Abstract
Large amounts of epigenomic data are generated under the umbrella of the International Human Epigenome Consortium, which aims to establish 1000 reference epigenomes within the next few years. These data have the potential to unravel the complexity of epigenomic regulation. However, their effective use is hindered by the lack of flexible and easy-to-use methods for data retrieval. Extracting region sets of interest is a cumbersome task that involves several manual steps: identifying the relevant experiments, downloading the corresponding data files and filtering the region sets of interest. Here we present the DeepBlue Epigenomic Data Server, which streamlines epigenomic data analysis as well as software development. DeepBlue provides a comprehensive programmatic interface for finding, selecting, filtering, summarizing and downloading region sets. It contains data from four major epigenome projects, namely ENCODE, ROADMAP, BLUEPRINT and DEEP. DeepBlue comes with a user manual, examples and a well-documented application programming interface (API). The latter is accessed via the XML-RPC protocol supported by many programming languages. To demonstrate usage of the API and to enable convenient data retrieval for non-programmers, we offer an optional web interface. DeepBlue can be openly accessed at http://deepblue.mpi-inf.mpg.de.
INTRODUCTION
Due to technical advances in the past decade, massive volumes of molecular data are now routinely generated by several epigenome mapping consortia, including the Encyclopedia of DNA Elements (ENCODE) (1), the NIH Roadmap Epigenomics Mapping Consortium (ROADMAP) (2), the BLUEPRINT Epigenome Project (3) and the German Epigenome Programme (DEEP) (http://www.deutsches-epigenom-programm.de).
The International Human Epigenome Consortium (IHEC) (http://ihec-epigenomes.org) serves as an umbrella organization that coordinates the production of reference epigenomes for cell types relevant to health and disease. IHEC has the ambitious goal of deciphering at least 1000 epigenomes by 2020. While these data are broadly useful for studying the human epigenome, their effective use is currently hindered by the lack of a simple data retrieval mechanism.
Epigenomic data are mainly composed of region-set files, in which a region corresponds to a specific contiguous segment of the genome for which experimental data are available. This includes, for example, the level of DNA methylation or gene expression as well as data on histone modifications. The two most common data types are signal values, i.e. continuous values across the genome, and peaks, i.e. discrete regions of the genome that result from transforming and filtering signal data. Such filtering reduces the amount of data to comparably few discrete regions of presumed biological relevance. The signal data type is typically represented in the WIG file format, while peaks are commonly provided as BED files. Region data are provided by the Data Distribution Center (DCC) portals of epigenome projects, for example the IHEC DCC portal (http://epigenomesportal.ca/ihec/).
Identifying, downloading and preprocessing these datasets is a manual process and can be tedious. Contributing to IHEC, we were interested in providing a simple data retrieval mechanism that could be used by other research groups and developers to locate and extract epigenomic data in a straightforward fashion. To this end, we have developed the DeepBlue Epigenomic Data Server, which facilitates interactive and programmatic access, selection, download and processing of epigenomic data with a focus on region sets.
DeepBlue (http://deepblue.mpi-inf.mpg.de) is a freely accessible data server that streamlines key steps that usually have to be repeated when retrieving epigenomic data: (i) selecting and obtaining regions that meet certain criteria, (ii) handling the associated metadata and (iii) filtering and data processing. DeepBlue aims at providing a comprehensive epigenomic resource with methods for storing, organizing, searching and retrieving data efficiently. We addressed the following software architecture challenges: (i) scalability: the system needs to be able to cope with the currently available data as well as with the growing volume of epigenomic data that will be generated over the next few years; (ii) metadata standardization: It is imperative to handle all data accessible in a standardized form to increase the efficiency of future epigenomic data analysis and software development; (iii) usability: DeepBlue provides a simple application programming interface (API) that users can access to operate on epigenomic data using any of a number of common programming languages. To guarantee widespread use of this web service, access is anonymous and does not require a login.
STATE OF THE ART
Existing tools for epigenomic data retrieval, such as the UCSC Genome Browser (4), Galaxy (5), the UCSC Table Browser (6), ENCODE Search (https://www.encodeproject.org/search/) and the epigenomic data portals of DEEP (http://deep.dkfz.de), BLUEPRINT (http://dcc.blueprint-epigenome.eu) and IHEC (http://epigenomesportal.ca/ihec/) provide user-friendly web interfaces. However, none of these tools offers easy programmatic data retrieval via a cross-platform API. DeepBlue fills this gap by allowing users to search, retrieve and, most importantly, to operate on epigenomic data from several epigenomic mapping consortia programmatically. A comparison of available web platforms for obtaining and working on epigenomic data is shown in Table 1.
Table 1. Comparison between web platforms for accessing and processing epigenomic data.
| Tool | Epigenomic data | Operations | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ENCODE | ROADMAP | BLUEPRINT | DEEP | Text search | Filter by | Count and summarize | Visualization | |||
| Metadata | Regions content | Overlap | ||||||||
| UCSC GB | ✓ | ✓ | ✓ | |||||||
| Galaxy | ✓ | ✓ | ✓ | ✓ | ||||||
| UCSC TB | ✓ | ✓ | ✓ | |||||||
| ENCODE portal | ✓ | ✓ | ✓ | ✓ | ||||||
| IHEC portal | ✓ | ✓ | ✓ | ✓ | ||||||
| DeepBlue | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
Abbreviations: UCSC GB: UCSC Genome Browser; UCSC TB: UCSC Table Browser.
SOFTWARE ARCHITECTURE AND IMPLEMENTATION
The data server has been developed in C++. It has three main components: (i) an XML-RPC server that receives and handles user requests; (ii) a processing engine that performs operations on the data; (iii) a database access point that is connected to a MongoDB server and allows for storing and retrieving data.
Commands are sent to DeepBlue via the XML-RPC protocol, which is supported by all major programming languages. We tested the DeepBlue API with Python, R, JavaScript, PHP and Java. We use more than 200 unit tests written in Python as the basis of our automated testing.
MongoDB (https://www.mongodb.org) was selected as the backend database due to its horizontal scalability (sharding) and its flexible data model, which makes it possible to include arbitrary metadata fields. This allows for the inclusion of standardized properties common to all data sources as well as additional information specific to one dataset. MongoDB was the best performing noSQL database system for insertion and retrieval of terabytes of compressed epigenomic data among the database systems that we inspected when starting the project.
DeepBlue currently holds more than 13 TB of epigenomic data. Optimizations were made in four main aspects: (i) compressing the region-set data; (ii) indexing the regions for fast retrieval; (iii) using efficient algorithms that can handle large data volumes; (iv) implementing the data operations using parallel processing code. We also made use of scalable solutions to cope with the increasing amount of data: (i) data processing: multiple instances of DeepBlue on different servers can be started when the processing load is high; (ii) data storage: MongoDB facilitates the inclusion of new computer nodes in the database clusters to increase capacity. MongoDB is known to handle volumes of hundreds of terabytes of data in business use cases. We thus expect that DeepBlue will be able to cope with very large amounts of epigenomic data.
DEEPBLUE DATA MODEL
The DeepBlue Epigenomic Data Server contains two types of region-set data, namely experiments and annotations. Examples of experimental data include results from experimental assays, for example, DNA methylation signals or histone mark peaks. Annotations include, for example, information related to associated genes, promoters and transcripts. Additional experiments and annotations can be easily uploaded by registered users with the necessary access rights, which can be granted by an administrator. The DeepBlue website contains a list of all experiments and annotations currently included in DeepBlue.
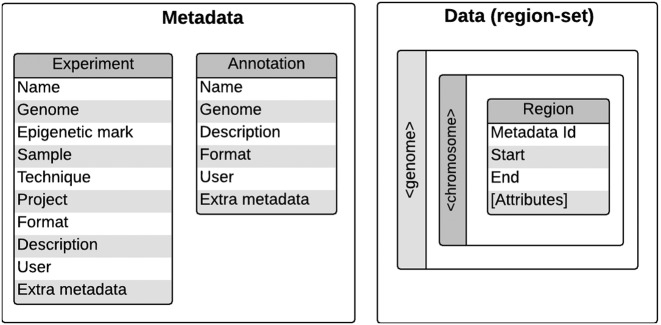
Each experiment consists of a region set and associated metadata (Figure 1), which includes five mandatory fields, as well as an arbitrary number of fields specific to an individual data source, referred to as extra metadata. The five mandatory fields present the genome assembly that was used, the common name of the epigenetic mark, the internal sample identifier (that references a biosource), the experimental technique used to collect the data and the name of the project. To enforce consistent naming throughout the database, each of these fields is restricted to terms that are registered in controlled vocabularies, which are collections of unique terms available for genome assemblies, epigenetic marks, biosources, experimental techniques and projects. As an example, the controlled vocabulary Genome contains the names of genome assemblies used by the epigenome mapping consortia, such as hg19, mm10 and GRCh38. We import the histone mark names from the HIstome database (7) and other epigenomic targets are imported from the targets page of the ENCODE Search (https://www.encodeproject.org/targets/). DeepBlue indexes the metadata and extra metadata fields for all experiments, annotations and controlled vocabularies, allowing users to locate information quickly via full-text search.
Figure 1.
DeepBlue Data Model: the experiments and annotation metadata are constituted by their names, controlled vocabulary terms and extra metadata. The experiments and annotations data form a set of regions. Each region is linked to the corresponding metadata and contains its start and end, as well as additional attributes found in the data files. The regions are stored in their respective genome and chromosome collections.
The DeepBlue Biosources terms are used in the samples to name the biological source (cell line, cell type, tissue or organ). They are imported from the Cell Type Ontology (8), Experimental Factor Ontology (9) and Uber Anatomy Ontology (10), following IHEC policy. The terms are imported together with their synonyms and hierarchy. The user can thus conveniently use biosources synonyms for finding experiments. It is also possible to use a biosource term to obtain all experiments related to it and its subterms.
DATA CONTENTS
Currently, the server comprises epigenomic datasets from four major contributors of IHEC, namely ENCODE, ROADMAP, BLUEPRINT and DEEP. At the time of writing, DeepBlue hosts peak files, signal files, as well as metadata for 33,061 experimental datasets across 2,566 different samples. Of these datasets, 15,950 are peak files and 17,092 are signal files. A list of all data available in DeepBlue is provided on the project website. In order to keep DeepBlue up to date, the data sources of the relevant consortia are checked periodically and automatically for changes and new data are imported.
PROGRAMMATIC INTERFACE
The DeepBlue Epigenomic Data Server enables users to operate on epigenomic data directly on the server. This is facilitated through an API specifically designed for region-set epigenomic data and metadata. The functionality of the API includes the following operations on regions: filtering by region attributes, detecting overlaps, aggregating summary statistics, searching for DNA motifs and counting regions. The result can be downloaded in a tabular or matrix format.
DeepBlue can be used anonymously with access to all public data. In addition, users can create their personal account, to have access to their command history, to upload their own data, to access or to share privately managed data in their DeepBlue workspace.
The DeepBlue API can be divided into six main command categories: information, list and search, selection, operation, result and request download of epigenomic data. Table 2 summarizes the main operations found in these categories. The DeepBlue website contains the full API list. DeepBlue entities, e.g. experiments, annotations, controlled vocabularies and requests, have a unique identifier, which can be used to obtain detailed information through the info command.
Table 2. DeepBlue commands to list, search, select, operate and retrieve epigenomic data.
| Category | Command | Description |
|---|---|---|
| Information | info | Obtain information about an entity |
| List and search | list_genomes | List all registered genomes |
| list_biosources | List all registered biosources | |
| list_samples | List all registered samples | |
| list_epigenetic_marks | List all registered epigenetic marks | |
| list_experiments | List all available experiments | |
| list_annotations | List all available annotations | |
| search | Perform a full-text search | |
| Selection | select_regions | Select regions from experiments |
| select_experiments | Select regions from experiments | |
| select_annotations | Select regions from annotations | |
| select_genes | Select genes as regions | |
| tiling_regions | Generate tiling regions | |
| input_regions | Upload and use a small region-set | |
| Operation | aggregate | Aggregate and summarize regions |
| filter_regions | Filter regions using their attributes | |
| flank | Generate flanking regions | |
| intersection | Filter overlapping regions | |
| merge_queries | Merge two regions set | |
| Result | count_regions | Count selected regions |
| score_matrix | Request a score matrix | |
| get_regions | Request the selected regions | |
| Request | get_request_data | Obtain the requested data |
The typical workflow starts with listing the experiments, followed by data selection. Optionally, the selected data can be processed by counting or retrieving the selected regions. The results can be downloaded as a formatted table or score matrix.
DeepBlue provides commands for listing and searching for entities, such as controlled vocabulary terms, annotations and experiments. It is possible to obtain all entities or to list those that match specified criteria. For example, the list_genomes command returns a list of all genome assemblies registered in DeepBlue. The same concept extends to other list commands, such as list_experiments, which is used to list the experiments corresponding to a given metadata field.
Listing commands are a practical way of learning what data is stored in DeepBlue but less appropriate for retrieving specific information. Instead, the command search can be used to find any terms in the controlled vocabularies, as well experiments and annotations, by their metadata and extra metadata content using full-text search.
DeepBlue operates on defined genomic region sets, which can be selected directly or retrieved based on experiments, annotations or genes. All of these commands return a request identifier, which can be used as input for region-set operation commands that enable users to aggregate regions, filter regions by their content, generate flanking regions, find overlapping regions or merge different regions based on additional criteria. These commands return a new request identifier, which in turn can be used as input for other operation commands. This allows users to perform complex operations by constructing a sequence of simple ones. Alternatively, request identifiers are used to download intermediate or final results.
Operations and requests for results are processed asynchronously. In other words, instead of blocking the connection to the client until the result is available, DeepBlue will immediately return a request identifier. This so-called request_id can be used in the info command to query the status and progress of the requested operation. When the processing is finished, the data can be downloaded using the get_request_data command.
USE CASES
To illustrate the wide range of applications in which DeepBlue can be utilized for the efficient retrieval of epigenomic data, we present three typical use cases that until now had to be performed manually in epigenetic studies: (i) identification of H3K27ac peaks overlapping promoters and transcription factor (TF) binding sites; (ii) calculating DNA methylation levels across H3K4me3 peak regions; (iii) obtaining mRNA expression levels for a set of genes. Here, we illustrate the central commands necessary to execute the first use case in a workflow diagram (Figure 2), while the comprehensive description of all three use cases can be found on the DeepBlue website.
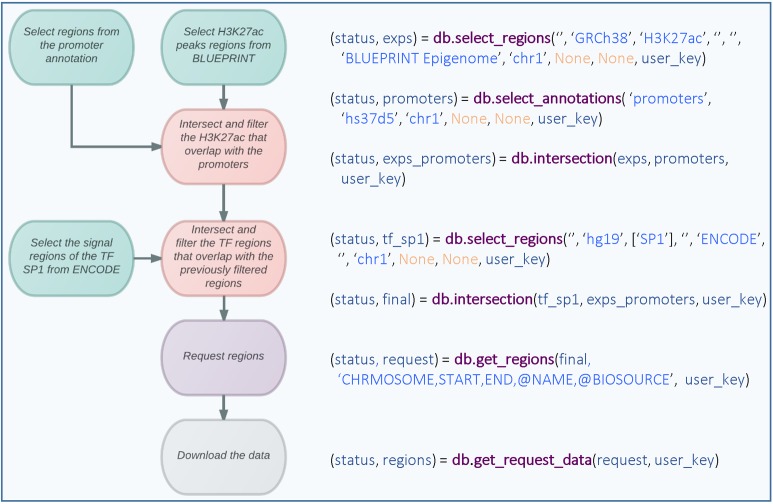
Figure 2.
Workflow diagram and source code for the identification of H3K27ac peaks that overlap with promoters in any of the BLUEPRINT datasets and subsequent identification of transcription factor peaks that overlap with these promoters in any of the ENCODE datasets. The different colors represent different types of commands, e.g. green for data selection, red for operations, purple for requests and gray for download.
Chromatin regulators exert control over cell type-specific gene expression. In particular, promoters and enhancer elements are known to be associated with certain histone marks (11). In the following use case, we demonstrate how DeepBlue can be used to obtain promoter-specific region sets, in which histone modification H3K27ac may potentially act in concert with a TF. To further show how DeepBlue can operate on several data sources in the same workflow, we first select BLUEPRINT datasets with H3K27ac peaks (line 1) overlapping with promoter regions (line 2) as well as TF binding sites of SP1 from the ENCODE datasets (line 3). Next, we filter the H3K27ac peaks that overlap with promoters (line 4) and filter for regions that also overlap with the selected TF regions (line 5). Finally, the resulting regions are requested (line 6) selecting only columns of interest. Columns starting with an ‘@’ are called meta-columns that include the associated region information. Finally, the data can be downloaded using the request ID (line 7).
CONCLUSION
Large volumes of epigenomic data are being generated, for instance, by the various IHEC members. These data hold the promise to revolutionize our understanding of cell regulation and of human diseases. However, studies aimed at fulfilling this promise are faced with the complexity of data acquisition and processing. These steps are time-consuming due to the lack of suitable programmatic data retrieval. Currently, epigenomic data is only retrieved through web portals, which are set up by the respective consortia. These portals offer access to region-set data and metadata but lack effective mechanisms for searching, filtering and processing of these data programmatically. Moreover, a lack of metadata standardization complicates the use of data from different sources in a single study. In a typical epigenomics analysis, such routine preprocessing steps are thus often more time-consuming than the following analyses and consequently hinder effective research in the field.
To mitigate this problem, we have developed DeepBlue as a data server that enables software developers and users of epigenomic data to select, preprocess and retrieve region-set data from several epigenomic mapping consortia, namely ENCODE, ROADMAP, BLUEPRINT and DEEP. DeepBlue hosts peaks files, signal files and the corresponding metadata, currently amounting to 33,061 datasets and annotations of various types of genomic regions. Programmatic data retrieval allows for selecting and aggregating data efficiently, making DeepBlue highly effective regarding answering specific research questions with comparably little effort.
We addressed the following software architecture challenges: (i) scalability: we store all data in a scalable database system and more instances of DeepBlue can be started when the workload is high; (ii) metadata standardization: we map all experiment metadata from different consortia to terms from the same controlled vocabularies. Terms that cannot be mapped are stored in key-value pairs; (iii) usability: DeepBlue has a powerful and well-documented API that enables users to programmatically search, list, select, operate and download epigenomic data. We chose the XML-RPC protocol for implementing the DeepBlue API to maximize compatibility with various programming languages. Requests that involve the aggregation of millions of region sets can typically be computed in a few minutes. The DeepBlue server is currently configured to use up to 16 GB per workflow, which is enough to hold more than 500 million regions. This value is set to prevent the abuse of computational resources and can be changed for individual users.
DeepBlue serves as a comprehensive online resource for the epigenomic community. It is unique in its ability to handle epigenomic data from different consortia in a single workflow. This is particularly advantageous for large-scale data analysis involving many different biosources. However, there are limitations to this type of analysis caused by the differences in the raw data processing pipelines. Such pipelines differ across and even within the epigenome mapping consortia. For instance, using a different set of tools or even the same tools with different versions or parameters can have a significant impact on the results and lead to batch effects. Similarly, the use of different reference genome assemblies will have an impact on the exact location of the regions, introducing bias in the results. While this is an issue that we cannot immediately address, it can be expected that the efforts of the IHEC consortium members to achieve a higher degree of data processing standardization will make these analyses more robust in the future.
DeepBlue has been under active development and extensive testing for more than three years. It has been openly available for users outside our institute since September 2015. Since that date, DeepBlue has processed more than 2,500 workflow processing requests, most of which involve the combination of data selection and intersection commands. 300 requests were made by registered users and 2,200 by anonymous users.
For the future, we plan to include additional dataset, comprising also other epigenome mapping projects and research groups. We will also extend the web interface to enable non-programmers not only to browse and download the data, but to perform more complex data operations in an intuitive way. Moreover, we plan to develop a Bioconductor package that acts as a DeepBlue client and will make it possible to integrate DeepBlue with existing epigenome data analysis tools in R, such as the LOLA package for region-set enrichment analysis (12).
The development of DeepBlue was motivated by our involvement in the DEEP and BLUEPRINT projects, which also allowed us to closely communicate with the epigenomics community. As a result, DeepBlue has already received substantial interest from several members of the IHEC community, who plan to use DeepBlue routinely in the future. We expect that DeepBlue has the potential to find widespread adoption as a tool for epigenomics data retrieval and processing both in software and in analysis pipelines used in future studies involving epigenomic data.
FUNDING
German Science Ministry (DEEP project) [01KU1216A]; EU (BLUEPRINT project) [HEALTH-F5-2011-282510]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 2.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Wang J., Ziller M.J., Whitaker J.W., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams D., Altucci L., Antonarakis S.E., Ballesteros J., Beck S., Bird A., Bock C., Boehm B., Campo E., Caricasole A., et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 4.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goecks J., Nekrutenko A., Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2003;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khare S.P., Habib F., Sharma R., Gadewal N., Gupta S., Galande S. HIstome–a relational knowledgebase of human histone proteins and histone modifying enzymes. Nucleic Acids Res. 2011;40:D337–D342. doi: 10.1093/nar/gkr1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bard J., Rhee S.Y., Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malone J., Holloway E., Adamusiak T., Kapushesky M., Zheng J., Kolesnikov N., Zhukova A., Brazma A., Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mungall C.J., Torniai C., Gkoutos G.V., Lewis S.E., Haendel M.A. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheffield N.C., Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]