Abstract
3Drefine is an interactive web server for consistent and computationally efficient protein structure refinement with the capability to perform web-based statistical and visual analysis. The 3Drefine refinement protocol utilizes iterative optimization of hydrogen bonding network combined with atomic-level energy minimization on the optimized model using a composite physics and knowledge-based force fields for efficient protein structure refinement. The method has been extensively evaluated on blind CASP experiments as well as on large-scale and diverse benchmark datasets and exhibits consistent improvement over the initial structure in both global and local structural quality measures. The 3Drefine web server allows for convenient protein structure refinement through a text or file input submission, email notification, provided example submission and is freely available without any registration requirement. The server also provides comprehensive analysis of submissions through various energy and statistical feedback and interactive visualization of multiple refined models through the JSmol applet that is equipped with numerous protein model analysis tools. The web server has been extensively tested and used by many users. As a result, the 3Drefine web server conveniently provides a useful tool easily accessible to the community. The 3Drefine web server has been made publicly available at the URL: http://sysbio.rnet.missouri.edu/3Drefine/.
INTRODUCTION
The spatial structure of proteins has been shown to have critical effects on protein function including protein structure-based drug design (1), protein docking (2) and prediction of biological functions based on protein structure (3). Therefore, protein structure modeling is important in determining such spatial properties. Protein structure properties such as back-bone positioning and its relation to side-chain conformation of a protein model (4) make simultaneous refinement of both the global topologies and local structural qualities of a protein structure necessary in modeling. Computational steps in the modeling process are necessary to address this problem to produce accurate model predictions. However, the deviation of predicted models from their experimentally derived true, native structures is one limitation of computational modeling. Thus, the problem of protein structure refinement remains to be solved despite attracting constant attention by researchers (5,6).
Here we present the 3Drefine web server, a website dedicated to the computationally efficient and reliable protein structure refinement method called 3Drefine and its iterative implementation, i3Drefine (7,8). The 3Drefine web server employs the iterative protocol based on a combination of two steps of minimization: Optimizing Hydrogen Bonding Network and Energy Minimization using a composite physics and knowledge based force fields, which is implemented within the MESHI (9) molecular modeling framework (7). The method has extensively been tested in Critical Assessment of Techniques for Protein Structure Prediction (CASP) datasets. 3Drefine demonstrated significant potential in atomic-level protein structure refinements in terms of both global and local measures of structural qualities compared to other refinement methods in CASP8, CASP9 and CASP10. The web server has been prepared to allow convenient use of the software and quick analysis of the results.
MATERIALS AND METHODS
User input
The 3Drefine web server only requires a job name and the protein structure for the refinement process. The job name is required to keep track of the refined structure and to provide a unique web page to display the results. The required protein structure can be uploaded via text insertion in PDB format or file upload of a PDB file. An example option is provided for testing the services. Optionally, users can also enter an email address for notification upon completion. Additionally, an optional post refinement analysis of refinement can be performed by two structure assessment tools: (1) RWplus (10), a pair-wise distance-dependent, atomic statistical potential function combined with side-chain packing orientation specificity that evaluates global qualities of the refined models; and (2) MolProbity (11), a log-weighted combination of the clashscore, percentage Ramachandran not favored and percentage bad side-chain rotamers that assesses physical realism and local errors of the refined models.
Server processing
Jobs sent from the web server are entered into a queuing system for all jobs being processed. Once ready, the given protein structure is received by the server which performs the i3Drefine algorithm (8). i3Drefine is basically an iterative version of 3Drefine refinement protocol. In i3Drefine, the starting model is refined using 3Drefine protocol (7) and the resulting refined model is again processed by the same method. This iteration is done five times in order to generate five refined models for the starting structure. Since 3Drefine employs restrained backbone flexibility during energy minimization, such an iterative scheme is effective to escape any local minima and move closer to the native structure. Upon completion, the results of the structure refinement are displayed on a unique web page and an email notification is sent, if specified.
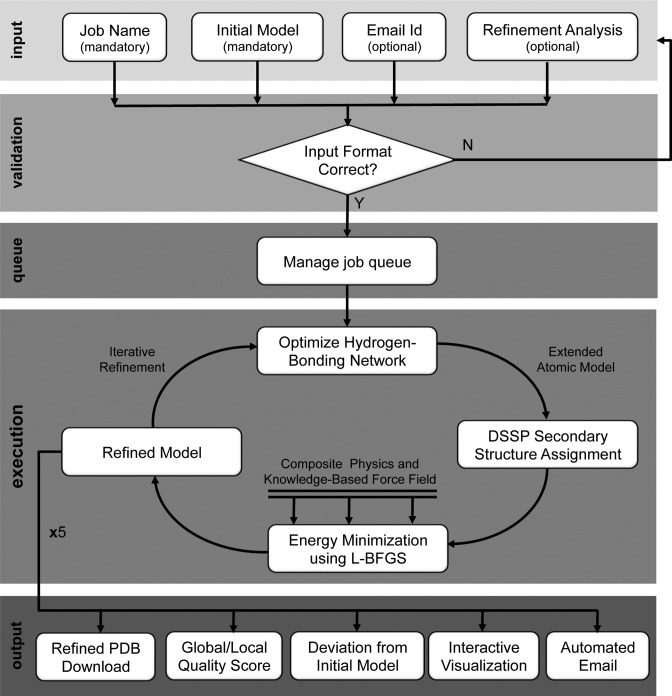
The i3Drefine refinement process involves an iterative implementation of two-steps: optimizing hydrogen bonding network and atomic-level energy minimization using a combination of physics and knowledge based force fields; implemented using the molecular modeling package MESHI (9). Given a starting structure for refinement, a combination of local geometry restraint and a conformational search is first performed to optimize the hydrogen bonding network. Subsequently, 200 000 steps of energy minimization is employed on the optimized model using highly convergent limited memory Broyden–Fletcher–Goldfarb–Shannon (L-BFGS) (12) algorithm or until convergence to machine precision using a customized all-atom force field. The force field consists of a combination of physics based and knowledge based terms. The physics-based terms include the energetic contributions of the bonded interactions described in ENCAD potential (13) (bond length, bond angle and torsion angle) along with a tethering term of the Cα and Cβ atoms (7) and the knowledge-based terms include the atomic pairwise potential of mean force (6) and explicit hydrogen bonding potential. The resulting energy-minimized model is the refined model. A detailed analysis of the relative importance of these energy terms and the i3Drefine algorithm has been presented in the published work of i3Drefine (7,8). Figure 1 visualizes the 3Drefine web server workflow.
Figure 1.
Flowchart of 3Drefine web server workflow depicting input, validation, queue, execution and output layers of the server.
Server output
Upon completion of the refinement process users are presented with a comprehensive set of results that include the following: (i) five different refined protein models derived from the initial input model are provided. Each model can be downloaded in PDB format or the entire folder can be downloaded. (ii) The potential energies of all five refined model after energy minimization are displayed for analysis. (iii) For each refined model, the similarities and deviations from the supplied input structure can be analyzed through provided metrics including GDT-TS (14), GDT-HA (15) and RMSD (16), as well as RWplus (10) and Molprobity (11) if selected by the user. (iv) Web based interactive visualization is also provided via JSmol, a JavaScript applet version of Jmol (17). Initially, the first refined model is represented in JSmol with rainbow coloring, spin enabled and cartoon structure style. Checkboxes allow the user to toggle visualizations of the refined structure and the initial structure, which is displayed in a white color. Additionally, through JSmol, users are enabled access to all the protein model analysis tools and features of Jmol within the applet (17). (v) A unique web URL is provided containing the above information. Users are able to bookmark the page for future reference. (vi) Automated status update of the job via email is provided if specified. Figure 2 depicts the web server workflow with a continuous screenshots of the process including a screenshot of the output page.
Figure 2.
Representative example of a refinement job processed through 3Drefine web server visually presented through various screenshots of the process starting from the home page through the queuing page, executing page and finally, output page.
Web server configuration
The 3Drefine service does not require registration and is freely available. The web server is hosted on a Linux server with an Intel Xeon 2.4GHz CPU with 16 cores and has 16 GB of memory. The web interface utilizes apache HTTP, HTML, cgi-bin, Perl, JavaScript and jQuery technologies for functionality and the JSmol web applet for protein analyzing and visualization (17). The underlining i3Drefine software uses the java programming language as well as MESHI (9) and DSSP libraries (18).
Performance of web server
The 3Drefine web server has been tested extensively to perform protein structure refinement in an efficient and consistent manner. The web server utilizes the i3Drefine protocol that typically requires <5 min of computation for a protein of typical length (300 residues) for a single iteration and therefore needs around 25 min to generate 5 refined models. The site has serviced several thousand users with >19 000 protein structure submissions since 2013.
The 3Drefine process itself has also been proven extensively to perform successful structure refinement. Performance of 3Drefine was benchmarked using refinement targets from CASP8 (12 targets) (19) and CASP9 (14 targets) refinement targets in a completely automated manner, without using the knowledge of additional information. The models refined by 3Drefine have shown improvement of the global topologies of the starting models as measured by GDT-TS, TM-score and Cα RMSD to native structures as well as the local structure qualities as measured by the MolProbity score (11). The overall results of 3Drefine were better than or comparable to other top methods worldwide (19,20). Additionally, the i3Drefine algorithm (8) was tested in complete blind mode during CASP10 (group name MULTICOM-CONSTRUCT) and exhibited consistent improvement over the initial structures in both global and local structural quality metrics as demonstrated by diverse quality metrics. i3Drefine was ranked as the best method in the server section (8) in CASP10 refinement experiment. The biggest advantage of i3Drefine method compared to other state-of-the-art refinement methods like KobaMin (21) and GalaxyRefine (22) is consistency. Like CASP10, in CASP11 refinement experiment, i3Drefine method (group name MULTICOM-CONSTRUCT) demonstrated consistent global quality improvement as measured by GDT-TS score with 31 out of 37 refinement targets being either improved or unaltered in terms of best of top five submissions. The degree of improvement, however, is marginal in nature with average GDT-TS points getting improved from 72.2 to 72.5 averaged over 37 CASP11 refinement targets. The users of 3Drefine web server should therefore, expect slight, albeit consistent, improvement of global qualities due to refinement.
CONCLUSION
The 3Drefine web server provides convenient access to efficient and consistent protein structure refinement through the i3Drefine protocol. The method achieves consistent performance through an iterative, two-step process involving optimization of hydrogen bonding network combined with atomic-level energy minimization on the optimized model using a composite physics and knowledge-based force fields (7,8). This post-tertiary structure prediction refinement software is packaged through the 3Drefine web server in an easy to use manner through text or file upload submission, example data, email notification and the absence of a registration system. Furthermore, results can conveniently be analyzed through the use of statistical feedback, multiple refinement results and interactive visualization and analysis tools through the JSmol web app (17). Therefore, the 3Drefine web server conveniently provides an efficient tool for protein model refinement to the community.
FUNDING
National Institutes of Health [R01GM093123 to J.C.]. Funding for open access charge: National Institutes of Health [R01GM093123].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wieman H., Tondel K., Anderssen E., Drablos F. Homology-based modelling of targets for rational drug design. Mini Rev. Med. Chem. 2004;4:793–804. [PubMed] [Google Scholar]
- 2.Mendez R., Leplae R., Lensink M.F., Wodak S.J. Assessment of CAPRI predictions in rounds 3-5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 3.Arakaki A.K., Zhang Y., Skolnick J. Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–1096. doi: 10.1093/bioinformatics/bth044. [DOI] [PubMed] [Google Scholar]
- 4.Keskin O., Bahar I. Packing of sidechains in low-resolution models for proteins. Fold. Des. 1998;3:469–479. doi: 10.1016/S1359-0278(98)00064-9. [DOI] [PubMed] [Google Scholar]
- 5.Kryshtafovych A., Venclovas C., Fidelis K., Moult J. Progress over the first decade of CASP experiments. Proteins. 2005;61(Suppl. 7):225–236. doi: 10.1002/prot.20740. [DOI] [PubMed] [Google Scholar]
- 6.Summa C.M., Levitt M. Near-native structure refinement using in vacuo energy minimization. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3177–3182. doi: 10.1073/pnas.0611593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya D., Cheng J. 3Drefine: consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization. Proteins. 2013;81:119–131. doi: 10.1002/prot.24167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya D., Cheng J. i3Drefine software for protein 3D structure refinement and its assessment in CASP10. PLoS One. 2013;8:e69648. doi: 10.1371/journal.pone.0069648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalisman N., Levi A., Maximova T., Reshef D., Zafriri-Lynn S., Gleyzer Y., Keasar C. MESHI: a new library of Java classes for molecular modeling. Bioinformatics. 2005;21:3931–3932. doi: 10.1093/bioinformatics/bti630. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Zhang Y. A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PLoS One. 2010;5:e15386. doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D.C., Nocedal J. On the limited memory BFGS method for large scale optimization. Math. Program. 1989;45:503–528. [Google Scholar]
- 13.Levitt M., Hirshberg M., Sharon R., Daggett V. Potential energy function and parameters for simulations of the molecular dynamics of proteins and nucleic acids in solution. Comput. Phys. Commun. 1995;91:215–231. [Google Scholar]
- 14.Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A. 1976;32:922–923. [Google Scholar]
- 17.Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L. JSmol and the next-generation web-based representation of 3D molecular structure as applied to Proteopedia. Isreal J. Chem. 2013;53:207–216. [Google Scholar]
- 18.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 19.MacCallum J.L., Hua L., Schnieders M.J., Pande V.S., Jacobson M.P., Dill K.A. Assessment of the protein-structure refinement category in CASP8. Proteins. 2009;77(Suppl. 9):66–80. doi: 10.1002/prot.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Liang Y., Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues J.P., Levitt M., Chopra G. KoBaMIN: a knowledge-based minimization web server for protein structure refinement. Nucleic Acids Res. 2012;40:W323–W328. doi: 10.1093/nar/gks376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heo L., Park H., Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41:W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]