Abstract
Next generation sequencing (NGS) allows investigating mitochondrial DNA (mtDNA) characteristics such as heteroplasmy (i.e. intra-individual sequence variation) to a higher level of detail. While several pipelines for analyzing heteroplasmies exist, issues in usability, accuracy of results and interpreting final data limit their usage. Here we present mtDNA-Server, a scalable web server for the analysis of mtDNA studies of any size with a special focus on usability as well as reliable identification and quantification of heteroplasmic variants. The mtDNA-Server workflow includes parallel read alignment, heteroplasmy detection, artefact or contamination identification, variant annotation as well as several quality control metrics, often neglected in current mtDNA NGS studies. All computational steps are parallelized with Hadoop MapReduce and executed graphically with Cloudgene. We validated the underlying heteroplasmy and contamination detection model by generating four artificial sample mix-ups on two different NGS devices. Our evaluation data shows that mtDNA-Server detects heteroplasmies and artificial recombinations down to the 1% level with perfect specificity and outperforms existing approaches regarding sensitivity. mtDNA-Server is currently able to analyze the 1000G Phase 3 data (n = 2,504) in less than 5 h and is freely accessible at https://mtdna-server.uibk.ac.at.
INTRODUCTION
Mitochondrial DNA (mtDNA) is maternally inherited in humans and present in thousands of copies per cell. Heteroplasmy describes a de novo mtDNA mutation often present in only a few copies. The differentiation between real mutational clones and sequencing artefacts can be complex, but is crucial in researching somatic mutations in cancer, neurodegenerative diseases and aging (1). Artefacts became even more evident with new and more sensitive sequencing technologies (2,3). Furthermore, the paradigm shift from analyzing few reliable long reads (400–800 bp) in Sanger based sequencing to millions of short reads (50–250 bp) in Next Generation Sequencing (NGS) requires new computational models and additional attention interpreting results. While higher error rates within NGS can be opposed with higher sequencing coverage for variant detection, interpretation of results still needs consideration when analyzing variant allele frequencies (VAF) below 10%, the detection limit for Sanger-based sequencing. While the role of such variants is acknowledged for some diseases (e.g. in mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) (4)) its origin and mechanisms to prevail as somatic mutations is largely unknown (1). Since the first description of analyzing mtDNA heteroplasmy on NGS devices in 2010 (5), several Unix command line pipelines have been presented (6–8). These pipelines facilitate the analysis of mtDNA data, but can be challenging to install. To eliminate these shortcomings, web servers were implemented (9–11), but they were limited to small input files, revealed shortcomings in usability, overloaded with parameter options, or generate poor and often unreliable results (see Supplementary Tables S1–3).
Here we present mtDNA-Server, a highly scalable Hadoop-based server (12) for mtDNA NGS data processing. For handling large studies (>100 samples), we implemented new parallel mechanisms to overcome limitations of local single node architectures. We efficiently parallelized workflow steps such as sequence alignment, per-base alignment scoring (BAQ) (13), and heteroplasmy and contamination detection. To avoid misinterpretation of data which can arise from sequencing errors as well as low-level contamination of samples, we introduced extensive QC checks. Furthermore, we provide a clean user interface to guide researchers through the various analysis steps. Additionally, we integrated the Maximum Likelihood (ML) heteroplasmy model presented in (14) and incorporated the haplogroup classifier HaploGrep (15,16) to check for sample contamination in an automated way. To ensure reproducibility and usability, we make use of the Hadoop workflow system Cloudgene (17). mtDNA-Server is currently able to analyze the 1000G Phase 3 BAM data in <5 h.
MATERIALS AND METHODS
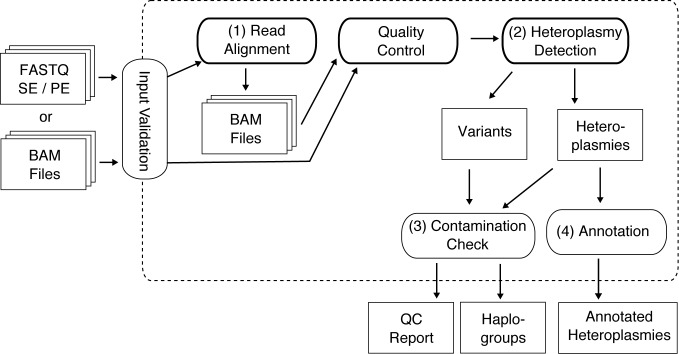
mtDNA-Server provides an mtDNA analysis workflow starting with raw data in FASTQ or BAM format and resulting in reliable detection of heteroplasmic sites, contamination estimates and numerous QC statistics (see Figure 1). To reach a high level of parallelism, mtDNA-Server supports the upload of several samples at once, whereby each input file is further split into independent chunks (intra-sample chunking). For parallelization, the Hadoop MapReduce framework (http://hadoop.apache.org/) is used. Within Hadoop, a cluster of nodes (each consisting of several cores) processes chunks in parallel (map), grouped and collected by the framework (shuffle) and further analyzed or directly returned to the application (reduce). The underlying Cloudgene framework handles the communication with the Hadoop cluster and provides a web interface for all job-related tasks (see section ‘Web Service’).
Figure 1.
Overall mtDNA-Server workflow for FASTQ and BAM input.
Input validation
The validation step verifies sample input first by automatic format detection. Currently input data in FASTQ (Single and Paired End) and SAM/BAM format is supported. Furthermore, a valid mitochondrial reference length tag (Yoruba reference NC_001807.4 with length 16571, rCRS (18) or RSRS (19) with length 16569) must be included in the BAM header. Since rCRS constitutes the standard (included in GRCh37 since patch release 2), input samples aligned to the mitochondrial Yoruba reference are realigned to the rCRS reference to avoid misleading results in post-processing. Depending on the detected file format, a specific set of workflow steps is executed in parallel.
Parallel read alignment
For FASTQ input, mtDNA-Server applies the sequence aligner BWA-MEM v. 0.7.5 (20) to each chunk and aligns the reads to the rCRS reference. For single-end reads (SE), BWA-MEM is executed on each chunk, storing all SAM records in the Hadoop Distributed File System (HDFS)). For paired-end (PE) reads, correct read pairs are identified by setting the Hadoop output key to the read name as suggested in (21). After pairs have been mapped, they are written to a BAM file by using the secondary sort mechanism of Hadoop MapReduce. All reads are sorted according to the start position.
Quality control
During the quality control step, statistics on the BAM input file are generated. This allows mtDNA-Server to provide real-time feedback to end users including several metrics on reads and bases, such as mapped reads, detected duplicates or reference issues. Moreover, it helps users to identify low quality input data (i.e. showing a high amount of unmapped reads), which will otherwise lead to incorrect results.
Parallel analysis of the BAM file and variant detection
For homoplasmic as well as heteroplasmic variant detection, HadoopBAM (22) is applied to split input BAM files. For each chunk, mtDNA-Server filters reads with a mapping quality score <20 (Phred score) and a read length <25 (9). BAM reads marked as duplicates are filtered within this step. Additionally, mtDNA-Server excludes all reads with an alignment Phred score ≤30 and applies per-BAQ to all reads by default. For this purpose, the GATK (23) BAQ implementation has been adapted to work with the circular nature of mitochondrial genomes. For each passed read, all bases with a quality Phred score <20 are filtered. Since mtDNA-Server detects point heteroplasmies, recalibration and re-alignment did not affect the result quality and has been excluded. Finally, all passed bases for each site are counted per strand (A, C, G, T, N (unknown base) or d (deletion)). For heteroplasmy detection, several filters and methods are applied: first, mitochondrial hotspots around 309 and 315 as well as 3107 according to the rCRS are excluded. Sites showing coverage <10 bases per strand are filtered. For all remaining sites showing (i) a VAF of ≥ 1% (strand independent) and (ii) an allele coverage of three bases per strand, an ML model is applied (14). The ML model takes sequencing errors per base into account and is applied to each strand. All sites with a log likelihood ratio (LLR) of ≥5 are tagged as heteroplasmic sites. Since strands are analyzed independently, mtDNA-Server can filter all heteroplasmic sites with a strand bias score <1 (24,25). Furthermore, the Wilson and the Agresti-Coull confidence interval is calculated for heteroplasmic variants (7). The assigned heteroplasmy level is the weighted mean of heteroplasmy of the forward and reverse strand. We further tag positions in low complexity regions (LCR) (9) and known polymorphic nuclear mitochondrial insertions (NumtS) (26). LCR tagging is particular necessary for IonTorrent samples, which show a per-base error increase in homopolymeric stretch >4 bases. Finally, the unfiltered pileup format file, annotated files for variants and the HaploGrep files are generated.
Intra-sample contamination check
The increased coverage of mtDNA NGS studies allows analyzing heteroplasmy levels down to 1% VAF. This poses the risk of interpreting low-level contamination or sample mix-up as real heteroplasmy (27,28). Contamination is thereby not limited to issues in the laboratory (cross-contamination or sample mix-up), but also carry-over contamination between NGS runs, or even issues in post-processing step for NGS (adapter removal, file merging). Therefore, mtDNA-Server performs a contamination check based on the current phylogeny in order to avoid misinterpretations and erroneous conclusions. As previously outlined (5,29), positions showing VAF on sample-level can be checked for haplogroup concordance and thereby hinting to intra-sample contamination: mtDNA-Server therefore generates two profiles based on the VAF; a minor (<50%) and major (>50%) profile which is used to perform haplogroup checks with HaploGrep. In case of contamination caused by different mtDNA sequences, the profiles lead to different valid haplogroups. We further take the homoplasmic variants for both profiles into consideration, rendering the classification more reliable, and helping tracing the origin of the sample mix-up. Thereby homoplasmic variants are shared haplogroup defining polymorphisms on the main branch from the rCRS to the mitochondrial Eve (mt-MRCA). Subsequently, heteroplasmic variants are divided into major and minor profiles, which are segregated into different branches in case of contamination (see Supplementary Figure S1).
Web service
mtDNA-Server uses Cloudgene as the underlying platform to build a scalable web service including a graphical user interface. Cloudgene describes a high-level workflow system for Apache Hadoop designed as a web application. The API provides methods for the execution and monitoring of MapReduce jobs and is conceived as an additional layer between Hadoop and the web client. mtDNA-Server has been integrated by using Cloudgene's workflow definition language and its plugin interface. Once integrated, mtDNA-server takes advantage of features provided by Cloudgene including user login, data security or real-time feedback. Moreover, it enables mtDNA-Server to assemble all possible heteroplasmies, QC statistics and contaminations into an interactive graphical report that can be viewed directly in the web browser or can be shared with collaborators.
RESULTS
Data import
The two major issues when designing publicly available web servers are (i) file upload limitations and (ii) data sensitivity/privacy concerns.
(i) The mtDNA-Server can analyze files from numerous locations including (a) local file upload via web or a command line tool and (b) import from sftp servers or https web-servers. Direct file uploads are especially convenient for small sample sizes (<100 MB). For large samples sizes, mtDNA-Server provides a sftp and https importing tool to upload large datasets from a specified web location in background (see Table 1).
Table 1. Possible data sources and files sizes for mtDNA-Server.
| mtDNA data source | Sample file size (BAM Format) | Mean coverage |
|---|---|---|
| Ancient DNA | <10 MB / sample | ≤100-fold |
| Whole exome sequencing | <20 MB / sample | ≤1,000-fold |
| Whole genome low coverage | 1-80 MB / sample | ≤3,000-fold |
| Whole genome high coverage | 200 MB / sample | ≤20,000-fold |
| Targeted mtDNA sequencing | Up to 1 GB / sample | ∼50,000-fold |
To support users with high upload demands even further, mtDNA-Server provides a command line upload tool which can be downloaded from the mtDNA-Server website. Before uploading files, the tool extracts the mitochondrial genome from large BAM files and executes QC validation checks locally. If all checks succeeded, a new analysis on mtDNA-Server is started automatically.
(ii) To address data privacy and sensitivity concerns, a wide array of security measures has been implemented: the complete interaction with the server is secured with HTTPS, input data is deleted automatically from our servers directly after job termination and final datasets are kept on the server for a period of 7 days. Thereafter all data are erased automatically. The data can also be deleted immediately by the user after downloading or inspecting the results online. Results derived from the public mode are protected by encrypted token URLs only accessible by the user.
Data output
The final output generated by mtDNA-Server provides several downloadable files and interactive reports. This includes (i) an interactive HTML report summarizing all findings, (ii) detected heteroplasmic and homoplasmic sites, (iii) the HaploGrep input and final classification file and (iv) the raw pileup file including base position counts per sample.
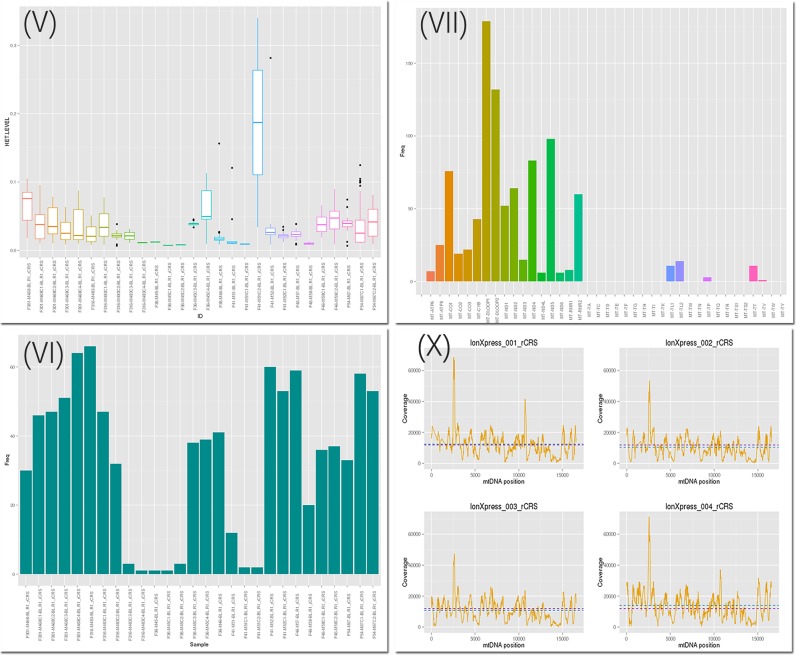
The HTML report itself presents several quality control (QC) measures: (I) a check for the selected base-quality based on the ratio of mapped to filtered reads; (II) a list of all samples with >20 heteroplasmic sites as ‘suspicious’; (III) an interactive table listing all heteroplasmic sites, which can be searched, filtered or sorted by the user in real time; (IV) a heteroplasmy frequency table including heteroplasmic sites found in more than two samples, which indicates potential artefacts; (V) a heteroplasmy boxplot summarizing the heteroplasmy levels per sample and (VI) a bar plot of the heteroplasmies found per sample as well as (VII) the map-loci of heteroplasmic sites; (VIII) a haplogroup interactive table of resulting haplogroups including their quality score and basic coverage information for each sample (the table includes the covered mtDNA in absolute numbers, which is important for ancient mtDNA or mtDNA data derived from exome sequencing). (IX) a contamination table indicating phylogenetic issues, based on 5,437 haplogroups present in Phylotree (17); (X) coverage-plots for each sample, which is especially important for targeted resequencing studies to detect issues with incorrect concentrations of the polymerase chain reaction products in the used fragments. (XI) Furthermore, an interactive table for homoplasmic sites is included in the report. It provides for each variant the haplogroup defining status and includes information regarding amino acid changes, phylogenetic weights as presented in HaploGrep 2 and pathogenicity scores. Figure 2 includes four plots from the final HTML report.
Figure 2.
Four plots of the final HTML report: Boxplot over heteroplasmic levels per sample (V), frequency of heteroplasmic variants as a bar plot (VI), locus of the heteroplasmic variants on the mitochondrial genome over all analyzed samples (VII) and coverage plots per sample (X).
Validation
We validated the heteroplasmy model based on four different sample-mix ups on Illumina HiSeq (mean coverage 50,000-fold) and IonTorrent PGM (mean coverage 5,000-fold). We mixed two samples in the laboratory as follows: M1—1:2 (50%), M2—1:10 (10%), M3—1:50 (2%), M4—1:100 (1%). For validation, we sequenced the two initial samples also independently. With sample-specific heteroplasmic mutations excluded, 27 heteroplasmic sites are expected in the mix-ups (see Supplemental Figure S1). By comparing expected to detected heteroplasmic sites, precision (positive predictive value), sensitivity (true positives) and specificity (true negatives) have been calculated. Since currently available web services were not capable of analyzing our mix-ups due to their file size (>50 MB), we compared mtDNA-Server with LoFreq (30), a software for ultra-sensitive variant detection. As Tables 2 and 3 show, both LoFreq and mtDNA-Server report heteroplasmies reliable. They show a similar level of specificity and precision, but LoFreq is slightly outperformed by mtDNA-Server regarding sensitivity on both NGS devices. Furthermore, mtDNA-Server classifies mix-ups into haplogroups as expected. For comparison with other web services, we analyzed two independent datasets with (i) mit-o-matic (10), (ii) MToolBox (7), (iii) LoFreq, (iv) Galaxy Naive Caller and (v) MitoSeek (6) (see Supplementary Tables S1–3). The evaluation shows that mtDNA-Server generates accurate results over all datasets, without reporting additional false positive hits.
Table 2. mtDNA-Server versus LoFreq on IonTorrent PGM.
| Sample mix-up IonTorrent PGM | mtDNA-Server | LoFreq | ||||
|---|---|---|---|---|---|---|
| Precision | Sensitivity | Specificity | Precision | Sensitivity | Specificity | |
| 1:2 | 100% | 88.89% | 100% | 100% | 81.48% | 100% |
| 1:10 | 100% | 81.48% | 100% | 100% | 81.48% | 100% |
| 1:50 | 100% | 77.78% | 100% | 100% | 55.56% | 100% |
| 1:100 | 100% | 59.26% | 100% | 100% | 11.11% | 100% |
Table 3. mtDNA-Server versus LoFreq on Illumina HiSeq.
| Sample mix-up Illumina HiSeq | mtDNA-Server | LoFreq | ||||
|---|---|---|---|---|---|---|
| Precision | Sensitivity | Specificity | Precision | Sensitivity | Specificity | |
| 1:2 | 100% | 100% | 100% | 93.10% | 100% | 99.98% |
| 1:10 | 100% | 92.6% | 100% | 89.29% | 92.6% | 99.98% |
| 1:50 | 100% | 92.6% | 100% | 82.76% | 88.9% | 99.99% |
| 1:100 | 100% | 85.2% | 100% | 83.87% | 96.3% | 99.99% |
DISCUSSION
The here presented mtDNA-Server describes a web server based on Hadoop, allowing to analyze human mitochondrial NGS data. All aspects from sequence alignment of FASTQ raw data to final results including heteroplasmic variants are covered. By hiding all complex workflow steps, we provide the mtDNA research community an easy to use web server, allowing the detection of heteroplasmic variants in a secure and reproducible way. The validation of the integrated heteroplasmy approach shows that mtDNA-Server detects heteroplasmic variants and sample contamination accurately. The within-sample contamination detection based on mitochondrial haplogroups has significant general potential to assess data from whole genome sequencing studies for potential contamination and is therefore not limited to target mtDNA sequencing. mtDNA-Server is conceived to handle large amount of mtDNA NGS data, by taking advantage of parallel data processing using the MapReduce framework, having the main focus on ease of use and reliability of the results to prevent erroneous conclusions.
Supplementary Material
Acknowledgments
We want to thank Gertraud Erhart for generating the mix-ups in the laboratory, Matthias Weiler and Michael Opitz for setting up the hardware infrastructure for mtDNA-Server, Margot Haun and Bernhard Rupp for helpful comments.
Footnotes
Present address: Dr Sebastian Schönherr, Division of Genetic Epidemiology, Department of Medical Genetics, Molecular and Clinical Pharmacology, Medical University of Innsbruck, Innsbruck 6020, Austria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU FP7 project [602133]; Austrian Science Fund (FWF) [J-3401 to C.F.]. Funding for open access charge: EU FP7 project [602133]; Austrian Science Fund (FWF) [J-3401].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wallace D.C., Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Med. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skonieczna K., Malyarchuk B.a, Grzybowski T. The landscape of mitochondrial DNA variation in human colorectal cancer on the background of phylogenetic knowledge. Biochim. Biophys. Acta. 2012;1825:153–159. doi: 10.1016/j.bbcan.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Bandelt H.-J., Salas A. Current Next Generation Sequencing technology may not meet forensic standards. Forensic Sci. Int. Genet. 2012;6:143–145. doi: 10.1016/j.fsigen.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M., Schönberg A., Schaefer M., Schroeder R., Nasidze I., Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y., Li J., Li C.-I., Shyr Y., Samuels D.C. MitoSeek: extracting mitochondria information and performing high throughput mitochondria sequencing analysis. Bioinformatics. 2013;29:1210–1211. doi: 10.1093/bioinformatics/btt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabrese C., Simone D., Diroma M.A., Santorsola M., Gutta C., Gasparre G., Picardi E., Pesole G., Attimonelli M. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics. 2014;30:3115–3117. doi: 10.1093/bioinformatics/btu483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickins B., Rebolledo-Jaramillo B., Su M.S.-W., Paul I.M., Blankenberg D., Stoler N., Makova K.D., Nekrutenko A. Controlling for contamination in re-sequencing studies with a reproducible web-based phylogenetic approach. Biotechniques. 2014;56:134–141. doi: 10.2144/000114146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhidkov I., Nagar T., Mishmar D., Rubin E. MitoBamAnnotator: A web-based tool for detecting and annotating heteroplasmy in human mitochondrial DNA sequences. Mitochondrion. 2011;11:924–928. doi: 10.1016/j.mito.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Vellarikkal S.K., Dhiman H., Joshi K., Hasija Y., Sivasubbu S., Scaria V. mit-o-matic: a comprehensive computational pipeline for clinical evaluation of mitochondrial variations from next-generation sequencing datasets. Hum. Mutat. 2015;36:419–424. doi: 10.1002/humu.22767. [DOI] [PubMed] [Google Scholar]
- 11.Santorsola M., Calabrese C., Girolimetti G., Diroma M.A., Gasparre G., Attimonelli M. A multi-parametric workflow for the prioritization of mitochondrial DNA variants of clinical interest. Hum. Genet. 2015;135:121–136. doi: 10.1007/s00439-015-1615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean J., Ghemawat S. MapReduce: simplified data processing on large clusters. Commun. ACM. 2004;51:107–113. [Google Scholar]
- 13.Li H. Improving SNP discovery by base alignment quality. Bioinformatics. 2011;27:1157–1158. doi: 10.1093/bioinformatics/btr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye K., Lu J., Ma F., Keinan A., Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1403521111. doi:10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kloss-Brandstätter A., Pacher D., Schönherr S., Weissensteiner H., Binna R., Specht G., Kronenberg F. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 16.Weissensteiner H., Pacher D., Kloss-Brandstätter A., Forer L., Specht G., Bandelt H.-J., Kronenberg F., Salas A., Schönherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw233. doi:10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schönherr S., Forer L., Weissensteiner H., Kronenberg F., Specht G., Kloss-Brandstätter A. Cloudgene: a graphical execution platform for MapReduce programs on private and public clouds. BMC Bioinformatics. 2012;13:200. doi: 10.1186/1471-2105-13-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 19.Behar D.M., van Oven M., Rosset S., Metspalu M., Loogväli E.-L., Silva N.M., Kivisild T., Torroni A., Villems R. A ‘Copernican’ reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013. 1303.3997 arXiv:
- 21.Pireddu L., Leo S., Zanetti G. SEAL: a distributed short read mapping and duplicate removal tool. Bioinformatics. 2011;27:2159–2160. doi: 10.1093/bioinformatics/btr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemenmaa M., Kallio A., Schumacher A., Klemelä P., Korpelainen E., Heljanko K. Hadoop-BAM: directly manipulating next generation sequencing data in the cloud. Bioinformatics. 2012;28:876–877. doi: 10.1093/bioinformatics/bts054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y., Cai Q., Samuels D.C., Ye F., Long J., Li C.-I., Winther J.F., Tawn E.J., Stovall M., Lähteenmäki P., et al. The use of next generation sequencing technology to study the effect of radiation therapy on mitochondrial DNA mutation. Mutat. Res. 2012;744:154–160. doi: 10.1016/j.mrgentox.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y., Li J., Li C.-I., Long J., Samuels D.C., Shyr Y. The effect of strand bias in Illumina short-read sequencing data. BMC Genomics. 2012;13:666. doi: 10.1186/1471-2164-13-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dayama G., Emery S.B., Kidd J.M., Mills R.E. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 2014;42:12640–12649. doi: 10.1093/nar/gku1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Just R.S., Irwin J.A., Parson W. Questioning the prevalence and reliability of human mitochondrial DNA heteroplasmy from massively parallel sequencing data. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E4546–E4547. doi: 10.1073/pnas.1413478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye K., Lu J., Ma F., Keinan A., Gu Z. Reply to Just et al.: mitochondrial DNA heteroplasmy could be reliably detected with massively parallel sequencing technologies. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4548–4550. doi: 10.1073/pnas.1415171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avital G., Buchshtav M., Zhidkov I., Tuval Feder J., Dadon S., Rubin E., Glass D., Spector T.D., Mishmar D. Mitochondrial DNA heteroplasmy in diabetes and normal adults: role of acquired and inherited mutational patterns in twins. Hum. Mol. Genet. 2012;21:4214–4224. doi: 10.1093/hmg/dds245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.