Abstract
RaptorX Property (http://raptorx2.uchicago.edu/StructurePropertyPred/predict/) is a web server predicting structure property of a protein sequence without using any templates. It outperforms other servers, especially for proteins without close homologs in PDB or with very sparse sequence profile (i.e. carries little evolutionary information). This server employs a powerful in-house deep learning model DeepCNF (Deep Convolutional Neural Fields) to predict secondary structure (SS), solvent accessibility (ACC) and disorder regions (DISO). DeepCNF not only models complex sequence–structure relationship by a deep hierarchical architecture, but also interdependency between adjacent property labels. Our experimental results show that, tested on CASP10, CASP11 and the other benchmarks, this server can obtain ∼84% Q3 accuracy for 3-state SS, ∼72% Q8 accuracy for 8-state SS, ∼66% Q3 accuracy for 3-state solvent accessibility, and ∼0.89 area under the ROC curve (AUC) for disorder prediction.
INTRODUCTION
The structure and function of a protein is determined partially by its local structural properties, such as 3/8-state protein secondary structure (SS3/SS8), solvent accessibility (ACC), and disordered regions (DISO) (1–3). Currently there are about 90 million sequences in TrEMBL (4), many of which do not have any structural and functional information. Since the experimental structure determination methods are laborious and expensive, as of January 2016 there are only ∼110 000 protein structures in PDB (5), and merely ∼37K of them are annotated in UniProt entries (6). Consequently, there is an urgent need for accurate and high-throughput methods that can predict protein structural properties from amino acid sequence alone, without using any template information (7).
However, the prediction accuracy of protein structural properties, while without exploiting experimentally-solved structures (i.e. templates), is still far away from satisfactory. Taking 3-state secondary structure prediction as an example, when template information is not used and only sequence profile is considered, so far the best Q3 accuracy is ∼80% obtained by a few predictors such as PSIPRED (8) and JPRED (7), which is significantly lower than the estimated prediction accuracy limit 88–90% (9). Such a gap motivates us to develop a better method to further improve SS prediction. A similar trend is observed on solvent accessibility prediction with three-state accuracy ∼60% obtained by SPINE-X (10) and SANN (11). To further increase prediction accuracy, we will need a more sophisticated method that can model the complex sequence-structure relationship in a much better way.
This paper presents RaptorX Property, a web server predicting protein structure property solely based on protein sequence or sequence profile. A profile is derived from multiple sequence alignment (MSA) of sequence homologs in a protein family (12). To predict structure properties, this server employs a new machine learning model DeepCNF (Deep Convolutional Neural Fields) (13), which embraces the advantages of both conditional neural fields (CNF) (14) and deep convolutional neural networks (DCNN) (15). This model captures not only complex sequence-structure relationship, but also models the property label correlation among adjacent residues. To deal with the imbalanced distribution of some property label, such as 8-state secondary structure and order/disorder, we train DeepCNF by maximizing area under the ROC curve (AUC), which is a good measure for class-imbalanced data (16). The experimental results show that our server greatly outperforms existing servers in protein structure property prediction.
The underlying reason why we develop an independent structure property prediction server instead of merging it with our 3D structure prediction server (17,18) is that structure property can be predicted much faster than 3D structure. By separating them, our server can quickly respond to those users who only want structure property prediction and this also makes it feasible to do genome-scale structure property prediction through our web server.
MATERIALS AND METHODS
Structure properties to be predicted
SS3/SS8
The 3- and 8-state protein secondary structure element is defined by DSSP (19). In particular, DSSP assigns three types for helix (G for 310 helix, H for alpha-helix, and I for pi-helix), two types for strand (E for beta-strand and B for beta-bridge) and three types for coil (T for beta-turn, S for high curvature loop, and L for irregular). The background distribution of the eight states H, E, L, T, S, G, B, I is around 34:21:20:11:9:4:1:0 (20). For 3-state secondary structure, we follow the definition in (21), i.e. E for beta-strand, H for alpha-helix and C for the others (i.e. G,I,B,T,S,L).
ACC
The relative solvent accessibility (RSA) for a given residue is defined as the absolute accessible surface area (calculated by DSSP) normalized by the maximum solvent accessibility of that residue (22). By RSA, we may have 3-state ACC: buried (B) with RSA from 0 to 10%, intermediate (I) with RSA from 10% to 40% and exposed (E) with RSA from 40% to 100%. The background distribution of B, I, E is around 1:1:1 (22).
DISO
Following the definition in (23), we label a residue as disordered (denoted as ‘*’) if it is in a segment of more than three residues missing atomic coordinates in the X-ray structure. The other residues are labeled as ordered (denoted as ‘.’). The background distribution of these two states is 94:6 (24).
Prediction algorithm
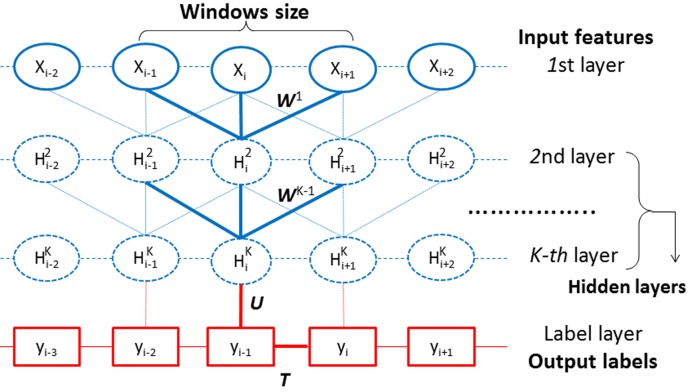
We use our in-house deep learning method DeepCNF to predict SS, ACC and DISO. Our technical details for SS prediction has been described in (13). We use a similar method to predict ACC and DISO. Here we briefly describe DeepCNF. As shown in Figure 1, DeepCNF has two modules: (i) the Conditional Random Fields (CRF) module consisting of the Kth layer and the label layer (shown in red), and (ii) the deep convolutional neural network (DCNN) module covering the input to the Kth layer (shown in blue). When only one hidden layer is used, DeepCNF becomes conditional neural fields (CNF) (14).
Figure 1.
Illustration of DeepCNF. Here, i is the position index and Xi the associated input features, Hk represents the kth hidden layer, and Y is the output label. All the layers from the 1st to the Kth form a DCNN with parameter Wk{k = 1,2,…,K}, which is shown in blue. The Kth layer and the label layer form a CRF (shown in red), in which the parameter U specifies the relationship between the Kth layer and the label layer, and T the binary relationship between adjacent labels.
To deal with the imbalanced distribution of some property label, we train DeepCNF by maximizing AUC. To fulfill this, we formulate AUC in a pairwise ranking framework, approximate it by a polynomial function and then apply a gradient-based procedure to optimize it.
See our supplemental file for DeepCNF details, the maximum-AUC training approach, and protein features.
RESULT
Servers to compare
For SS3/SS8 prediction, we compare our server with PSIPRED (8), JPRED (7) and SSpro (25). We tested PSIPRED with and without using sequence profile, denoted as PSIPREDp and PSIPREDa, respectively. JPRED is a profile-based method. SSpro supports 3-state and 8-state secondary structure prediction, and uses two prediction strategies: profile-based and template-based (i.e. using a solved structure in PDB as template). The former mode is denoted as SSprop and the latter as SSproT, respectively.
For ACC prediction, we compare our server with SOLVPRED (26), SPINE-X (10), SANN (11) and ACCpro (25). We test SOLVPRED without using sequence profile, denoted as SOLVPREDa. SPINE-X is a profile-based method, while SANN utilizes template information. Again, ACCpro has two prediction modes: ACCprop that utilizes profile information and ACCproT that employs template information. Since SOLVPRED, SPINE-X and ACCpro predict real-valued relative solvent accessibility, we use the same 10%/40% threshold as our ACC definition to re-label their output.
For DISO, we compare with IUpred (27) and DisoPred3 (28). IUpred does not use sequence profile. We use DisoPred3p and DisoPred3T to denote the profile-based and template-based prediction modes for DisoPred, respectively.
Performance on CASP and CAMEO
We tested RaptorX Property using 228 CASP test proteins (123 CASP10 and 105 CASP11 targets) and 179 CAMEO test proteins (from December 5, 2014 to May 29, 2015). Note that all these targets share <25% sequence identity with the training data CullPDB (29). See Supplemental for the definition of performance metric: Q3, Q8, Matthews correlation coefficient (Mcc), and AUC. In short, the Q3 (Q8) accuracy is defined as the percentage of residues for which the predicted labels are correct (13).
As shown in Table 1, when sequence profile is used, our server obtains ∼84% Q3 and ∼72% Q8 accuracy on CASP and CAMEO datasets, which significantly exceeds the others and breaks the long-lasting ∼80% Q3 accuracy barrier for 3-state secondary structure prediction. When sequence profile is not used, our server obtains ∼74% Q3 and ∼59% Q8 accuracy, respectively, much better than PSIPRED without using sequence profile.
Table 1. Q3/Q8 accuracy of secondary structure prediction on CASP and CAMEO targets.
| Methods | Q3 (%) | Q8 (%) | ||
|---|---|---|---|---|
| CASP | CAMEO | CASP | CAMEO | |
| PSIPREDa | 71.3 | 70.5 | – | – |
| PSIPREDp | 80.9 | 80.1 | – | – |
| JPREDp | 81.0 | 79.7 | – | – |
| SSprop | 78.0 | 77.5 | 65.3 | 63.5 |
| SSproT | 82.3 | 78.9 | 71.8 | 65.7 |
| RaptorX-Propertya | 74.4 | 73.2 | 60.2 | 58.6 |
| RaptorX-Propertyp | 84.6 | 84.4 | 72.0 | 72.1 |
Table 2 shows the result of the Q3 accuracy of 3-state solvent accessibility. Our server obtains ∼66% (∼57%) on CASP and CAMEO test proteins when sequence profile is used (not used). For disorder region prediction, our server obtains ∼0.89 (∼0.85) AUC when profile is used (not used), much better than their peers.
Table 2. Q3 accuracy of solvent accessibility prediction on CASP and CAMEO targets.
| Methods | Q3 (%) | |
|---|---|---|
| CASP | CAMEO | |
| SOLVPREDa | 49.7 | 49.0 |
| SPINE-Xp | 57.7 | 56.9 |
| SANNT | 61.6 | 60.7 |
| ACCprop | 57.8 | 57.2 |
| ACCproT | 60.1 | 58.6 |
| RaptorX-Propertya | 57.5 | 56.7 |
| RaptorX-Propertyp | 66.3 | 66.7 |
Table 3. Per-residue performance of disorder prediction on CASP and CAMEO targets.
| Methods | CASP | CAMEO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| bAcc | Sens | Spec | Mcc | AUC | bAcc | Sens | Spec | Mcc | AUC | |
| IUpreda | 0.66 | 0.36 | 0.95 | 0.33 | 0.68 | 0.69 | 0.46 | 0.93 | 0.37 | 0.79 |
| DisoPred3p | 0.67 | 0.36 | 0.98 | 0.47 | 0.84 | 0.70 | 0.46 | 0.95 | 0.42 | 0.83 |
| DisoPred3T | 0.68 | 0.39 | 0.99 | 0.54 | 0.85 | 0.70 | 0.43 | 0.96 | 0.44 | 0.83 |
| RaptorX-Propertya | 0.72 | 0.47 | 0.97 | 0.51 | 0.86 | 0.72 | 0.48 | 0.95 | 0.45 | 0.84 |
| RaptorX-Propertyp | 0.76 | 0.53 | 0.98 | 0.55 | 0.89 | 0.75 | 0.53 | 0.97 | 0.49 | 0.88 |
The evaluation criteria are balanced accuracy (bAcc), sensitivity (Sens), specificity (Spec), the Matthews correlation coefficient (Mcc) and area under the ROC curve (AUC).
In conclusion, when sequence profile is used, our server outperforms all the other methods, including those using profile and even template information. When profile is not used, our server also performs well, much better than those without using profile, especially for disorder region prediction. Although prediction accuracy without using profile is worse than that using profile, prediction without profile is still useful especially for genome-scale prediction since it runs much faster.
SERVER IMPLEMENTATION
Overall description
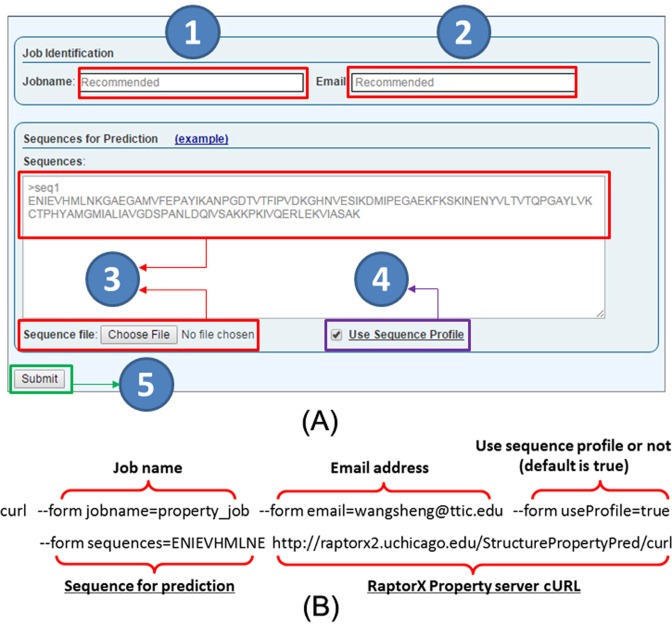
Our server predicts 3/8-state protein secondary structure (SS3/SS8), 3-state solvent accessibility (ACC), and disordered regions (DISO). Users can submit sequences through our web interface or using a publicly available program curl (see Figure 2). When the web interface is used, users may submit a batch of ≤100 sequences at a time. Our server employs two prediction modes: sequence profile used and not used. When sequence profile is not used, only residue-related features are used for prediction. Otherwise, sequence profiles generated by PSI-BLAST (30) (with three iterations and E-value set to 0.001) are also applied. The predicted results are shown as (i) strings and (ii) label distribution at each residue (see Figure 3).
Figure 2.
RaptorX Property server job submission. (A) Web interface for job submission. Job name (1) and user email address (2) are optional. Sequences in FASTA format can be submitted through the text box or a file (3). Click on the example link to see an example. Users can specify if sequence profile shall be used or not (4). Submit a job by clicking on the submit button (5). (B) An example for submission by a publicly available program Curl. Only ‘sequences’ and the submission URL (shown in underlines) are required and the others are optional. A job URL will be returned on screen after submission.
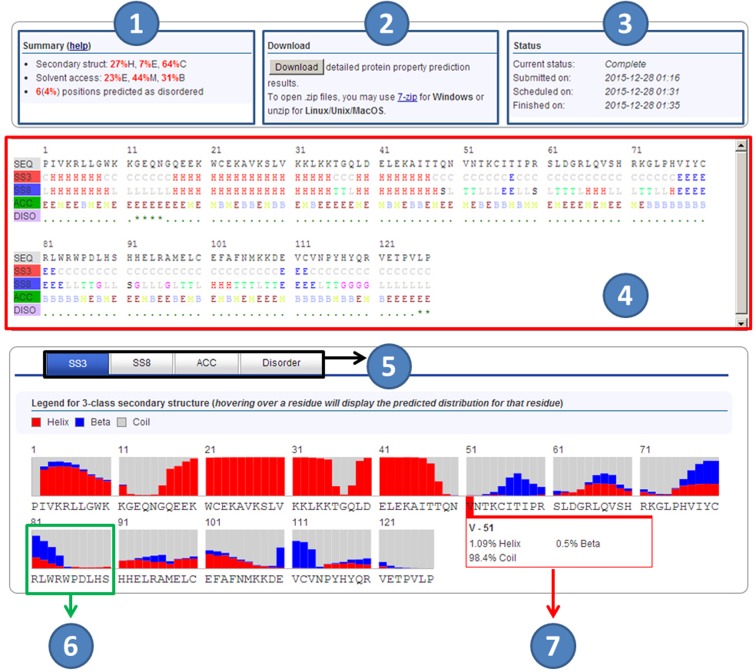
Figure 3.
RaptorX Property result page. The first section shows a summary of the prediction results (1), downloading button (2), and the job status (the submitted, scheduled, and finished time) (3). The second section shows predicted results (4), with the first row showing input sequence, and the remaining rows showing the prediction of 3-state secondary structure (SS3), 8-state secondary structure (SS8), 3-state solvent accessibility (ACC), and order/disorder regions (DISO) with ‘*’ indicating disorder, respectively. The third section shows more detailed prediction results of SS3, SS8, ACC, and DISO (5), visualizing the predicted distribution at each residue (6). Hovering over a residue will display the predicted label distribution for that residue (7).
Input
The only required input to the server is one (or batch of) protein sequence(s). Users may optionally provide a jobname and an email address, which can be used to retrieve the job results.
Output
For each submission, one unique job ID and one URL are assigned to track the job results. When an email is provided in submission, users will be notified by email once the jobs are done. Specifically, the result page has three sections. The first section includes (a) a summary of prediction results, (b) the result download button and (c) job status. The second section shows the prediction results in strings, with the first row showing input sequence, and the remaining four rows showing the predicted SS3, SS8, ACC and DISO. The third section shows the predicted distribution at each residue, which will be displayed when hovering mouse over a residue.
Processing time
The running time of our server depends on two factors: (i) prediction mode and (ii) sequence length. Typically, when sequence profile is not used, it takes only 5–10 s to finish one sequence after it is scheduled to run. When sequence profile is used, the average processing time for a protein of 300 residues is ≤10 min, most of which is spent to generate sequence profile. Currently our server schedules a job every 1 min, so each job needs at least 1 min to finish after submission.
Documentation
The documentation of RaptorX Property is available by the ‘Docs’ link at the web page. It includes some details about the server, descriptions of input and output, explanations of prediction results, and a sample prediction result. Further, RaptorX Property also provides an example input at the submission page.
CONCLUSION AND FUTURE WORK
We have presented RaptorX Property, a novel server for predicting structure property of a protein sequence without using any template information. It outperforms other servers especially for proteins without close homologs in PDB or with very sparse sequence profile.
Currently, this server is able to predict 3/8-state secondary structure (SS3/SS8), solvent accessibility (ACC) and disordered regions (DISO) simultaneously, making use of an emerging machine learning model DeepCNF (Deep Convolutional Neural Fields).
In the future, we may further improve prediction accuracy by extending our DeepCNF into a multi-task-enabled deep learning model (31). To reduce the running time for sequence profile construction, we may run HHblits (32) or allow users to upload MSA directly (7).
However, it should be noted that the accuracy of our server is tied to secondary structure and solvent accessibility assignment by DSSP and disorder definition employed by us. Currently we determine disordered regions using all missing atoms in crystal structures. Both sources are well established, but have their own shortcomings. For instance, atoms may be missing in PDB structures due to technical problems instead of a disordered state (33). Also different annotation criteria, such as STRIDE and DSSP, may have different secondary structure assignment (34). Thus, the quality of our SS or DISO predictions is somehow impacted by the quality of training data.
Supplementary Material
Acknowledgments
The authors are also grateful to the computing power provided by the UChicago Beagle and RCC allocations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM0897532 to J.X.]; National Science Foundation [DBI-0960390 to J.X.]. Funding for open access charge: National Institutes of Health [R01GM0897532 to J.X.]; National Science Foundation [DBI-0960390 to J.X.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Dill K.A. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 2.Myers J.K., Oas T.G. Preorganized secondary structure as an important determinant of fast protein folding. Nat. Struct. Mol. Biol. 2001;8:552–558. doi: 10.1038/88626. [DOI] [PubMed] [Google Scholar]
- 3.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 4.Bairoch A., Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucleic Acids Res. 1997;25:31–36. doi: 10.1093/nar/25.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium U. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 9.Faraggi E., Yang Y., Zhang S., Zhou Y. Predicting continuous local structure and the effect of its substitution for secondary structure in fragment-free protein structure prediction. Structure. 2009;17:1515–1527. doi: 10.1016/j.str.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faraggi E., Zhang T., Yang Y., Kurgan L., Zhou Y. SPINE X: improving protein secondary structure prediction by multistep learning coupled with prediction of solvent accessible surface area and backbone torsion angles. J. Comput. Chem. 2012;33:259–267. doi: 10.1002/jcc.21968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joo K., Lee S.J., Lee J. Sann: solvent accessibility prediction of proteins by nearest neighbor method. Proteins: Struct. Funct. Bioinform. 2012;80:1791–1797. doi: 10.1002/prot.24074. [DOI] [PubMed] [Google Scholar]
- 12.Ma J., Wang S., Wang Z., Xu J. MRFalign: protein homology detection through alignment of Markov random fields. PLoS Comput. Biol. 2014;10:e1003500. doi: 10.1371/journal.pcbi.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S., Peng J., Ma J., Xu J. Protein secondary structure prediction using deep convolutional neural fields. Scientific Rep. 2016;6:18962. doi: 10.1038/srep18962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J., Peng J., Wang S., Xu J. A conditional neural fields model for protein threading. Bioinformatics. 2012;28:i59–i66. doi: 10.1093/bioinformatics/bts213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 16.Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 17.Källberg M., Margaryan G., Wang S., Ma J., Xu J. RaptorX server: a resource for template-based protein structure modeling. Protein Struct. Predict. 2014:17–27. doi: 10.1007/978-1-4939-0366-5_2. [DOI] [PubMed] [Google Scholar]
- 18.Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen‐bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 20.Pollastri G., Przybylski D., Rost B., Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins: Struct. Funct. Bioinform. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 21.Cuff J.A., Barton G.J. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins: Struct. Funct. Bioinform. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Ma J., Wang S. AcconPred: Predicting solvent accessibility and contact number simultaneously by a multitask learning framework under the conditional neural fields model. BioMed Res. Int. 2015;2015:1. doi: 10.1155/2015/678764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monastyrskyy B., Fidelis K., Moult J., Tramontano A., Kryshtafovych A. Evaluation of disorder predictions in CASP9. Proteins: Struct. Funct. Bioinform. 2011;79:107–118. doi: 10.1002/prot.23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S., Weng S., Ma J., Tang Q. DeepCNF-D: predicting protein order/disorder regions by weighted deep convolutional neural fields. Int. J. Mol. Sci. 2015;16:17315–17330. doi: 10.3390/ijms160817315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnan C.N., Baldi P. SSpro/ACCpro 5: almost perfect prediction of protein secondary structure and relative solvent accessibility using profiles, machine learning and structural similarity. Bioinformatics. 2014;30:2592–2597. doi: 10.1093/bioinformatics/btu352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones D.T., Singh T., Kosciolek T., Tetchner S. MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics. 2015;31:999–1006. doi: 10.1093/bioinformatics/btu791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dosztányi Z., Csizmok V., Tompa P., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 28.Jones D.T., Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G., Dunbrack R.L. PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heffernan R., Paliwal K., Lyons J., Dehzangi A., Sharma A., Wang J., Sattar A., Yang Y., Zhou Y. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Sci. Rep. 2015;5:11476. doi: 10.1038/srep11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 33.Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Heinig M., Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32:W500–W502. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.