Abstract
Interactive Tree Of Life (http://itol.embl.de) is a web-based tool for the display, manipulation and annotation of phylogenetic trees. It is freely available and open to everyone. The current version was completely redesigned and rewritten, utilizing current web technologies for speedy and streamlined processing. Numerous new features were introduced and several new data types are now supported. Trees with up to 100,000 leaves can now be efficiently displayed. Full interactive control over precise positioning of various annotation features and an unlimited number of datasets allow the easy creation of complex tree visualizations. iTOL 3 is the first tool which supports direct visualization of the recently proposed phylogenetic placements format. Finally, iTOL's account system has been redesigned to simplify the management of trees in user-defined workspaces and projects, as it is heavily used and currently handles already more than 500,000 trees from more than 10,000 individual users.
INTRODUCTION
Phylogenetics and phylogenetic trees are important concepts and tools in a wide variety of biological and other scientific studies. Various tree visualization tools have been developed through the years (1–4), covering many aspects of classical tree visualization. iTOL (5) was one of the first tools to enable the annotation of trees with various types of additional data. Nowadays, several software packages, both standalone and online, offer tree annotation features (6–8). As both the number of features and the handling of the data are important aspects in user acceptance of tools, we report here on a redesigned version of iTOL with greatly expanded and streamlined functionality, making it faster and simpler to use.
FEATURES
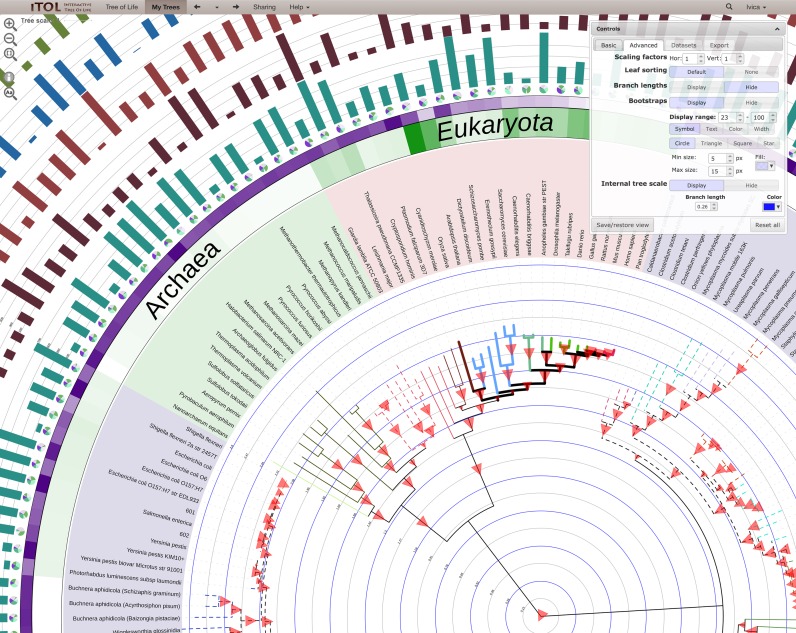
iTOL is an online tool, accessible with any recent web browser (Figure 1). The display engine for the current version was completely re-written, implemented in pure Javascript and now uses the HTML5 Canvas element. Most of the engine calculations and functionality are performed by the client web browser, allowing instant response to user interactions and fine grained control over various display parameters.
Figure 1.
Tree of Life (15) annotated with various datasets, highlighting several novel features, along with iTOL's interactive user interface. Tree branches are colored and displayed with varying styles and widths. Support values are visualized as triangles of varying size. An internal tree scale is displayed, with user-defined colors and level values. A text label dataset was precisely positioned above the colored strips using interactive controls. Several dataset types can include user-defined scales.
Input types
iTOL works with commonly used phylogenetic tree formats, Newick, Nexus (9) and phyloXML (10). A new, recently proposed file format for phylogenetic placements, i.e. mappings of environmental sequence data (e.g. short reads) into a phylogenetic tree (11) is also supported, making iTOL the first tool which can visualize these files generated by EPA (12) and pplacer (13).
All additional data used for various types of tree annotation are provided in plain text files, and can be simply dragged and dropped onto the trees to be instantly visualized in the user's web browser.
Basic functions
Like in version 2 (14), iTOL provides most common functions available in any phylogenetic tree viewer. Various tree display formats are supported: phylograms or cladograms, rooted or unrooted, rectangular or radial. Trees can be manipulated in various ways, and basic editing functions allow users to interactively delete or move single nodes or whole clades. Clades can also be pruned or collapsed, either manually or automatically, based on various parameters (such as associated bootstrap values or average branch length distances). Trees can be re-rooted manually on any node, or automatically using the midpoint rooting method.
ANNOTATING TREES
Ever since the initial release of iTOL in 2006 (5,15), our primary goal was to offer various ways of annotating phylogenetic trees with external data. The current version introduces various new annotation features, which simplify the process by greatly expanding user control over the individual display elements.
There are three major groups of tree annotations in iTOL:
Direct interactive annotations of colors and styles for individual nodes and text labels
Automatic annotations based on intrinsic node features (bootstrap values, branch lengths)
User provided annotations associated with different nodes (datasets and other information)
Tree style and colors
iTOL 3 supports individual styles and colors for each node and label in the tree. Color handling was significantly improved, and now fully supports the use of the alpha channel (enabling transparency). Branches can be visualized as solid or dashed lines of varying widths. The new version supports the display of different fonts and font styles, with each label's size, color and style individually adjustable. Label content can also be interactively changed directly on the tree, together with all other parameters.
Displaying support values
Clade support values (bootstraps) offer important clues regarding the quality of the tree and allow simple identification of problematic areas. iTOL 3 increases the display variety of bootstrap visualization, which can now can be classified into four options:
Direct display of numeric values above the branches in the tree, with adjustable font style and size
Display of symbols whose size correlates with the bootstrap values. Four different symbol types are available, and their colors and sizes are fully adjustable.
Coloring of tree branches using user-specified two- or three-color gradients, with actual colors calculated from the associated bootstrap value
Changing the branch display width according to the underlying bootstrap value. Widths associated with minimum and maximum values are fully adjustable.
Internal tree scales
iTOL 3 introduces a novel feature allowing users to display custom internal tree scales with adjustable colors and levels tied to different branch length values (Figure 1). These allow easy estimation of distances between various clades, and can be particularly useful if branch lengths are calibrated to correspond to known evolutionary timespans.
Incorporation of external datasets
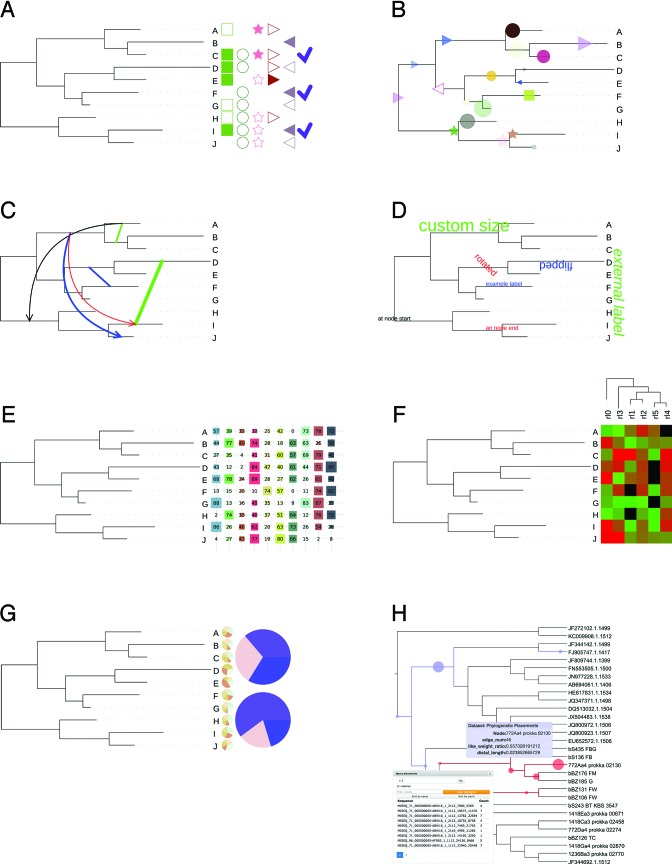
At the moment, iTOL supports 14 different dataset types (Figure 2). All datasets are uploaded as plain text files, and use simple predefined templates which are available through iTOL's help pages. As opposed to previous iTOL versions, most dataset parameters do not have to be specified during upload, and can be interactively adjusted through the web interface. The new version of iTOL removes any restrictions on the number of datasets to be displayed. Datasets are automatically separated, allowing simultaneous visualization of many different sets which can be manually adjusted directly on the tree. Detailed description and information on required formats for each dataset is available in iTOL's online help pages, together with example data provided for testing.
Figure 2.
New and significantly changed dataset types in iTOL version 3. Each tree in iTOL can be annotated with an unlimited number of datasets. Dataset files are drag-dropped onto the trees displayed in the browser, and visualized automatically. (A) Binary: multiple columns are now supported, with different colors and shapes. (B) Symbols: Various shapes of user-defined size and color can be displayed anywhere along tree branches. (C) Connections: supported in any tree display mode, they are visualized as straight or curved links between any two nodes, with or without arrows heads. (D) Text labels: any text of individually adjustable size, color, style, and rotation angle can be positioned anywhere along branches, or on the outside of the tree. (E) Shape plots: multiple values associated with nodes are displayed as geometric shapes of different sizes in consecutive columns outside the tree, optionally with value labels. (F) Heatmap: an additional tree can be displayed above the heatmap, and will be used to automatically sort the data columns. (G) External pie charts: pie charts can now also be displayed outside the tree, with adjustable sizes and positions. (H) Phylogenetic placements: generated automatically from user uploaded .jplace files (11) created by EPA (12) or pplacer (13). Placements can be visualized individually or as clade summaries, and queried through an integrated search engine.
USER ACCOUNTS
iTOLs freely available personal account system is being heavily used, with currently more than 10’000 users who stored more than half a million uploaded trees. The current version increases the support for this access modus, with a complete redesign of user account pages, making them more responsive and easier to use in modern browsers. A simple search engine has been implemented, allowing users to quickly access any of their trees from any iTOL page.
DOCUMENTATION AND VIDEO TUTORIALS
iTOL version 3 includes updated documentation, in which the novel features are explained in detail. The creation of user tree annotations is now much simpler because of the predefined template files. All templates include individual documentation and explanation of available features for various annotation types. In addition, an example tree with a set of predefined data files has been designed to illustrate and to explore the various annotation possibilities.
To facilitate the use of iTOL and to help users get going quickly, several tutorial videos have been developed that complement the static help pages. Thus, it should be easy for new users to get an overview of different functions and to understand the tree annotation process in detail.
Export
One of iTOL's main goals is the creation of high quality figures for publication or inclusion into other documents. The current version includes a complete WYSIWYG (What-You-See-Is-What-You-Get) export functionality, allowing users to replicate exactly what is displayed on their screen and export it into various graphical formats, both as vector graphics and as bitmaps. Currently supported formats are Scalable Vector Graphics (svg), Portable Network Graphics (png), Encapsulated Postscript (eps), Postscript (ps) and Portable Document Format (pdf). Trees can be also exported in all supported field-specific plain text formats (e.g. Newick, Nexus or phyloXML). In addition, interactively defined colors and styles associated with a tree can be exported as plain text annotation files, which can be re-used to automatically apply the same annotations to other trees.
Taken together, iTOL version 3 adds a number of frequently user requested features, incorporates full support for new browser generations and streamlines the tree annotation process, enabling the handling of larger and more complex datasets.
Funding
Funding for open access charge: Biobyte Solutions GmbH; European Molecular Biology Laboratory; de.NBI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Huson D.H., Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 2.Chevenet F., Brun C., Banuls A.L., Jacq B., Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zmasek C.M., Eddy S.R. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 4.Procter J.B., Thompson J., Letunic I., Creevey C., Jossinet F., Barton G.J. Visualization of multiple alignments, phylogenies and gene family evolution. Nat Methods. 2010;7:S16–S25. doi: 10.1038/nmeth.1434. [DOI] [PubMed] [Google Scholar]
- 5.Letunic I., Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 6.Jordan G.E., Piel W.H. PhyloWidget: web-based visualizations for the tree of life. Bioinformatics. 2008;24:1641–1642. doi: 10.1093/bioinformatics/btn235. [DOI] [PubMed] [Google Scholar]
- 7.Huerta-Cepas J., Dopazo J., Gabaldon T. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asnicar F., Weingart G., Tickle T.L., Huttenhower C., Segata N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilgenbusch J.C., Swofford D. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinformatics. 2003 doi: 10.1002/0471250953.bi0604s00. Chapter 6, Unit 6.4. [DOI] [PubMed] [Google Scholar]
- 10.Han M.V., Zmasek C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsen F.A., Hoffman N.G., Gallagher A., Stamatakis A. A format for phylogenetic placements. PLoS One. 2012;7:e31009. doi: 10.1371/journal.pone.0031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger S.A., Krompass D., Stamatakis A. Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 2011;60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsen F.A., Kodner R.B., Armbrust E.V. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letunic I., Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]