Abstract
The PSI/TM-Coffee web server performs multiple sequence alignment (MSA) of proteins by combining homology extension with a consistency based alignment approach. Homology extension is performed with Position Specific Iterative (PSI) BLAST searches against a choice of redundant and non-redundant databases. The main novelty of this server is to allow databases of reduced complexity to rapidly perform homology extension. This server also gives the possibility to use transmembrane proteins (TMPs) reference databases to allow even faster homology extension on this important category of proteins. Aside from an MSA, the server also outputs topological prediction of TMPs using the HMMTOP algorithm. Previous benchmarking of the method has shown this approach outperforms the most accurate alignment methods such as MSAProbs, Kalign, PROMALS, MAFFT, ProbCons and PRALINE™. The web server is available at http://tcoffee.crg.cat/tmcoffee.
INTRODUCTION
Building accurate multiple sequence alignments (MSAs) is one of the cornerstones of computational biology, as judged by the number of citations collected by MSA publications over the last 30 years (1). One of the most important developments in MSA methods over these last 10 years has been the introduction of the concept of template-based MSAs. Template-based MSAs make it possible to enrich the sequences one wishes to align with selected information. The first reports of template-based alignments describe the use of structural templates associated with the sequences of interest (2,3). Consistency-based methods provide an ideal framework for such template based analysis and over the years, novel data sources have been turned into sequence templates, including RNA secondary structures (4) and profiles (5) using a procedure known as homology extension (6). Although several reports suggest a significant increase in accuracy when using homology extension, this approach has seen its systematic usage limited by an often prohibitive computational cost and relatively complex deployment that requires tight integration between the aligner, BLAST and the databases.
We introduce here a new web server designed to provide users with a very efficient implementation of homology extension. This server features the T-Coffee homology extension procedure for both regular proteins (Position Specific Iterative T-Coffee, PSI-Coffee) (7) and transmembrane proteins (TM-Coffee) (8). Homology extension strategies use database searches to collect homologous sequences and generate a representative sequence profile for each input sequence. Relative to the individual sequences the profiles are enriched with evolutionary information, thus allowing informative position-specific scoring schemes to be derived for each sequence. The subsequent alignment of profiles provides more accurate alignments that capture specific conservation patterns. These patterns often reflect the structural and functional constraints of a protein family. Therefore, the use of profiles can achieve reasonable accurate alignments for highly divergent sequences (i.e. BAliBASE RV11 data set < 25% sequence identity) without the use of structural information (7). The notion of homology extension was initially pioneered in PROMALS-3D (9) and was also developed in a slightly different way in the PRALINE series (9,10). None of these associated analyses did, however, explore the consequence of database redundancy levels when doing homology extension and the first analysis of this type was published in (8) showing that low redundancy databases allow very fast homology extension at a limited cost in terms of accuracy. For transmembrane proteins (TMPs), TM-Coffee takes this approach further by using reduced databases containing only TMPs. The databases available for homology searching within the web server range from UniRef50 (fast/rough) to the full UniRef100 non-redundant set (slow/accurate). To the best of our knowledge this is the only web server offering users the choice of the databases against which homology extension is to be carried out.
The web server addresses an important need in the community by providing an accessible method for performing homology extension-based alignments. Our benchmarking on the BAliBASE2-ref7 α-helical TMPs shows a significant improvement over the most accurate methods (8). In these benchmark reports, we show that our approach outperforms the most accurate alternative procedures (MSAProbs, Kalign, PROMALS, MAFFT, ProbCons and PRALINE™) while carrying out significantly faster homology extension thanks to the use of non-redundant databases.
By using reduced databases containing only transmembrane annotated proteins, TM-Coffee is a specific alignment procedure for TMPs that obtains similar results at a significantly reduced computational cost over full protein databases. TM-Coffee also incorporates transmembrane topology prediction using generated BLAST profiles as input to the HMMTOP package (Hidden Markov Model for TOpolgy Prediction) (11). HMMTOP utilizes a previously trained hidden markov model to classify the amino acid distribution in localized protein segments (12). TM-Coffee maps these predictions back to the input sequences. Both the homology extension and transmembrane topology prediction are implemented in the web server, as well as in the T-Coffee package (13). The HMMTOP predictions do not contribute to the alignment procedure and are merely a post-processing prediction meant to yield a more informative output.
ALGORITHM
PSI/TM-Coffee incorporates homology information into T-Coffee library construction. In T-Coffee, a library is a collection of paired residues, as obtained in an all-against-all comparison of all the sequences within the dataset. This library is used to derive a position-specific scoring scheme through a process known as extension during which the cost for matching two residues is set by combining the score of all the residue pairs supporting a triplet linking these two residues (i.e. pair X-Z and pair Z-Y to support matching X-Y where X, Y and Z are three residues from three different sequences). Of course, the higher the accuracy of the pairwise alignments used to populate the library, the higher the expected accuracy. In this respect, the main strength of T-Coffee is its capacity to incorporate pairs from various sources. In PSI/TM-Coffee these pairs are obtained through the profile comparison we described in the next paragraph and therefore are more likely to be correct. The extended library scoring scheme is used by T-Coffee in the same way as a standard scoring scheme used in a progressive alignment while sequences are being incorporated following the guide tree order.
In order to achieve this, the first computational step is homology extension where individual sequences are replaced with a set of multiply aligned homologs. The purpose of homology extension is to reveal the evolutionary variability associated with each site of the considered sequences thus producing a more accurate T-Coffee library for the alignment stage. This procedure involves performing BLAST (blast+ version 2.2.25) for each query sequence against the selected database. The default database is UniRef50, a non-redundant database derived from UniProt where no two sequences have an identity >50%. For transmembrane inputs, UniRefXX-TM is an even smaller database produced by filtering the corresponding UniRef dataset with the query string: ‘keyword:transmembrane’. These TMP-specific databases are typically 80% smaller than the database they are extracted from. The server uses static databases (UniProt release 2011_02), so as to insure stability with respect to the original TM-Coffee publication (8). At this point, it is not entirely clear how regular updates should be so as to yield a positive impact on accuracy. Such update would also impact reproducibility. We therefore intend to keep this database static, using it in a way somehow similar to a substitution matrix (i.e. PAM or BLOSUM matrices are not updated on larger datasets). We nonetheless intend to monitor accuracy variation with newer database versions and will eventually proceed to regular updates if this proves suitable.
BLAST hits with an identity level between 50 and 90% and a coverage higher than 70% with respect to the query sequence are kept. The BLAST alignments of these hits are then stacked onto the query sequence, thus generating a one to all MSA. This MSA is eventually turned into a profile (*.prf, ‘Template Profile’ in the ‘Result files’ section) by removing all columns corresponding to positions unaligned to the query (i.e. gaps in the query) and by filling unmatched query positions with gaps.
A T-Coffee library is then produced by aligning every pair of profiles with a pair-HMM adapted from the ProbCons method (14) in order to deal with profiles. The parameters and biphasic gap penalties (two distinct sets of gap opening and extension penalties for short and longer gaps) are the same as described by the ProbCons authors. Every pair of matched columns with a posterior probability of being aligned higher than 0.99 are included into the library. In this procedure, the matching cost of a pair of profile sequences is set to the average cost measured on the vectorized columns, as described in (15).
This library of potential matching pairs across the considered sequences is eventually fed to the T-Coffee consistency progressive alignment algorithm. T-Coffee uses its standard heuristic approach in an attempt to deliver an MSA having the highest possible level of consistency with the library. When the TMP toggle is checked, the transmembrane topology of each sequence will be predicted by HMMTOP using its associated profile (*.tmp, ‘Template Protein Transmembrane Topology’ in the ‘Result files’ section). This extra prediction procedure is only used for display purposes (see next ‘Output’ section) and is not required by the alignment procedure.
PSI/TM-COFFEE WEB SERVER
The PSI/TM-Coffee web server is part of the T-Coffee web platform; its access is free and unrestricted, without login procedure. The server is accessible from http://tcoffee.crg.cat/tmcoffee with any standard web browser (Mozilla Firefox 5+, Google Chrome, Internet Explorer 8+, Safari 6+ and Opera 11+). All the functions of the server are also available through the command line version of the T-Coffee available at http://tcoffee.crg.cat/. Detailed documentation for the T-Coffee package is available at www.tcoffee.org (http://www.tcoffee.org/Projects/tcoffee/#DOCUMENTATION).
Input
The server receives as input sequences in FASTA format. Sequences can be input via the input field or alternatively provided using the upload link. The sequence size limit is 1000 sequences and the sequence length limit is 5000. The homology extension is determined by the homology search options. By default the transmembrane sequence type toggle is off and the database is UniRef50. When the transmembrane toggle button is checked, the source databases will be further reduced by only keeping transmembrane-annotated entries. These TMP-specific databases are 80% smaller than their sources (8). Besides performing alignment, the prediction of the transmembrane topology will be conducted for each sequence. The output will be tm_html instead of score_html (see more details in the ‘Output’ interpretation section). The homology extension databases available are:
UniRef50, very fast, rough (default): built by clustering UniRef100 sequences at the 50% sequence identity level (16).
UniRef90, fast, approximate: built by clustering UniRef100 sequences at the 90% sequence identity level.
UniRef100, slow, accurate: combines identical sequences and sub-fragments from any source organism into a single UniRef entry (i.e. cluster).
Advanced output options are also available as explained in the ‘Output’ section below. Users can provide an email address to be notified when the job is complete.
Computation
The server performs a BLAST search for each query sequence against the selected database and turns the BLAST outputs into a profile that is used to produce the T-Coffee library. Computation time is highly dependent on the number of sequences and the chosen homology extension database. For regular proteins, aligning 10 sequences using UniRef50 takes ∼5 min on the web server whilst 200 sequences with the same database takes ∼2 h (Table 1). In comparison, aligning the same 10 sequences using the full UniRef 100 database takes ∼20 min (Table 2). When the TMP toggle is selected, the profile is utilized to predict TMPs topology by HMMTOP. Whilst the database BLAST step is more efficient using the reduced transmembrane only databases, the topology prediction step adds to the computation cost. During and after the computation, job details and results can be retrieved using the permanent URL http://tcoffee.crg.cat/apps/tcoffee/result?rid=jobid, where jobid is the ID given at the time of submission. The server history is also kept in a cookie on the user's browser enabling it to be accessed at any moment using the History link on the main menu.
Table 1. Running time (in seconds) of PSI/TM-Coffee on the web server (default mode, UniRef50 for homology extension) as a function of the number of sequences in the input dataset (sequences randomly extracted from the PFAM family PF00001, corresponding to 7-transmembrane receptors).
| Number of sequences | |||||||
|---|---|---|---|---|---|---|---|
| 10 | 20 | 30 | 40 | 50 | 100 | 200 | |
| Mode/Average length | 257 | 262 | 261 | 260 | 261 | 262 | 262 |
| PSI-Coffee | 354 | 714 | 1010 | 1330 | 1721 | 3780 | 8880 |
| TM-Coffee | 125 | 243 | 362 | 604 | 664 | 1573 | 4740 |
Table 2. Running time (in seconds) of PSI/TM-Coffee for 10 sequences (same dataset used in Table 1) as a function of the database used for the homology extension.
| Database | |||
|---|---|---|---|
| T-Coffee mode | UniRef50 | UniRef90 | UniRef100 |
| PSI-Coffee | 354 | 671 | 1109 |
| TM-Coffee | 125 | 189 | 349 |
Output
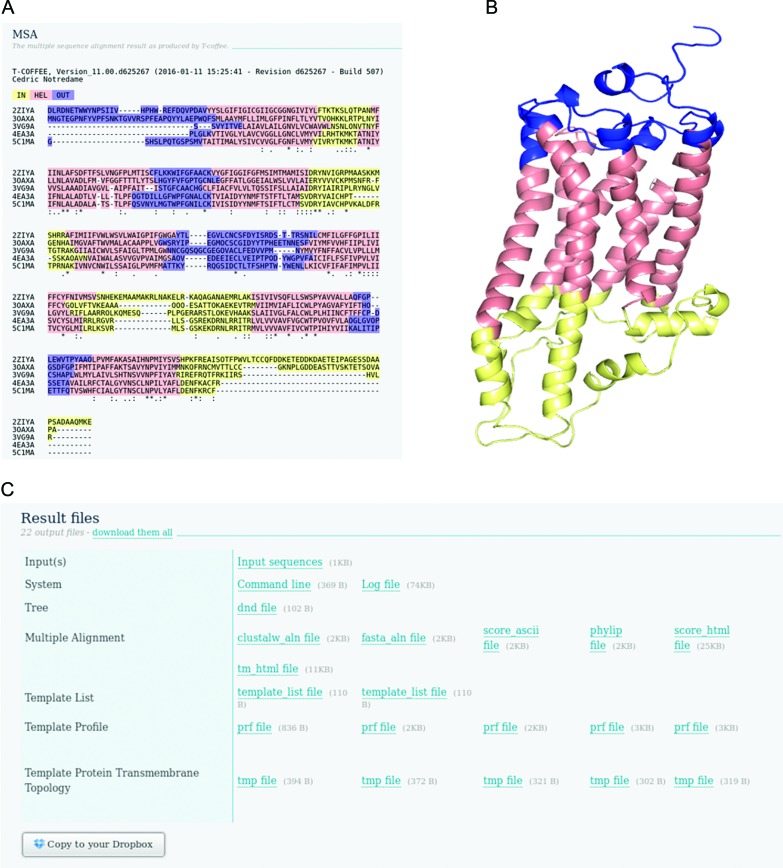
The final output page contains a graphical version of the MSA (Figure 1A), the raw result files, run parameters, citations and links for forwarding the results to other online tools. The format of outputs presented is dependent on whether the transmembrane mode was activated and any advanced output options.
Figure 1.
(A) The graphical MSA output coloured according to transmembrane topology prediction where yellow residues are predicted to be in the inner loop, red in the transmembrane helix and blue in the outer loop. (B) 3D structure of PDB ID: 2ZIY with the HMMTOP predicted transmembrane topology colouring. (C) The raw result files.
For regular proteins, the result page contains the following sections:
MSA: the MSA is coloured according to the T-Coffee TCS scheme (19). Dark pink blocks are very reliable, while blue and green bits are unreliable based on the T-Coffee library. The colour scheme has been designed to be easily visualized by colour-blind people.
Citation: the article for citing PSI/TM-Coffee.
- Result files:
- Tree is the guide tree used during the alignment procedure.
- Multiple Alignment subsection provides the MSAs in CLUSTAL, FASTA and Phylip formats. score_html is the html format of the above MSA section and its plain text format, score_ascii.
- Template List is the list of the homology profiles for the input sequences.
- Template Profile contains the homology profile of each sequence from the BLAST search procedure.
For TMPs the result page contains the following sections:
MSA: the MSA coloured according to transmembrane topology prediction by HMMTOP (11), where yellow is in loop, red is TM helix and blue is out loop (Figure 1A).
Citation: the article for citing PSI/TM-Coffee.
- Result files:
- Tree is the guide tree used during the alignment procedure.
- Multiple Alignment subsection provides the MSAs in CLUSTAL, FASTA and Phylip formats. score_html the html format of the T-Coffee TCS score and its plain text format, score_ascii. tm_html the html format of the above MSA section indicates the transmembrane topology prediction.
- Template List contains the list of homology profiles and topology predictions for the input sequences.
- Template Profile contains the homology profile of each sequence from the BLAST search procedure.
- Template Protein Transmembrane Topology is the transmembrane topology prediction of each sequence, where I is in loop, H is TM helix and O is out loop.
For both modes, all files (Figure 1C) can be downloaded as a single zip file, copied to the user's Dropbox account or forwarded to other online tools for downstream visualization and analysis. The info panel provides information such as elapsed time and a replay function allows re-running of the job with modifications to input options and data.
DISCUSSION
Here we describe the PSI/TM-Coffee web server, a tool for performing homology extension based MSAs. As far as we know, the web server is the only online tool allowing for the choice of different homology extension databases. The inclusion of specific transmembrane databases reduces the overall computational time and automated topology predictions give users a highly relevant and useful annotation (Figure 1B). Future updates will involve an on the fly computation of suitable NR databases using Pfam as a starting point to select the most suitable sequence sets.
FUNDING
Plan Nacional [BFU2011-28575 to C.N., P.D.]; Center for Genomic Regulation (CRG); ‘Fundació Obra Social la Caixa’ (to E.W.F, M.C); Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’ [SEV-2012–0208]; Center for Genomic Regulation (CRG). Funding for open access charge: The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Van Noorden R., Maher B., Nuzzo R. The top 100 papers. Nature. 2014;514:550–553. doi: 10.1038/514550a. [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan O., Suhre K., Abergel C., Higgins D.G., Notredame C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 3.Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilm A., Higgins D.G., Notredame C. R-Coffee: a method for multiple alignment of non-coding RNA. Nucleic Acids Res. 2008;36:e52. doi: 10.1093/nar/gkn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei J., Tang M., Grishin N.V. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:W30–W34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatzou M., Magis C., Chang J.-M., Kemena C., Bussotti G., Erb I., Notredame C. Multiple sequence alignment modeling: methods and applications. Brief. Bioinform. 2015;2015:bbv099. doi: 10.1093/bib/bbv099. [DOI] [PubMed] [Google Scholar]
- 7.Kemena C., Notredame C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics. 2009;25:2455–2465. doi: 10.1093/bioinformatics/btp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang J.-M., Di Tommaso P., Taly J.-F., Notredame C. Accurate multiple sequence alignment of transmembrane proteins with PSI-Coffee. BMC Bioinformatics. 2012;13(Suppl. 4):S1. doi: 10.1186/1471-2105-13-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pei J., Grishin N.V. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 10.Simossis V.A., Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tusnády G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 12.Tusnády G.E., Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 13.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 14.Do C.B., Mahabhashyam M.S.P., Brudno M., Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H., UniProt Consortium UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang J.-M., Di Tommaso P., Notredame C. TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol. Biol. Evol. 2014;31:1625–1637. doi: 10.1093/molbev/msu117. [DOI] [PubMed] [Google Scholar]