Abstract
The LassoProt server, http://lassoprot.cent.uw.edu.pl/, enables analysis of biopolymers with entangled configurations called lassos. The server offers various ways of visualizing lasso configurations, as well as their time trajectories, with all the results and plots downloadable. Broad spectrum of applications makes LassoProt a useful tool for biologists, biophysicists, chemists, polymer physicists and mathematicians. The server and our methods have been validated on the whole PDB, and the results constitute the database of proteins with complex lassos, supported with basic biological data. This database can serve as a source of information about protein geometry and entanglement-function correlations, as a reference set in protein modeling, and for many other purposes.
INTRODUCTION
Entanglement in proteins is an important phenomenon, inspiring multidisciplinary research involving biology, biophysics, chemistry and mathematics. Proteins with knots and slipknots, intensively studied over last years, provide an important example of this phenomenon. While proteins with knots and slipknots still challenge our understanding of complexity of proteins, nowadays their analysis is supported by several servers and databases (1–4).
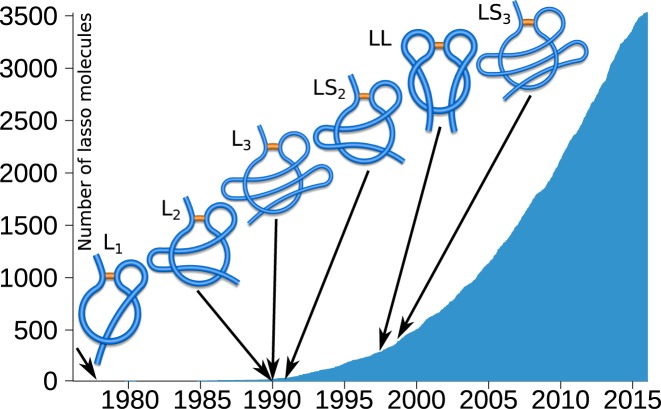
This work is devoted to another important family of entangled structures discovered more recently, which are referred to as complex lassos. While such structures have been analyzed so far in proteins (5–7), the server presented in this work enables analysis of lassos in all types of biopolymers. Lasso structures arise when a polymer possesses a closed loop (closed e.g., by a cysteine bridge in case of proteins), which is pierced by at least one chain terminus, as shown schematically in Figure 1. Currently it is known that around 18% of proteins with cysteine bridges (in a non-redundant set) possess lassos. The number of complex lasso proteins deposited in the Protein Data Bank (PDB) grows exponentially (Figure 1).
Figure 1.
The number of lasso structures deposited in the PDB over years. The first occurrence of a given lasso type is shown by an arrow. Note that the complexity of discovered structures grows with time. The schemes represent lasso types L1, L2, L3, LS2, LL and LS3 (from left to right), which are characterized in the text.
There are many challenging questions concerning proteins with lassos, motivated also by preliminary in vivo experiments: which lasso configurations are possible, what is the role of lassos for biological function and stability of proteins, how those structures fold, etc. In addition, it is well known that therapeutic properties of some proteins can be designed by the presence of cysteine knots (8), therefore presumably complex lassos may provide new tools to steer folding and functions of proteins.
Due to a large number of proteins with cysteine loops, and also because of their geometric complexity, it is impossible to identify systematically lasso proteins without analytical tools (by a naked eye). Moreover the impact of entanglement on the stability or the folding/unfolding mechanism can be understood only via analysis of protein dynamics (9), which requires analysis of lasso structures in many steps of whole time trajectories. In response to those needs we created the LassoProt server (http://lassoprot.cent.uw.edu.pl/), the server to detect and analyze complex lasso structures. The server is based on the technique implemented in a simplified form in (5–7), which we developed to detect different kinds of loop closing bridges (e.g. amide, ester or any other kind of bridge), and to analyze dynamics (trajectories). The method is also extended to parse: PDB files, polymer structures, or chemical compounds. The LassoProt offers a bunch of accessories to facilitate interpretation of entanglement and to perform its comprehensive analysis.
The LassoProt server and our methods have been validated on the whole PDB (over 200 000 chains). All protein chains with detected lassos are stored and are publicly available, thereby providing a unique database, which comprises comprehensive information about entanglement, supported with basic structural, sequential, and biological data. The closed loop proteins are categorized according to the number and directions of piercings. Currently, five major classes of lasso proteins are known: L1, L2 and L3 in which one tail pierces the covalent loop once, twice or three times respectively, LLi, j class where both tails pierce the surface i and j times respectively, and the ‘supercoiling’ class, LSn, where one tail winds around the closed loop, piercing the surface at least twice in a row from the same direction (performing n piercings in total). The LassoProt is self-updating every week based on new structures deposited in the PDB.
The LassoProt is a powerful tool that could support typical biophysical methods to study proteins and other biopolymers. The accompanying database of proteins with lassos can be used to study e.g., correlations between entanglement and function. Geometric characteristics of lassos (e.g. location of piercings) provide new reaction coordinates, useful especially in the analysis of protein folding. Apart from studies of proteins, the LassoProt can be also used to study threaded ring polymers (10–12), to study the topological mechanism of site-specific recombination enzymes, etc.
MATERIALS AND METHODS
Detection of lassos
The general idea of lasso identification follows earlier works (5–7) and consists of four consecutive steps: (i) selection of the (appropriate) covalent loop; (ii) spanning a (minimal) surface on this loop; (iii) detection of surface piercings; (iv) reduction of artificial piercings. Closed loops are detected based on the PDB file or a file provided by a user. The algorithm relies on coordinates of Cα atoms (from the PDB file) or any atoms/monomers (in XYZ file). The triangulated surface of minimal area (called a minimal surface) is spanned on the closed loop following techniques in computer graphics (13); our algorithm, optimized for proteins and taking into account surface orientation (to detect supercoiling), gives the same results as established software (e.g. Surface Evolver (14)). The piercings and their orientation are found as the intersections of vectors joining the monomers (Cα atoms) with oriented triangles. Piercings suspected to be artifacts, or to arise from thermal fluctuations, are ignored (for details see on-line help). The whole algorithm is implemented in C++.
Accompanying database
The database accompanying the server comprises all protein chains deposited in the PDB, including non-Xray or gapped structures. The gaps are modeled as straight segments. The server detects automatically disulfide and other types of linkages based on a header of a PDB file. The PDB file parser (gap filling, conversion of non-coded amino acids, etc.) is written in Python.
Graphical content and structure smoothing
The plots are generated using matplotlib and D3 libraries. The dynamic plots are obtained using Highchart (JavaScript) library. Structures and surfaces are visualized using JSmol (HTML5/JavaScript version). The barycentric plot is calculated according to the algorithm by Tutte (15), with a hyperbolic transformation shifting the vertices of triangles towards the center of the plot. The code for Mathematica and VMD (16) software is translated from data calculated by our algorithm (triangulated surface, barycentric surface, coordinates of smoothed protein). The smoothed structure (with the same lasso type) is obtained based on running average of the positions of three consecutive atoms.
Server and database technicalities
The Server is written in Python with Flask framework dynamically generating HTML pages, uses Apache2 with WSGI. The data are stored using SQLite 3 database. Information about proteins are downloaded from PDB using RESTful services, Pfam and EC data using SIFTS service. The whole service is installed on multicore Linux nodes.
SERVER DESCRIPTION
The LassoProt consists of two parts: the server and the database. In the server part structures can be uploaded either as single frames, or in sets of frames (e.g. the whole MD trajectories). In the latter case, the overall entanglement in a trajectory can be detected and then analyzed more precisely via single frame method. The output can be compared with entries in the database (supported with biological data).
Server input files
The server is designed to be intuitive and easy to use. An input data can be uploaded as an original PDB file (with a header), a file in the PDB format (e.g. a single frame from a trajectory), or in XYZ format (positions of atoms in Cartesian coordinates). Due to its versatility, the last option is suitable for the analysis of various polymers.
The simplest option is to upload the original PDB file, or to fetch it directly from the RCSB database. With the default server settings (‘automatic detection of closed loops’), the LassoProt detects automatically the geometry in all closed loops (see ‘bridge type’ section). A user can additionally a) choose the type of chemical bonds closing the loop (‘choose type of loop closing’) or b) enter the indices of loop closing residues manually (‘choose your own loop closing bridge’). In the former option the strongest, most common bonds are specified, i.e. Disulfide, Amide, Ester and Thioester, responsible for protein dynamics. Additionally, there is also ‘Other’ class containing among others C–C bonds.
The trajectories can be uploaded in multimodel PDB, XYZ or Gromacs XTC files. In this case the loop closing residues are chosen manually by the user. Optionally the output complexity (‘more detailed output file’) and detailed level of analysis (‘more detailed algorithm’) can be adjusted. Finally, to speed up the analysis, the user can define how many frames should be skipped (‘step’). All details and sample input files are given in the on-line manual.
Advanced options
The strength of the server is its versatility. Although the server is optimized for proteins, the user has several options to adjust it to his/her own needs (e.g. to study polymer chains with different flexibility).
The user can modify the minimal (sequential) distance between (i) piercings; (ii) piercing and the chain terminus; (iii) bridge and the piercing. Moreover, the user can turn off the filters validating the protein structure (distance between consecutive Cα atoms, bridge length, etc.).
Single structure presentation
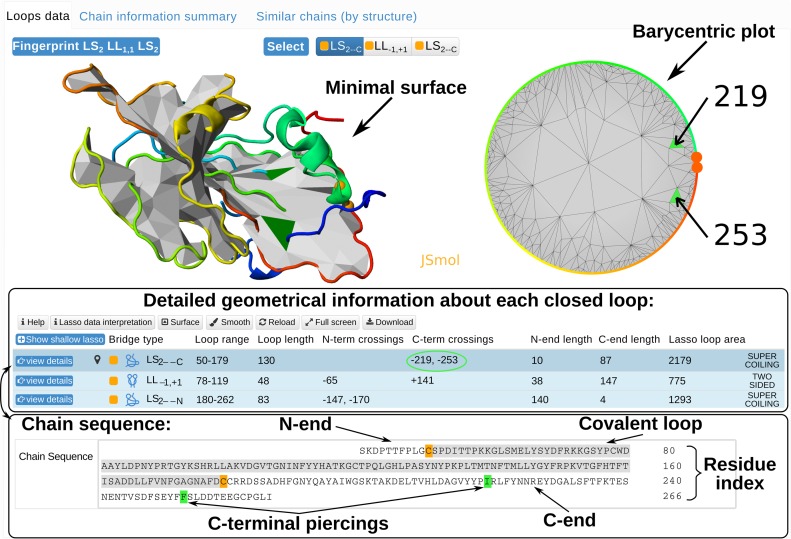
Each structure (uploaded by a user or stored in the database) is presented in the same way (Figure 2). For each structure the server displays: (i) all detected lasso motifs (lasso fingerprint - top left corner of Figure 2); (ii) detailed geometrical and chemical information for each closed loop (if possible); (iii) its various graphical representations; (iv) comprehensive biological and structural information (for proteins deposited in the database). The lasso fingerprint is the concatenation of lasso type symbols for each pierced closed loop, e.g., L2 L1 (in analogy to the knot fingerprint (17)), describing the overall entanglement pattern of the chain. This information is important in the analysis of properties of the whole chain, e.g. its stability, the search of protein with similar lasso pattern or correlations between non-trivial geometry and active sites.
Figure 2.
The main panel presenting detected lasso structures (supercoiling LS2, two sided LL1, 1) in protein with cysteine bridges.
Detailed information about each (also trivial) closed loop (as in Figure 2) is presented in the table in the bottom of the page (see Figure 2). The table contains color-coded chemical types of bridges, lasso types, a list of signed piercings by N- and C-terminus, the area of the (triangulated) surface, and other geometrical information. Upon clicking on ‘show shallow lasso’ button the reduced (shallow) piercings can be displayed.
The entanglement of each closed loop is shown explicitly in the 3D plot, featuring a colored closed loop and the minimal surface with colored pierced triangles. Additionally, to localize easily the surface piercings, the barycentric plot (mapping the surface into a circle) is shown. The colors are compatible with the 3D visualization. The barycentric plot is especially useful in analyzing the self-intersecting surfaces, where identification of piercings in 3D structure is hindered. Moreover, such visualization provides an easy method for selecting surfaces with spatially close piercings. The examples are given in SI. Clicking on ‘view details’ in each row of the table reloads graphical content presenting entanglement of the corresponding covalent loop. Furthermore, the ‘smooth’ option smooths the protein chain exposing its geometry (Figure 3A). The visualization provides also built-in JSmol options. In case of proteins, in the very bottom of the page the lasso motif is projected on the chain sequence, showing its sequential position and types of amino acids piercing the surface, e.g. C-terminal piercings in Figure 2).
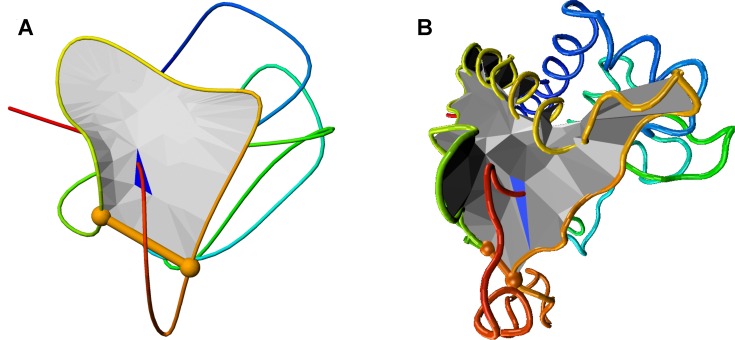
Figure 3.
Comparison between (A) visualization of a smoothed protein and (B) visualization in VMD of the same protein.
All the graphical content and data used to detect geometry is downloadable for further analysis. In particular, the structure (original and smoothed) can be downloaded along with the triangulated surface in Mathematica and TCL script, applicable directly to VMD (Figure 3B), as well as the barycentric plot as SVG file or Mathematica script.
Finally, for structures deposited in the database comprehensive biological and structural data is provided (top of Figure 2), including the EC number (for enzymes), and Pfam (18) accession code. Also sequentially (Restful services RCSB) and structurally similar structures (CATH (19) database) are shown, which is important, e.g. in checking if lasso motifs are conserved among other family members.
Trajectory analysis
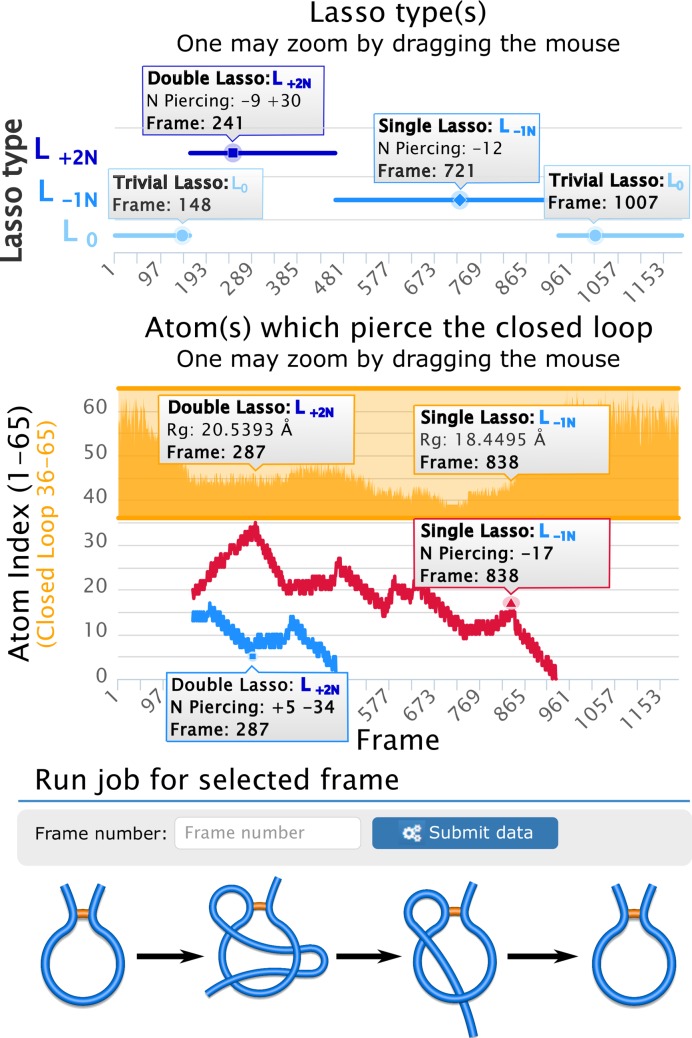
The trajectory analysis consists of a repeated single frame analysis, which gives a global and then detailed description of the entanglement, see Figure 4. As a result, one can study the dynamical behavior of the lasso type and surface piercings. These are presented as two interactive zoomable plots (‘Lasso type(s)’ and ‘Atom(s) which pierce the closed loop’), displaying the detailed information for every frame while pointer is above the selected frame (Figure 4). Moreover, to present evolution of piercings along the folding pathway, we show each piercing (i.e. the same piece of a backbond piercing the same loop) using the same color in different frames. For example, to understand how a double lasso changes to a single lasso in Figure 4, we present two piercings of a double lasso in blue and red, and then the surviving piercing is shown in red. Such presentation has a pivotal role in understanding of dynamics (e.g. to recognize how slipknot conformations change to knots when protein fold). The shallow piercings are shown in dotted lines, so that a full overview about the topological landscape is provided. For comparison, in the second plot the radius of gyration is shown (e.g. to inform about compactness of a protein when its topology changes), as well as positions of closed loops.
Figure 4.
A sample trajectory output. The charts present L0 → L+2N → L−1N → L0 process shown schematically in the bottom. The top plot presents changes in lasso type. The bottom plot shows changes of crossing residue indices, with Rgyr dependence for comparison (orange plot). For each frame an additional information is displayed upon pointing the cursor in an appropriate position (small gray frames). Each frame can be analyzed in detail upon entering its number.
The trajectory analysis is fully compatible with a single frame analysis. The user can choose individual frames and display them in the single frame analysis mode, as described above. The server also detects automatically conformations with the most complex lasso and presents them in the single frame mode. All presented data (with the static plots) are downloadable.
DATABASE—VALIDATION METHOD
The method underlying the server has been validated on all proteins deposited in the PDB (over 200 000 chains). Only less than 800 structures were rejected, which shows that the LassoProt is a powerful server suitable to analyze even most entangled structures.
Proteins with detected lassos are collected and constitute a unique database, updated each week with structures newly deposited in the PDB. Combination of geometrical, biological and structural information presented in the database enables analysis of proteins from different viewpoints. Due to various search options, the LassoProt is a perfect tool for statistical analysis.
There are three main ways the user can access the data from the database. The easiest one is entering the PDB code of the structure of interest. Second, all the structures with non-trival loops (lassos) can be listed in the ‘Browse’ mode. Finally, the ‘Search’ mode can be used. In the search mode the user can choose to display the structures with a given lasso type or chemical nature of loop-closing bonds. In the main view the general types (Ln, LLi, j or LSn) can be chosen. The search can also be restricted to various subclasses.
Moreover, basic length statistics are given, enabling to select structures with chosen loop or tail length. In the bottom of the page the possible lasso fingerprints are given. As the function is strictly correlated with protein conformation, the fingerprint summarizing the topology may be biologically relevant. The user can also perform more ‘biologically oriented’ search and use the ‘Molecule keywords’, ‘Molecule tags’, ‘Pfam family identifier’, ‘EC nomenclature’, ‘CATH classification’ or ‘Organism’ tabs, where analyzed chains are sorted according to a classification used in a given method.
All data is accessible based either on all PDB entries, or on the non-redundant set. Moreover the data can be also filtered based on the type of chemical bridges. This option enables to detect unique lasso motifs or to perform statistical analysis.
Artifact class
To avoid misinterpretation of lasso configurations due to unresolved (not determined experimentally) parts of protein chains, several conditions on mutual distances between Cα atoms are imposed (described in detail in the on-line documentation). If the structure does not meet our criteria it is still analyzed, but it is assigned to the ‘Artifact’ class.
ON-LINE DOCUMENTATION
All server documentation is provided on-line and includes: detailed description of lasso type assignment, piercing reduction rules, surface orientation assignment, lasso type determination, database (search and browse) tutorial, proteins structure validation, data interpretation, example of input files, examples of single frame and trajectory analysis, troubleshooting, and applications. Database statistics, such as lists of non-trivial structures, or the newest complex lasso protein deposited in the PDB, are also included.
APPLICATIONS
The LassoProt server is based on protein analysis, therefore it has many biological applications. Nevertheless, its versatility makes it useful in other areas of research.
It is still unknown how proteins with lassos fold/unfold (under oxidative condition) or what is the role of entanglement for biological function. Presumably the constraints imposed by the geometry of lassos should influence stability, or can lead to the mechanical clamps under mechanical tension. As shown, e.g. for cystein knots, geometrical analysis is necessary for complete understanding of mechanical manipulation results (20). The LassoProt provides a unique tool to this end.
On the other hand, the function is strongly correlated with the protein structure, in particular with its entanglement pattern (e.g. in therapeutically active miniproteins (8)). Thus, the LassoProt database is a useful tool for studying function-entanglement dependence. Moreover, the output from LassoProt can provide new reaction coordinates (5) useful in analysis of folding of knotted proteins (21,22).
Furthermore, a preliminary analysis of CASP data indicates that the LassoProt can be useful in structure prediction (23).
The LassoProt may also be helpful in designing proteins with desired properties imposed by topology and bridges (e.g. stability (24)). Moreover, bridges can serve as molecular switches, as the entanglement (and thus the function) could be turned on and off by changing the oxidizing potential (25).
Lassos should also be detected and analyzed in other biopolymers, e.g. circular DNA (26,27). This opens a new field in studying conformational effects in polymer physics and fluid dynamics concentrated on ring structures (28). The LassoProt can be used by chemists to design materials with topology-induced stability, and theorists in analyzing, e.g. Monte Carlo simulations of ring polymers (10–12).
Comparison with other servers
To our knowledge, this is the first server and database analyzing biopolymers with lassos.
SUMMARY
The LassoProt server analyzes proteins and other biopolymers with lassos. The server can analyze both single structures and whole trajectories. It is the first tool of this kind and should be valuable for a broad community of scientists. We hope that the server versatility will stimulate new discoveries and methods in various areas of research.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre [2012/07/E/NZ1/01900 to J.I.S.]; European Molecular Biology Organization [2057 to J.I.S.]; Foundation for Polish Science [130/UD/SKILLS/2015 to W.N.]; [120000-501/86-DSM-110200 to P.D.-T.]. Funding for open access charge: Foundation for Polish Science [130/UD/SKILLS/2015].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jamroz M., Niemyska W., Rawdon E.J., Stasiak A., Millett K.C., Sułkowski P., Sulkowska J.I. KnotProt: a database of proteins with knots and slipknots. Nucleic Acids Res. 2014;43:D306–D314. doi: 10.1093/nar/gku1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolesov G., Virnau P., Kardar M., Mirny L.A. Protein knot server: detection of knots in protein structures. Nucleic Acids Res. 2007;35(Suppl. 2):W425–W428. doi: 10.1093/nar/gkm312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai Y.-L., Yen S.-C., Yu S.-H., Hwang J.-K. pKNOT: the protein KNOT web server. Nucleic Acids Res. 2007;35(Suppl. 2):W420–W424. doi: 10.1093/nar/gkm304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai Y.-L., Chen C.-C., Hwang J.-K. pKNOT v. 2: the protein KNOT web server. Nucleic Acids Res. 2012;40:W228–W231. doi: 10.1093/nar/gks592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haglund E., Sułkowska J.I., He Z., Feng G.-S., Jennings P.A., Onuchic J.N. The unique cysteine knot regulates the pleotropic hormone leptin. PLoS One. 2012;7:e45654. doi: 10.1371/journal.pone.0045654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haglund E., Sulkowska J.I., Noel J.K., Lammert H., Onuchic J.N., Jennings P.A. Pierced lasso bundles are a new class of knot-like motifs. PLoS Comput. Biol. 2014;10:e1003613. doi: 10.1371/journal.pcbi.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niemyska W., Dabrowski-Tumanski P., Kadlof M., Haglund E., Sułkowski P., Sulkowska J.I. Complex lasso: new entangled motifs in proteins. Scient. Rep. doi: 10.1038/srep36895. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Zirah S., Rebuffat S. Lasso Peptides: Bacterial Strategies to Make and Maintain Bioactive Entangled Scaffolds. Springer; 2014. [Google Scholar]
- 9.Noel J.K., Onuchic J.N., Sulkowska J.I. Knotting a protein in explicit solvent. J. Phys. Chem. Lett. 2013;4:3570–3573. [Google Scholar]
- 10.Smrek J., Grosberg A.Y. Understanding the dynamics of rings in the melt in terms of the annealed tree model. J. Phys. Cond. Matter. 2015;27:064117. doi: 10.1088/0953-8984/27/6/064117. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki J., Takano A., Deguchi T., Matsushita Y. Dimension of ring polymers in bulk studied by Monte-Carlo simulation and self-consistent theory. J. Chem. Phys. 2009;131:144902. doi: 10.1063/1.3247190. [DOI] [PubMed] [Google Scholar]
- 12.Michieletto D., Marenduzzo D., Orlandini E., Alexander G.P., Turner M.S. Threading dynamics of ring polymers in a gel. ACS Macro Lett. 2014;3:255–259. doi: 10.1021/mz500060c. [DOI] [PubMed] [Google Scholar]
- 13.Chen W., Cai Y., Zheng J. Constructing triangular meshes of minimal area. Comput.-Aided Des. App. 2008;5:508–518. [Google Scholar]
- 14.Brakke K.A. The surface evolver. Exper. Math. 1992;1:141–165. [Google Scholar]
- 15.Tutte W.T. How to draw a graph. Proc. London Math. Soc. 1963;13:743–768. [Google Scholar]
- 16.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 17.Sułkowska J.I., Rawdon E.J., Millett K.C., Onuchic J.N., Stasiak A. Conservation of complex knotting and slipknotting patterns in proteins. Proc. Nat. Acad. Sci. U.S.A. 2012;109:E1715–E1723. doi: 10.1073/pnas.1205918109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sillitoe I., Lewis T.E., Cuff A., Das S., Ashford P., Dawson N.L., Furnham N., Laskowski R.A., Lee D., Lees J.G., et al. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015;43:D376–D381. doi: 10.1093/nar/gku947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikora M., Sulkowska J.I., Cieplak M. Mechanical strength of 17 134 model proteins and cysteine slipknots. PLoS Comput. Biol. 2009;5:e1000547. doi: 10.1371/journal.pcbi.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virnau P., Mallam A., Jackson S. Structures and folding pathways of topologically knotted proteins. J. Phys. Cond. Matter. 2010;23:033101. doi: 10.1088/0953-8984/23/3/033101. [DOI] [PubMed] [Google Scholar]
- 22.Dabrowski-Tumanski P., Jarmolinska A., Sulkowska J. Prediction of the optimal set of contacts to fold the smallest knotted protein. J. Phys. Cond. Matter. 2015;27:354109. doi: 10.1088/0953-8984/27/35/354109. [DOI] [PubMed] [Google Scholar]
- 23.Khatib F., Weirauch M.T., Rohl C.A. Rapid knot detection and application to protein structure prediction. Bioinformatics. 2006;22:e252–e259. doi: 10.1093/bioinformatics/btl236. [DOI] [PubMed] [Google Scholar]
- 24.Clarke J., Fersht A.R. Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochem. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 25.Haglund E. Engineering covalent loops in proteins can serve as an on/off switch to regulate threaded topologies. J. Phys. Cond. Matter. 2015;27:354107. doi: 10.1088/0953-8984/27/35/354107. [DOI] [PubMed] [Google Scholar]
- 26.Reith D., Cifra P., Stasiak A., Virnau P. Effective stiffening of DNA due to nematic ordering causes DNA molecules packed in phage capsids to preferentially form torus knots. Nucleic Acids Res. 2012;40:5129–5137. doi: 10.1093/nar/gks157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tesi M., Van Rensburg E.J., Orlandini E., Sumners D., Whittington S. Knotting and supercoiling in circular DNA: a model incorporating the effect of added salt. Phys. Rev. E. 1994;49:868. doi: 10.1103/physreve.49.868. [DOI] [PubMed] [Google Scholar]
- 28.Laing C.E., Ricca R.L., De Witt S.L. Conservation of writhe helicity under anti-parallel reconnection. Scien. Rep. 2015;5:9224. doi: 10.1038/srep09224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.