Abstract
Identification of genomic regions associated with a phenotype of interest is a fundamental step toward solving questions in biology and improving industrial research. Bulk segregant analysis (BSA) combined with high-throughput sequencing is a technique to efficiently identify these genomic regions associated with a trait of interest. However, distinguishing true from spuriously linked genomic regions and accurately delineating the genomic positions of these truly linked regions requires the use of complex statistical models currently implemented in software tools that are generally difficult to operate for non-expert users. To facilitate the exploration and analysis of data generated by bulked segregant analysis, we present EXPLoRA-web, a web service wrapped around our previously published algorithm EXPLoRA, which exploits linkage disequilibrium to increase the power and accuracy of quantitative trait loci identification in BSA analysis. EXPLoRA-web provides a user friendly interface that enables easy data upload and parallel processing of different parameter configurations. Results are provided graphically and as BED file and/or text file and the input is expected in widely used formats, enabling straightforward BSA data analysis. The web server is available at http://bioinformatics.intec.ugent.be/explora-web/.
INTRODUCTION
Bulk segregant analysis (BSA) coupled with high-throughput sequencing is a widely used method to associate genomic regions to complex traits (1,2). Identifying these regions, commonly known as quantitative trait loci (QTL), is a fundamental step toward understanding the genomic component of phenotypic variation (3–6). BSA relies on crossing two parents which differ in a trait of interest, having one parent with a desired selectable trait. Segregants displaying the desired phenotype are pooled, the pooled DNA is extracted and subjected to pool sequencing. The sequencing data is mapped to a reference strain (usually the known parent not exhibiting the desired phenotype) and the allele frequency is calculated for all polymorphic marker sites. Genomic regions for which the allele frequency of the marker sites significantly deviates from random segregation are prioritized as QTL for the trait (7,8). Current availability of whole genome sequencing (WGS) provides the capacity to infer population allele frequencies for nearly every site that is polymorphic between the parental lines. Hence, the quality and length of the identified QTL is only affected by the number of crossovers during the development of the population which on its turn is determined by the recombination rate (9,10). BSA has been particularly useful in identifying trait-linked alleles in eukaryotes with small genomes such as yeast, mainly because the smaller number of base pairs per centimorgan reduces the number of candidate genes within each predicted QTL (11–13). However, this method has also been successfully applied in rice (14,15) and other plants (16,17).
To efficiently differentiate true from spuriously linking QTLs, several advanced BSA data analysis protocols have been proposed (1,4,18–20). Previous work has shown that one of the approaches that largely contributes to the power of BSA data analysis is to exploit the properties of linkage disequilibrium (LD) (10,21,22). The restricted number of recombination events cause proximal marker sites to be co-inherited, resulting in LD between neighboring marker sites: in a BSA set up, a causative mutation will therefore be embedded in a larger region of marker sites for which the variant counts all display a similar deviation from the allelic distribution one would expect if loci were not segregating with the trait. LD thus produces deviations of variant counts toward the alleles of the parent with the desired phenotype, not only at the genetic marker site(s) causative to the phenotype of interest, but also in genetic marker sites closely located to these causative marker sites. Although different algorithms for BSA exploiting LD have recently been implemented (10,21,22) in open source scripts and software tools, these tools remain difficult to operate for users.
Therefore we present EXPLoRA-web, an intuitive web server that facilitates users performing data analysis of BSA experiments. EXPLoRA-web is wrapped around our previously published BSA data analysis method that was shown to maximize power and accuracy in detecting QTLs by exploiting the properties of LD. The details of the Hidden Markov Model (HMM) and benchmarking with other tools are discussed in (10). The web service is available at http://bioinformatics.intec.ugent.be/explora-web/.
EXPLoRA WEB
EXPLoRA web is accessible using any internet browser (Google Chrome, Microsoft Edge and Firefox, among others). The web server's help pages http://bioinformatics.intec.ugent.be/explora-web/help provide detailed guidelines on how to perform the analysis, tune the parameters and interpret the results. The EXPLoRA web server is freely accessible and does not require login, although an optional account can be created to have easy access to the results of previously analyzed experiments.
EXPLoRA, which is the algorithm implemented in this web service, navigates the variable sites of the genome using an HMM that calculates the probability of allele frequencies at each marker site to be emitted by two possible states: a phenotype-linked state and a neutral state. While markers linked to the phenotype are expected to show predominantly the allele of the parent with the desired phenotype, neutral markers are expected to show the alleles of the two parents at a ratio that reflects random segregation.
The effect of LD is modeled by the transition probabilities between two neighboring marker sites. The transition probability models the chance of a change of state between two neighbor sites. Its distribution is described by a negative exponential function of the recombination rate and the physical distance between neighboring marker sites following Haldane's recombination model (23). The model captures the fact that neighboring marker sites are likely to be in LD and hence the probability of a state change between them is small.
Emission probabilities of marker site states are modeled by two β binomial distributions, one for the emission probabilities in phenotype linked states and another for emission probabilities in neutral states. The β-distribution for the neutral states is automatically estimated from the read count data. The distribution describing the expected frequencies of the phenotype-linked variants is defined by the user selecting the α and β parameters. The ratio between α and β defines the degree to which the relative variant frequency at a marker site needs to differ from the one expected based on random segregation in order to be called ‘linked to the phenotype’.
For full description of the model and algorithmic details we refer to (10).
Input
The data necessary to run EXPLoRA consists of the information on the experimental setup, the allele counts from the pooled sequencing and the selection of the method's parameters.
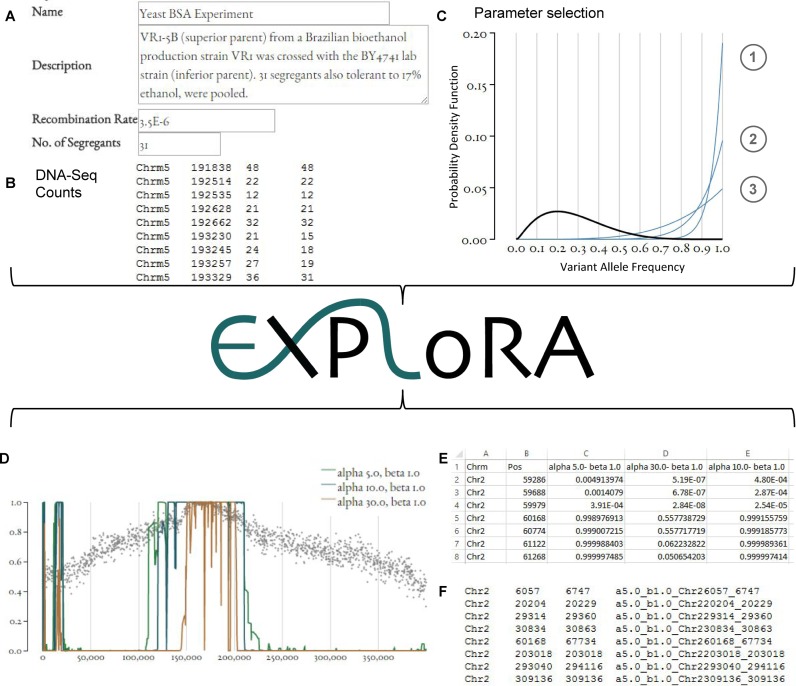
Information related to the experimental setup consists of the number of segregrants that were pooled prior to sequencing and an approximation of the average recombination rate of the organism under study. The latter is required to take into account the effect of LD between neighboring markers. Although we realize recombination rates are not constant across a genome, the average recombination works as a good approximation for our model: as we have shown in our previous work, small deviations from the recombination rate do not interfere with the performance of the algorithm in accurately detecting truly linked regions (10). Additional fields to name and describe the experiment are also provided (Figure 1A).
Figure 1.
Overview of the web service. (A) Input experimental information. (B) Upload count data. (C) Parameter selection. The black line corresponds to the cumulative distribution of allele frequencies (alternative read count/total read count) derived from the uploaded data and is used to estimate the probability distribution that models the emission probability of the neutral state allele distribution. The blue lines represent the β-distributions model for each of the three different ratios of α/β where line 1 corresponds to a setting reflecting high specificity and low sensitivity, line 2 medium specificity and sensitivity and line 3 low specificity and high sensitivity. (D) Visual Output. The X-axis corresponds to the chromosomal positions and the Y-axis to the posterior probabilities obtained for each marker site. (E) Posterior distributions of the marker sites for each parameter setting. (F) BED file indicating the regions linked to the phenotype.
The allele count at the marker sites derived from the pooled DNA sequencing should be uploaded as a variant call format (VCF) file or as a simple tab delimited file in which rows correspond to the different marker sites (Figure 1B). For the simple text tab delimited file the columns correspond respectively to the chromosome at which the marker sites are located (marker chromosome), the genomic position of the marker sites (marker site position) and two columns describing the read counts containing respectively the total read count at a marker site and the alternative allele read count (which usually corresponds to the allele of the parent with the desired phenotype). VCF files containing these allele counts are produced from raw reads by analysis pipelines for high-throughput sequencing data such as NGSEP (24) and GATK (25), so the outputs of these pipelines can be directly uploaded to EXPLoRA-web. Alleles refer to polymorphisms relative to a reference genome and are thus defined when generating the VCF or counts file prior to using EXPLoRA.
Besides the data and experimental information, EXPLoRA also requires specifying the parameters that control the model. The main parameter to be selected is the α/β ratio that determines the shape of the β distribution that models the emission probability for the phenotype-linked states (Figure 1C). Changing the α/β ratio affects the probability with which an observed relative variant allele frequency is interpreted by the model as representative for a region linked to the trait of interest. Increasing the α/β ratio makes the identification of phenotype-linked regions more stringent, meaning that a higher deviation of the relative allele frequency from the one expected under random segregation is needed before the region is considered linked. The higher the α/β ratio, the less phenotype-linked markers are called and the smaller the size of the called regions: identifying only the most pronouncedly linked regions results in more precisely pinpointing the linked region. However, this comes at the expense of potentially missing some truly linked markers/regions (lower sensitivity). By default, the web server proposes three different α/β ratio's corresponding to different trade-offs between sensitivity and specificity. A region that is identified with a stringent threshold and that thus corresponds to the most reliable signal in the data will by definition also be identified by the less stringent α/β ratio. Providing the results obtained with different thresholds thus allows the user to assess the reliability of the predictions based on the stringency of the threshold at which the linked region was first detected. The effect of changes in the α/β ratio can be observed visually in the graphical interface.
The emission probability of markers to be in the neutral state (i.e. not linked to the trait of interest) is directly estimated from the count data uploaded by the user, hereby assuming that most of the markers are not linked to the phenotype and their count data thus display the allele frequency distribution expected under random segregation. The web server also displays the distribution inferred from the real counts, allowing the user to select a set of running parameters in agreement with the data to be analyzed.
Output
The EXPLoRA web service determines per α/β ratio the posterior probability of each marker site to be linked to the phenotype of interest. Because of LD, neighboring markers on the chromosome will together display either high or low posterior probabilities. Consecutive sets of neighboring markers displaying high posterior probabilities are thus used to identify the QTL. The server displays the marker-specific posterior probability scores graphically (Figure 1D) along the chromosome for each α/β combination together with the observed allele frequencies at the variant sites and accordingly derives the genomic regions covered by the identified phenotype-linked markers. Notice that when parameter settings are chosen too stringent the resulting posterior probabilities of finding a site being linked to the phenotype will become zero for all marker sites and no results can be found. Results can be downloaded in two formats: (i) as a comma separated file, listing the posterior distributions per marker and parameter setting, and (ii) as a BED file containing the genomic information to identify the regions found to be linked to the phenotype. The BED file can be imported into other tools such as the Integrative Genomic Viewer (26).
Implementation
The web service was implemented in Java and connects to a MySQL database for information storage. The software was built using the model view controller pattern implemented via Tapestry 5.2. The access to the database was achieved using Hibernate for object-relational mapping. The graphical interface was made using bootstrap, jQuery and D3js. The server runs on a 16-core, 64bit CentOS 6.2 system with 128GB of memory.
DISCUSSION
We implemented EXPLoRA web, a novel web service for the identification of QTL from whole genome resequencing data in BSA experiments. The web server relies on a highly accurate model developed previously (10) that maximizes the power to identify promising candidate genomic regions for further study of causal relationships with complex traits. EXPLoRA was compared and shown to have superior accuracy to other recently published command line executable methods for QTL analysis on BSA experiments (18,21). The use of standard formats such as the VCF allows users to upload directly the outputs of software pipelines to obtain allele counts and genotype calls from WGS data such as NGSEP (24) or GATK (25) as inputs for QTL analysis. With EXPLoRA we anticipate on the increasing use of BSA in combination with pooled sequencing both for fundamental and applied purposes and on the concomitant need for user friendly applications to facilitate its complex data analysis activities.
FUNDING
Ghent University Multidisciplinary Research Partnership ‘Bioinformatics: from nucleotides to networks’; Fonds Wetenschappelijk Onderzoek-Vlaanderen (FWO) [G.0329.09, 3G042813, G.0A53.15N]; Agentschap voor Innovatie door Wetenschap en Technologie (IWT) [NEMOA]; Katholieke Universiteit Leuven [PF/10/010] (NATAR); International Center for Tropical Agriculture (CIAT). Funding for open access charge: Ghent University Multidisciplinary Research Partnership ‘Bioinformatics: from nucleotides to networks’; Fonds Wetenschappelijk Onderzoek-Vlaanderen (FWO) [G.0329.09, 3G042813, G.0A53.15N].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ehrenreich I.M., Torabi N., Jia Y., Kent J., Martis S., Shapiro J.A., Gresham D., Caudy A.A., Kruglyak L. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneeberger K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014;15:662–676. doi: 10.1038/nrg3745. [DOI] [PubMed] [Google Scholar]
- 3.Steinmetz L.M., Sinha H., Richards D.R., Spiegelman J.I., Oefner P.J., McCusker J.H., Davis R.W. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 4.Swinnen S., Schaerlaekens K., Pais T., Claesen J., Hubmann G., Yang Y., Demeke M., Foulquié-Moreno M.R., Goovaerts A., Souvereyns K., et al. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 2012;22:975–984. doi: 10.1101/gr.131698.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liti G., Louis E.J. Advances in quantitative trait analysis in yeast. PLoS Genet. 2012;8:e1002912. doi: 10.1371/journal.pgen.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkening S., Lin G., Fritsch E.S., Tekkedil M.M., Anders S., Kuehn R., Nguyen M., Aiyar R.S., Proctor M., Sakhanenko N.A., et al. An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae. Genetics. 2014;196:853–865. doi: 10.1534/genetics.113.160291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michelmore R.W., Paran I., Kesseli R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winzeler E.A., Richards D.R., Conway A.R., Goldstein A.L., Kalman S., McCullough M.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., et al. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 9.Parts L., Cubillos F.A., Warringer J., Jain K., Salinas F., Bumpstead S.J., Molin M., Zia A., Simpson J.T., Quail M.A., et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011;21:1131–1138. doi: 10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duitama J., Sánchez-Rodríguez A., Goovaerts A., Pulido-Tamayo S., Hubmann G., Foulquié-Moreno M.R., Thevelein J.M., Verstrepen K.J., Marchal K. Improved linkage analysis of Quantitative Trait Loci using bulk segregants unveils a novel determinant of high ethanol tolerance in yeast. BMC Genomics. 2014;15:1–15. doi: 10.1186/1471-2164-15-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijnen J.-P., Randazzo P., Foulquié-Moreno M.R., van den Brink J., Vandecruys P., Stojiljkovic M., Dumortier F., Zalar P., Boekhout T., Gunde-Cimerman N., et al. Polygenic analysis and targeted improvement of the complex trait of high acetic acid tolerance in the yeast Saccharomyces cerevisiae. Biotechnol. Biofuels. 2016;9:1–18. doi: 10.1186/s13068-015-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pais T.M., Foulquié-Moreno M.R., Hubmann G., Duitama J., Swinnen S., Goovaerts A., Yang Y., Dumortier F., Thevelein J.M. Comparative polygenic analysis of maximal ethanol accumulation capacity and tolerance to high ethanol levels of cell proliferation in yeast. PLoS Genet. 2013;9:e1003548. doi: 10.1371/journal.pgen.1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pomraning K.R., Smith K.M., Freitag M. Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Eukaryot. Cell. 2011;10:724–733. doi: 10.1128/EC.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagi H., Tamiru M., Abe A., Yoshida K., Uemura A., Yaegashi H., Obara T., Oikawa K., Utsushi H., Kanzaki E., et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015;33:445–449. doi: 10.1038/nbt.3188. [DOI] [PubMed] [Google Scholar]
- 15.Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., Matsumura H., Yoshida K., Mitsuoka C., Tamiru M., et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012;30:174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- 16.Steuernagel B., Taudien S., Gundlach H., Seidel M., Ariyadasa R., Schulte D., Petzold A., Felder M., Graner A., Scholz U., et al. De novo 454 sequencing of barcoded BAC pools for comprehensive gene survey and genome analysis in the complex genome of barley. BMC Genomics. 2009;10:1–15. doi: 10.1186/1471-2164-10-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trick M., Adamski N.M., Mugford S.G., Jiang C.-C., Febrer M., Uauy C. Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 2012;12:1–17. doi: 10.1186/1471-2229-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magwene P.M., Willis J.H., Kelly J.K. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput. Biol. 2011;7:e1002255. doi: 10.1371/journal.pcbi.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun H., Schneeberger K. SHOREmap v3.0: fast and accurate identification of causal mutations from forward genetic screens. Methods Mol. Biol. 2015;1284:381–395. doi: 10.1007/978-1-4939-2444-8_19. [DOI] [PubMed] [Google Scholar]
- 20.Austin R.S., Vidaurre D., Stamatiou G., Breit R., Provart N.J., Bonetta D., Zhang J., Fung P., Gong Y., Wang P.W., et al. Next-generation mapping of Arabidopsis genes. Plant J. 2011;67:715–725. doi: 10.1111/j.1365-313X.2011.04619.x. [DOI] [PubMed] [Google Scholar]
- 21.Leshchiner I., Alexa K., Kelsey P., Adzhubei I., Austin-Tse C.A., Cooney J.D., Anderson H., King M.J., Stottmann R.W., Garnaas M.K., et al. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22:1541–1548. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards M.D., Gifford D.K. High-resolution genetic mapping with pooled sequencing. BMC Bioinformatics. 2012;13:1–11. doi: 10.1186/1471-2105-13-S6-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haldane J. The combination of linkage values, and the calculation of distance between the loci of linked factors. J. Genet. 1919;8:299–309. [Google Scholar]
- 24.Duitama J., Quintero J.C., Cruz D.F., Quintero C., Hubmann G., Foulquié-Moreno M.R., Verstrepen K.J., Thevelein J.M., Tohme J. An integrated framework for discovery and genotyping of genomic variants from high-throughput sequencing experiments. Nucleic Acids Res. 2014;42:e44. doi: 10.1093/nar/gkt1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;11:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]