as regulation of gene expression programs in immune cells promises a better understanding of how altered stress responsivity connects to a wide range of somatic and psychiatric disorders. We suggest that childhood adversity leads to alterations in transcriptional control of stress responsive pathways, which—when chronically or repeatedly activated—might predispose individuals to stress-related psychopathology.
FUNDING AND DISCLOSURE
The authors declare no conflict of interest.
Acknowledgments
The study was supported by a research grant from the Deutsche Forschungsgemeinschaft (DFG, KU 2479/3-1) to RK and the BMBF projects LivSys and SysDT.
GEO ACCESSION
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE70603 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70603).
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Antoni MH, Lutgendorf SK, Blomberg B, Carver CS, Lechner S, Diaz A et al (2012). Cognitive-behavioral stress management reverses anxiety-related leukocyte transcriptional dynamics. Biol Psychiatry 71: 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister D, Akhtar R, Ciufolini S, Pariante CM, Mondelli V (2015). Childhood trauma and adulthood inflammation: a meta-analysis of peripheral C-reactive protein, interleukin-6 and tumour necrosis factor-alpha. Mol Psychiatry 21: 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57: 289–300. [Google Scholar]
- Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T et al (2003). Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl 27: 169–190. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Irwin MR, Arevalo JMG, Cole SW (2010). Fatigue and gene expression in human leukocytes: increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain Behav Immun 25: 147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD, Vermetten E, Mazure CM (2000). Development and preliminary psychometric properties of an instrument for the measurement of childhood trauma: the Early Trauma Inventory. Depress Anxiety 12: 1–12. [DOI] [PubMed] [Google Scholar]
- Cacioppo JT, Cacioppo S, Capitanio JP, Cole SW (2015). The neuroendocrinology of social isolation. Annu Rev Psychol 66: 733–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter L, Carvalho J, Tyrka A, Wier L, Mello A, Mello M et al (2007). Decreased adrenocorticotropic hormone and cortisol responses to stress in healthy adults reporting significant childhood maltreatment. Biol Psychiatry 62: 1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter LL, Gawuga CE, Tyrka AR, Lee JK, Anderson GM, Price LH (2010). Association between plasma IL-6 response to acute stress and early-life adversity in healthy adults. Neuropsychopharmacology 35: 2617–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DP, Whitfield CL, Felitti VJ, Dube SR, Edwards VJ, Anda RF (2004). Adverse childhood experiences and the risk of depressive disorders in adulthood. J Affect Disord 82: 217–225. [DOI] [PubMed] [Google Scholar]
- Chen E, Miller GE, Kobor MS, Cole SW (2010). Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol Psychiatry 16: 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Miller GE, Walker HA, Arevalo JM, Sung CY, Cole SW (2008). Genome-wide transcriptional profiling linked to social class in asthma. Thorax 64: 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV et al (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW (2014). Human social genomics. PLoS Genet 10: e1004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT (2007). Social regulation of gene expression in human leukocytes. Genome Biol 8: R189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT (2011). Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci USA 108: 3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Yan W, Galic Z, Arevalo J, Zack JA (2005). Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics 21: 803–810. [DOI] [PubMed] [Google Scholar]
- Dantzer R, O'Connor JC, Freund GG, Johnson RW, Kelley KW (2008). From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci 9: 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis NP, Cohen H, Cai G, Buxbaum JD, Yehuda R (2014). Expression profiling associates blood and brain glucocorticoid receptor signaling with trauma-related individual differences in both sexes. Proc Natl Acad Sci USA 111: 13529–13534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga BM, Roelofs K, Tollenaar MS, Bakvis P, van Pelt J, Spinhoven P (2008). Diminished cortisol responses to psychosocial stress associated with lifetime adverse events a study among healthy young subjects. Psychoneuroendocrinology 33: 227–237. [DOI] [PubMed] [Google Scholar]
- Franke GH (2000) BSI. Brief Symptom Inventory von L.R. Derogatis (Kurzform der SCL-90-R)—Deutsche Version. Beltz: Göttingen. [Google Scholar]
- Fredrickson BL, Grewen KM, Coffey KA, Algoe SB, Firestine AM, Arevalo JMG et al (2013). A functional genomic perspective on human well-being. Proc Natl Acad Sci USA 110: 13684–13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R, Widom CS, Browne K, Fergusson D, Webb E, Janson S (2009). Burden and consequences of child maltreatment in high-income countries. Lancet (London, England) 373: 68–81. [DOI] [PubMed] [Google Scholar]
- Heim C, Newport DJ, Heit S, Graham YP, Wilcox M, Bonsall R et al (2000). Pituitary-adrenal and autonomic responses to stress in women after sexual and physical abuse in childhood. JAMA 284: 592–597. [DOI] [PubMed] [Google Scholar]
- Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R et al (2010). Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 115: e10–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin MR, Cole SW (2011). Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol 11: 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Prescott CA (1999). Causal relationship between stressful life events and the onset of major depression. Am J Psychiatry 156: 837–841. [DOI] [PubMed] [Google Scholar]
- Kirschbaum C, Pirke KM, Hellhammer DH (1993). The ‘Trier Social Stress Test'—a tool for investigating psychobiology stress responses in a laboratory setting. Neuropsychobiology 28: 76–81. [DOI] [PubMed] [Google Scholar]
- Lovallo WR, Farag NH, Sorocco KH, Cohoon AJ, Vincent AS (2011). Lifetime adversity leads to blunted stress axis reactivity: studies from the Oklahoma Family Health Patterns Project. Biol Psychiatry 71: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Chen E, Fok A, Walker H, Lim A, Nicholls E et al (2009). Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci USA 106: 14716–14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Sze J, Marin T, Arevalo JM, Doll R et al (2008). A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry 64: 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Zhou ES (2007). If it goes up, must it come down? Chronic stress and the hypothalamic-pituitary-adrenocortical axis in humans. Psychol Bull 133: 25–45. [DOI] [PubMed] [Google Scholar]
- Miller GE, Cole SW (2012). Clustering of depression and inflammation in adolescents previously exposed to childhood adversity. Biol Psychiatry 72: 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Murphy ML, Cashman R, Ma R, Ma J, Arevalo JM et al (2014). Greater inflammatory activity and blunted glucocorticoid signaling in monocytes of chronically stressed caregivers. Brain Behav Immun 41: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslock R, Miller GE (2015). Early-life adversity and physical and emotional health across the lifespan: a neuro-immune network hypothesis. Biol Psychiatry (e-pub ahead of print 4 June 2015). [DOI] [PMC free article] [PubMed]
- O'Donovan A, Sun B, Cole S, Rempel H, Lenoci M, Pulliam L et al (2011). Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Dis Markers 30: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace TW, Mletzko TC, Alagbe O, Musselman DL, Nemeroff CB, Miller AH et al (2006). Increased stress-induced inflammatory responses in male patients with major depression and increased early life stress. Am J Psychiatry 163: 1630–1633. [DOI] [PubMed] [Google Scholar]
- Powell ND, Sloan EK, Bailey MT, Arevalo JMG, Miller GE, Chen E et al (2013). Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via β-adrenergic induction of myelopoiesis. Proc Natl Acad Sci USA 110: 16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, Thomas C, Li L, Hertzman C (2012). Childhood psychosocial adversity and adult cortisol patterns. Br J Psychiatry 201: 199–206. [DOI] [PubMed] [Google Scholar]
- Repetti RL, Taylor SE, Seeman TE (2002). Risky families: family social environments and the mental and physical health of offspring. Psychol Bull 128: 330–366. [PubMed] [Google Scholar]
- Schumacher J, Leppert K, Gunzelmann T (2005). Die Resilienzskala—Ein Fragebogen zur Erfassung der psychischen Widerstandsfähigkeit als Personmerkmal. Zeitschrift für Klinische Psychologie, Psychiatrie und Psychotherapie 53: 16–39. [Google Scholar]
- Smyth GK, Michaud J, Scott HS (2005). Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075. [DOI] [PubMed] [Google Scholar]
- Wingenfeld K, Driessen M, Mensebach C, Rullkoetter N, Schaffrath C, Spitzer C et al (2011). Die deutsche Version des, Early Trauma Inventory” (ETI). Diagnostica 57: 27–38. [Google Scholar]
- Wingenfeld K, Spitzer C, Mensebach C, Grabe HJ, Hill A, Gast U et al (2010). The German version of the Childhood Trauma Questionnaire (CTQ): preliminary psychometric properties. Psychother Psychosom Med Psychol 60: 442–450. [DOI] [PubMed] [Google Scholar]
- Wittchen H-U, Wunderlich U, Gruschwitz S, Zaudig M (1996) Strukturiertes Klinisches Interview für DSM-IV (SKID). Beltz: Göttingen. [Google Scholar]
- Zhang TY, Bagot R, Parent C, Nesbitt C, Bredy TW, Caldji C et al (2006). Maternal programming of defensive responses through sustained effects on gene expression. Biol Psychol 73: 72–89. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Meaney MJ (2010). Epigenetics and the environmental regulation of the genome and its function. Annu Rev Psychol 61: 439–466 C431-433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.