Abstract
Recent advances in next-generation sequencing technologies and genome assembly algorithms have enabled the accumulation of a huge volume of genome sequences from various species. This has provided new opportunities for large-scale comparative genomics studies. Identifying and utilizing synteny blocks, which are genomic regions conserved among multiple species, is key to understanding genomic architecture and the evolutionary history of genomes. However, the construction and visualization of such synteny blocks from multiple species are very challenging, especially for biologists with a lack of computational skills. Here, we present Synteny Portal, a versatile web-based application portal for constructing, visualizing and browsing synteny blocks. With Synteny Portal, users can easily (i) construct synteny blocks among multiple species by using prebuilt alignments in the UCSC genome browser database, (ii) visualize and download syntenic relationships as high-quality images, (iii) browse synteny blocks with genetic information and (iv) download the details of synteny blocks to be used as input for downstream synteny-based analyses, all in an intuitive and easy-to-use web-based interface. We believe that Synteny Portal will serve as a highly valuable tool that will enable biologists to easily perform comparative genomics studies by compensating limitations of existing tools. Synteny Portal is freely available at http://bioinfo.konkuk.ac.kr/synteny_portal.
INTRODUCTION
Recent advances in next-generation sequencing technologies and genome assembly algorithms, together with the results of various large-scale genome projects, such as the Genome 10K Project (1), the Bird 10K Project (2) and i5K (3), have enabled the accumulation of a huge volume of genomic sequences from various species. These genome sequences are usually stored in public databases, such as the NCBI Reference Sequence database (4), the Genome OnLine Database (5), the UCSC genome browser database (6) and Ensembl Genomes (7); they have been used to uncover genomic similarities and differences between different species and the functional consequences of such similarities and differences. Comparative genomics studies have contributed to a better understanding of the molecular-level mechanisms of species diversity and genome evolution (8–12).
Synteny blocks, which are genomic regions that are conserved among multiple species, play a pivotal role in comparative genomics. Various algorithms and tools have been introduced for the construction and utilization of synteny blocks, such as CYNTENATOR (13), DRIMM-Synteny (14), i-ADHoRe 3.0 (15), inferCars (16), OSfinder (17) and Sibelia (18). They are stand-alone and command-line tools, and users need to download and install them on a users’ machine. Therefore, the construction of synteny blocks using such tools requires computer resources, and advanced bioinformatics skills to properly prepare input data and running command-line programs.
To alleviate these difficulties, more user-friendly tools for utilizing and visualizing synteny blocks have been developed. For example, GenomeMatcher (19), Mauve (20), MizBee (21) and SyMap (22) are stand-alone synteny browsers that support graphical user interface. Gbrowse_syn (23) and Sybil (24) are web-based software packages for comparative genomics that need to be installed and configured in users’ own machines. GSV (25) and mGSV (26) are web-based synteny viewers that help to visualize user-provided synteny information. Cinteny (27) is a web server for identifying synteny blocks and analyzing genome rearrangements. CoGe (28), VISTA (29) and Ensembl SyntenyView (30) are web-based platforms that perform various analyses for comparative genomic studies. Genomicus (31) is a web browser that displays the homologous genomic contexts of different species. C-Sibelia (32) is a web server for comparing assemblies to a reference genome together with the annotation of variants in genes. GRIMM-Synteny (33) is a web server for analyzing genome rearrangements by using the list of genomic markers represented by numbers with the annotation of variants in genes. However, different tools still have different weaknesses, such as only supporting pairwise comparison, requiring the generation of synteny information by users, limited features for visualization and not supporting the saving or downloading of high-quality images (Supplementary Data for tool comparison).
Here, we present Synteny Portal, a versatile web-based application portal that allows for construction, visualization and browsing of synteny blocks by compensating limitations of existing tools. With Synteny Portal, users can easily (i) construct synteny blocks among multiple species by using prebuilt alignments in the UCSC genome browser database (6), (ii) visualize and download syntenic relationships as high-quality images, (iii) browse synteny blocks together with genetic information and (iv) download the details of synteny blocks that can be used as input for downstream synteny-based analyses, all in an intuitive and easy-to-use web interface.
MATERIALS AND METHODS
Data preparation
Data collection
We collected genomic information on well-studied reference species, including human, mouse, cow, dog, horse, pig, rat, rhesus, chicken and zebra fish, from the UCSC genome browser database (6), which includes genome assembly sequences, gene annotation, whole-genome pairwise alignments of the reference species in the chain and net format (34), cytobands and assembly sizes of the reference species. In total, 458 whole-genome pairwise alignments for 67 different species, including 124 different assembly versions and 33 assembly sequences for the ten reference species with different assembly versions are available on our web server. Gene/protein identifiers annotated with seven different criteria, such as Wiki Genes (35), RefSeq genes and proteins (4), Entrez genes (36), Ensembl genes, transcripts and proteins (7), allow for searching and browsing user-provided gene/protein identifiers. The mapping information using different identifiers was obtained from the Uniprot website (http://www.uniprot.org/) and the biomaRt package from Bioconductor (37).
Synteny block construction and visualization
Synteny blocks were built using the inferCars program package (16) with four different resolutions (150, 300, 400 and 500 Kbp). Specifically, pairwise collinear genome segments (PCGSs) of two species (i.e. a reference species and another species) are first collected from pairwise whole-genome alignments. Then, to create multiple collinear genome segments (MCGSs), two PCGSs are merged by splitting the PCGSs of one species at positions corresponding to the boundaries of the PCGSs of the other species based on the coordinates of the reference genome. This merging process repeats until the PCGSs of all species are merged, and the final MCGSs, which are larger than the given resolution, are reported as synteny blocks. If outgroup species are provided, which is optional, genomic regions of the outgroup species matched to each of the calculated synteny blocks are added. Syntenic relationships are visualized through the Circos program (38) and converted to an interactive version on the website with JavaScript. The BLAST program (39) is used to search for a top-matched region in the reference genome based on the user-provided DNA or protein sequence with default parameter values (Supplementary Data).
Implementations
The Synteny Portal website is implemented with the HTML5 and JavaScript libraries, including jQuery (http://jquery.com), d3.js (http://d3js.org/) and GenomeD3Plot (40) for the client-side user interface, and by PHP scripts (5.3.3) for interactive data processing. The server-side processes are generated by Perl scripts (v5.22.0).
RESULTS
Synteny Portal overview
Synteny Portal provides four main web applications: SynCircos, SynBrowser, SynSearcher and SynBuilder. SynCircos visualizes synteny blocks against a user-selected reference and target species in an interactive Circos plot. SynBrowser displays more details of synteny blocks with annotated reference genes. SynSearcher finds syntenic regions in other species based on query sequences. Finally, SynBuilder constructs synteny blocks among multiple chosen species. These four applications generate the high-quality Circos plots or records of synteny blocks that can be downloaded in various formats, including SVG, PNG, JPEG and PDF.
SynCircos
SynCircos depicts synteny blocks in an interactive Circos plot. Users can select a reference species, which is the reference of pairwise whole-genome alignment, multiple target species, target chromosomes and a resolution for the minimum size of a homologous reference block. The syntenic relationships among chosen species are visualized as an interactive Circos plot (Figure 1A), which can be downloaded in various formats. Ribbons connect homologous blocks in different species that belong to the same synteny block and users can highlight a specific syntenic relationship by placing a mouse pointer on a specific ribbon.
Figure 1.
Examples of SynCircos and SynBrowser outputs. (A) A representative Circos plot generated by SynCircos, which compared three species and their specific chromosomes (Human: chr1, chr3, chr5, chr6, chr8, chr12, chr15, chr19; Mouse: chr1, chr3, chr7, chr17, chr18; and Cow: chr3, chr9, chr10, chr14, chr17, chr22). The highlighted ribbons represent syntenic relationships between human chromosome 1 and the chromosomes of the other species. (B) A representative image generated by SynBrowser which shows the relationships between human chromosome 3 and the related mouse chromosomes. The coordinates of homologous blocks in each synteny block can be seen when a specific genomic region is selected (the khaki-colored box in the center of the image). The red triangle indicates the position of the user-queried gene. (C) A representative gene browser produced by SynBrowser which shows the genes of the reference species (human in this image) in synteny blocks. Details of the annotated genes are provided via links to the UCSC genome browser.
Inputs to SynCircos
SynCircos requires user-selected reference and target species, genome assembly version of the selected species, chromosome number, a resolution for the synteny blocks and an image format for visualization in the Circos plot.
Outputs of SynCircos
The outputs of SynCircos is an interactive Circos plot. Tracks and ribbons in the Circos plot represent the chromosomes and cytobands (if any) of the chosen species and the syntenic relationships among different species, respectively. Figure 1A shows a highlighted synteny block in human, mouse and cow.
SynBrowser
SynBrowser displays more details of syntenic relationships between two species (i.e. a reference and a target species), whereas SynCircos shows a global view of syntenic relationships among multiple species. SynBrowser presents homologous blocks between the two species in an interactive graphical form (Figure 1B and C). Users can easily highlight specific homologous blocks via exact genomic coordinates in a floating window. SynBrowser also provides two ways of navigating synteny blocks: (i) chromosome-level navigation, by clicking on a specific reference chromosome in a chromosome legend on the left of a webpage, and (ii) region-level navigation, by specifying a coordinate of a reference chromosome. Genes within a synteny block can also be navigated using a gene browser through a direct search for a specific gene with a variety of gene identifiers, such as RefSeq, Wiki Gene and Entrez Gene. The images in SynBrowser can also be downloaded in multiple formats.
Inputs to SynBrowser
To visualize the pairwise synteny blocks, SynBrowser needs only four user-selected parameters: a reference species, a reference chromosome, a target species and a synteny block resolution. Synteny blocks containing a specific gene or protein can be found by searching for a gene or protein identifier. Users can directly move to a chosen position in the reference genome by providing specific coordinates.
Outputs of SynBrowser
The outputs of SynBrowser are a plot showing connections between homologous blocks between two species and a gene browser displaying reference genes within synteny blocks. Figure 1B and C show highlighted homologous blocks between human and mouse in the same synteny block, along with their coordinates and reference genes within the synteny block. The red triangle in Figure 1B indicates the location of a gene searched for by users.
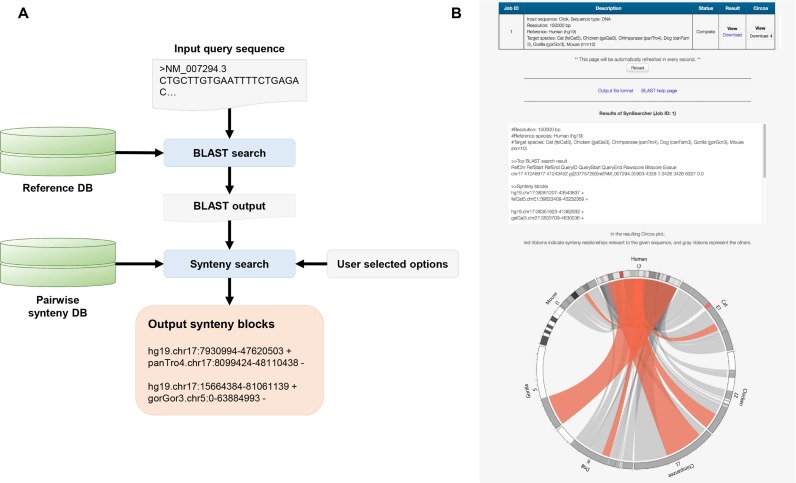
SynSearcher
SynSearcher searches for a reference genome sequence and finds the best-matched region based on a user-provided DNA or protein sequence by BLAST query; synteny blocks encompassing the top-matched reference region are retrieved (Figure 2A). The results are provided to users in two different formats: (i) the details of BLAST search results and the coordinates of synteny blocks as a text file, and (ii) an interactive Circos plot highlighting the retrieved syntenic relationships (Figure 2B). The Circos plot can be downloaded in various formats, and the text file can also be downloaded for downstream analyses with minor modifications.
Figure 2.
Flowchart and outputs of SynSearcher. (A) Given a DNA or protein sequence in the FASTA format, SynSearcher finds matching regions in a reference species using a BLAST search and returns pairwise synteny blocks containing the matched regions of the reference species. (B) The outputs of SynSearcher (white box, BLAST search results and the coordinates of pairwise synteny blocks; below, the Circos plot) for seven species (human, chimpanzee, gorilla, mouse, cat, dog and chicken) given a user-provided sequence.
Inputs to SynSearcher
A user-provided DNA or protein sequence can be directly inserted in a text box or uploaded as a file in a FASTA format. Users can also select a reference and target species with a chosen resolution.
Outputs of SynSearcher
The results consist of (i) the details of the BLAST output for the top-matched alignment result, (ii) the coordinates of identified pairwise synteny blocks and (iii) a Circos plot visualizing the synteny blocks. Figure 2B shows synteny blocks obtained from seven species (human, chimpanzee, mouse, gorilla, chicken, cat and dog) given a user-provided sequence.
SynBuilder
SynBuilder creates synteny blocks for user-selected species (a reference, target and optional outgroup species) with a chosen resolution using the inferCars program (16). Specifically, pairwise whole-genome alignments between the reference and the target species are first collected and then homologous genomic regions are identified, and, finally, co-linear genomic regions among the chosen species are grouped into synteny blocks (Figure 3A). If outgroup species are selected, matched genomic regions between the outgroup species and a reference species are added to the synteny blocks. The results of SynBuilder consist of (i) the coordinates of the synteny blocks as a text file, and (ii) an interactive Circos plot depicting syntenic relationships (Figure 3B). The Circos plot can be downloaded in various formats, and the text file can also be downloaded and used for downstream analyses with minor modifications.
Figure 3.
Flowchart and outputs of SynBuilder. (A) Given options (a reference species, target species, and optional outgroup species, and a resolution) selected by users, SynBuilder creates synteny blocks using pairwise whole-genome alignments between the reference and the other species. (B) The output of SynBuilder (white box, coordinates of synteny blocks; below, the Circos plot) for four species (human, chimpanzee, gorilla and mouse) with three outgroup species (cat, dog and chicken).
Inputs to SynBuilder
SynBuilder requires the selection of a reference, target and optional outgroup species, and a resolution for the synteny blocks.
Outputs of SynBuilder
The results of SynBuilder are (i) the coordinates of synteny blocks and (ii) an interactive Circos plot depicting syntenic relationships among the chosen species.
CONCLUSION
Synteny Portal facilitates studies on comparative genomics through easy construction of synteny blocks, intuitive graphical representation with a high-quality image format and easy-to-use querying and browsing functionalities. Synteny Portal enables a user with a lack of computational skills to perform comparative genomic analyses. The current version only works with prebuilt whole-genome alignments because of long computational time and high computational resource requirements for online creation of whole-genome sequence alignments. However, whole-genome alignment data will be periodically updated via mirroring of data in the UCSC genome browser database. In addition, our website will be updated to support the whole-genome alignment of user-provided genome sequences with additional reference species in future. We believe that Synteny Portal will serve as a highly valuable tool that will enable biologists to easily perform comparative genomics studies.
AVAILABILITY
The Synteny Portal web server is available at http://bioinfo.konkuk.ac.kr/synteny_portal/.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning [2014M3C9A3063544, 2013R1A1A2008183, 2011-619-E0002]. Funding for open access charge: Ministry of Science, ICT & Future Planning of Korea [2014M3C9A3063544].
Conflict of interest statement. None declared.
REFERENCES
- 1.Koepfli K.P., Paten B., O'Brien S.J. The Genome 10K Project: a way forward. Annu. Rev. Anim. Biosci. 2015;3:57–111. doi: 10.1146/annurev-animal-090414-014900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang G., Rahbek C., Graves G.R., Lei F., Jarvis E.D., Gilbert M.T. Genomics: bird sequencing project takes off. Nature. 2015;522:34. doi: 10.1038/522034d. [DOI] [PubMed] [Google Scholar]
- 3.i5K Consortium. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J. Hered. 2013;104:595–600. doi: 10.1093/jhered/est050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy T.B., Thomas A.D., Stamatis D., Bertsch J., Isbandi M., Jansson J., Mallajosyula J., Pagani I., Lobos E.A., Kyrpides N.C. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015;43:D1099–D1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersey P.J., Allen J.E., Armean I., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Falin L.J., Grabmueller C., et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44:D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiliano D.B., Hall N., Jones S.J., Clark L.N., Corton C.H., Barrell B.G., Blaxter M.L. Conservation of long-range synteny and microsynteny between the genomes of two distantly related nematodes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0057. RESEARCH0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowers J.E., Chapman B.A., Rong J., Paterson A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 10.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 11.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 12.Alfoldi J., Lindblad-Toh K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013;23:1063–1068. doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodelsperger C., Dieterich C. CYNTENATOR: progressive gene order alignment of 17 vertebrate genomes. PLoS One. 2010;5:e8861. doi: 10.1371/journal.pone.0008861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pham S.K., Pevzner P.A. DRIMM-Synteny: decomposing genomes into evolutionary conserved segments. Bioinformatics. 2010;26:2509–2516. doi: 10.1093/bioinformatics/btq465. [DOI] [PubMed] [Google Scholar]
- 15.Proost S., Fostier J., De Witte D., Dhoedt B., Demeester P., Van de Peer Y., Vandepoele K. i-ADHoRe 3.0–fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012;40:e11. doi: 10.1093/nar/gkr955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J., Zhang L., Suh B.B., Raney B.J., Burhans R.C., Kent W.J., Blanchette M., Haussler D., Miller W. Reconstructing contiguous regions of an ancestral genome. Genome Res. 2006;16:1557–1565. doi: 10.1101/gr.5383506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hachiya T., Osana Y., Popendorf K., Sakakibara Y. Accurate identification of orthologous segments among multiple genomes. Bioinformatics. 2009;25:853–860. doi: 10.1093/bioinformatics/btp070. [DOI] [PubMed] [Google Scholar]
- 18.Minkin I., Patel A., Kolmogorov M., Vyahhi N., Pham S. Sibelia: a scalable and comprehensive synteny block generation tool for closely related microbial genomes. In: Darling A, Stoye J, editors. Algorithms in Bioinformatics. Berlin Heidelberg: Springer; 2013. pp. 215–229. [Google Scholar]
- 19.Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer M., Munzner T., Pfister H. MizBee: a multiscale synteny browser. IEEE Trans. Vis. Comput. Graph. 2009;15:897–904. doi: 10.1109/TVCG.2009.167. [DOI] [PubMed] [Google Scholar]
- 22.Soderlund C., Bomhoff M., Nelson W.M. SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011;39:e68. doi: 10.1093/nar/gkr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKay S.J., Vergara I.A., Stajich J.E. Using the Generic Synteny Browser (GBrowse_syn) Curr. Protoc. Bioinformatics. 2010;31:9.12.1–9.12.25. doi: 10.1002/0471250953.bi0912s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crabtree J., Angiuoli S.V., Wortman J.R., White O.R. Sybil: methods and software for multiple genome comparison and visualization. Methods Mol. Biol. 2007;408:93–108. doi: 10.1007/978-1-59745-547-3_6. [DOI] [PubMed] [Google Scholar]
- 25.Revanna K.V., Chiu C.C., Bierschank E., Dong Q. GSV: a web-based genome synteny viewer for customized data. BMC Bioinformatics. 2011;12:316. doi: 10.1186/1471-2105-12-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revanna K.V., Munro D., Gao A., Chiu C.C., Pathak A., Dong Q. A web-based multi-genome synteny viewer for customized data. BMC Bioinformatics. 2012;13:190. doi: 10.1186/1471-2105-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha A.U., Meller J. Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinformatics. 2007;8:82. doi: 10.1186/1471-2105-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyons E., Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 29.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clamp M., Andrews D., Barker D., Bevan P., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., et al. Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res. 2003;31:38–42. doi: 10.1093/nar/gkg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louis A., Muffato M., Roest Crollius H. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 2013;41:D700–D705. doi: 10.1093/nar/gks1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minkin I., Pham H., Starostina E., Vyahhi N., Pham S. C-Sibelia: an easy-to-use and highly accurate tool for bacterial genome comparison. F1000Res. 2013;2:258. doi: 10.12688/f1000research.2-258.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pevzner P., Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008;40:1047–1051. doi: 10.1038/ng.f.217. [DOI] [PubMed] [Google Scholar]
- 36.Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R., et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43:D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laird M.R., Langille M.G., Brinkman F.S. GenomeD3Plot: a library for rich, interactive visualizations of genomic data in web applications. Bioinformatics. 2015;31:3348–3349. doi: 10.1093/bioinformatics/btv376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.