Abstract
The structural modeling of protein–protein interactions is key in understanding how cell machineries cross-talk with each other. Molecular docking simulations provide efficient means to explore how two unbound protein structures interact. InterEvDock is a server for protein docking based on a free rigid-body docking strategy. A systematic rigid-body docking search is performed using the FRODOCK program and the resulting models are re-scored with InterEvScore and SOAP-PP statistical potentials. The InterEvScore potential was specifically designed to integrate co-evolutionary information in the docking process. InterEvDock server is thus particularly well suited in case homologous sequences are available for both binding partners. The server returns 10 structures of the most likely consensus models together with 10 predicted residues most likely involved in the interface. In 91% of all complexes tested in the benchmark, at least one residue out of the 10 predicted is involved in the interface, providing useful guidelines for mutagenesis. InterEvDock is able to identify a correct model among the top10 models for 49% of the rigid-body cases with evolutionary information, making it a unique and efficient tool to explore structural interactomes under an evolutionary perspective. The InterEvDock web interface is available at http://bioserv.rpbs.univ-paris-diderot.fr/services/InterEvDock/.
INTRODUCTION
Computational methods aiming to predict the structures of macromolecular assemblies provide key insights for the characterization of macromolecular cross-talks. Models of protein–protein complex structures have useful applications in guiding mutagenesis at the complex interface, in complementing integrative structural biology (1) and in understanding the roles of disease-related mutations at the surface of proteins (2). To disentangle the complexity of protein–protein interaction networks under a structural perspective, two major approaches have been developed, template-based modeling and template-free docking methods, both of them bringing complementary insights in the structural space of complexes (3).
In the field of template-free protein docking, a number of servers are already available either to perform generic rigid-body docking (4–12) or to implement more specific tasks such as symmetry (SymmDock (13) or Z-DOCK (11) servers) or data-driven docking under restraints (HADDOCK (14) and pyDockSAXS (15)). Docking servers distinguish themselves and complement one another on three major aspects: (i) in the sampling strategy which can rely on Monte Carlo searches (as in RosettaDock (6)) or on grid-based representation of proteins combined with 3D Fast-Fourier transforms (Z-DOCK (11), GRAMM-X (5)), some tools using spherical harmonics to accelerate the rotational search (Hex (8)); (ii) in the way flexibility is taken into account, from rigid body as in ClusPro (4) to the integration of various degrees of freedoms in loops and along normal modes (ATTRACT (12) or SwarmDock (10) servers); (iii) in the scoring strategies, relying on physical potentials as in pyDockWEB (9), or on a combination of both physics and statistical potentials as in Z-DOCK server (11). There have recently been a number of developments in the field of co-evolution-based contact prediction, leading in particular to the EVcomplex (16) and GREMLIN (17) servers that predict contacts in protein–protein complexes. However, to date, no docking server directly takes into account co-evolutionary constraints accessible from the multiple sequence alignments (MSA) of the binding partners. The InterEvDock server proposes to fill this gap through an integrated solution to perform rigid-body docking, accounting for co-evolution information.
We have previously shown that beyond conservation, incorporation of co-evolution constraints could significantly increase the recognition of correct protein–protein complex decoys (18). The InterEvScore potential was developed as a residue-based statistical potential, scoring the likelihood of contacts between two to three residues across an interface and for all the sequences in the joint alignments of both partners. The InterEvDock server predicts the structures of protein–protein complexes, using first the FRODOCK rigid-body docking program to generate a large body of potential decoys (19) and next running the InterEvScore potential (18) combined with other complementary scoring methods provided by the FRODOCK (19) and SOAP-PP potentials (20) to propose a limited set of 10 most likely models. The success rates of InterEvDock server was benchmarked on the 85 complexes of the Weng Benchmark 4 (BM4) (21) for which a pair of MSAs (with no fewer than 10 sequences) can be generated. 49% of the complexes in the rigid-body category have an acceptable or better solution in the top10 consensus of InterEvDock server. In this category, performances are higher than published benchmarks for the SwarmDock (10) or Zdock3.0.2 (11) servers that reach 42% and 28% on the same set, respectively. For the cases in the Weng BM4 difficult category, the InterEvDock success rate is lower than the one of the rigid-body cases, consistent with the sampling strategy which incorporates no flexibility in the docking process. InterEvDock server also predicts a set of 10 residues most likely part of the interface with at least one residue properly predicted in 91% of the 85 test cases.
THE InterEvDock SERVER
Web interface
Users can either upload their own PDB files or retrieve the chain of interest by typing the PDB code and chain id for automatic retrieval. The sequence used for MSA construction is automatically extracted from the PDB. MSAs can also be uploaded by the user provided their first sequence matches that of the PDB file and that they respect the same order of species as in the automatic mode. Uploading alignments is for instance required when one of the input partners is a fusion of more than one protein chain. In case not enough sequences are found by InterEvDock or in case of a protein family with many paralogs, users can also generate the MSAs using the InterEvolAlign server that additionally implements a reciprocal blast search procedure but is more time consuming (22) (http://biodev.cea.fr/interevol/interevalign.aspx). The structures of the best selected decoys can be explored thanks to the PV applet (M. Biasini), a WebGL-based viewer for proteins and other macromolecular structures. Coordinates of the complexes and alignments are available for further off-web exploration. A demonstration test is accessible from InterEvDock page. The InterEvDock server is implemented in the Mobyle framework in which data privacy is ensured by two means. First, anonymous runs are isolated in separate directories depending on a 16 character key randomly generated, and are kept on the server for one month. User-logged sessions are in addition protected by a password and data are kept permanently. Note there is no restriction for anybody to request a user session.
Rigid-body docking
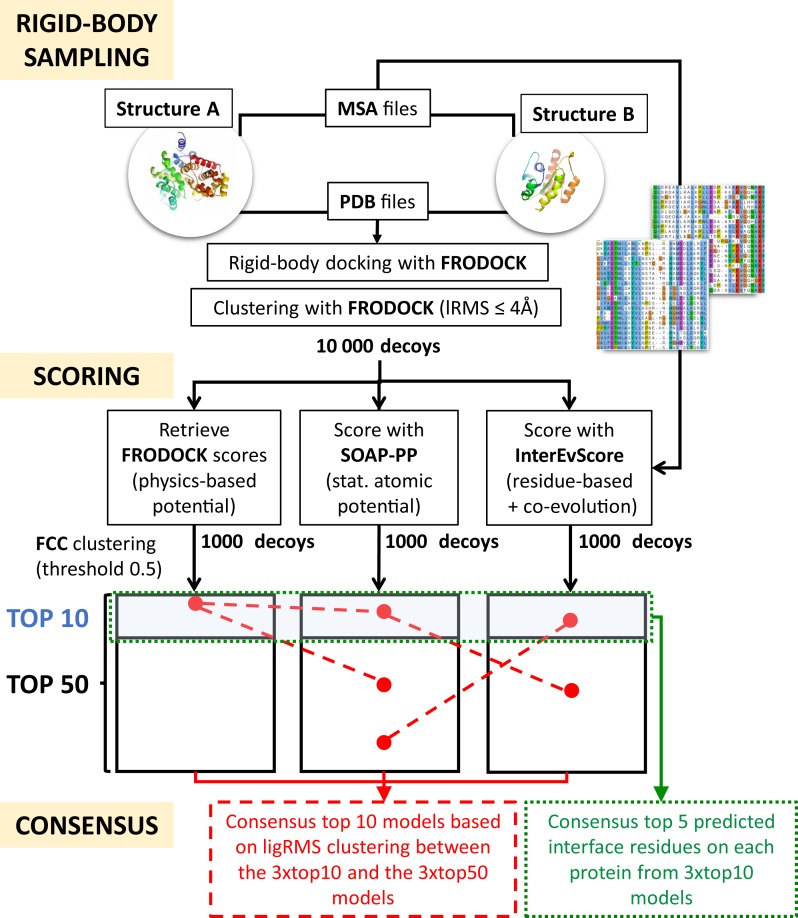
The server first performs an exhaustive rigid-body docking search between two input structures using FRODOCK (19) which combines a search algorithm based on spherical harmonics and an energy-based scoring function including van der Waals, electrostatics and desolvation terms. Recently challenged (23) on Weng BM4, the version currently running in InterEvDock was found to generate successful models with ligand Root Mean Square Deviation (RMSD) below 10 Å in up to 81.8% of the top2000 models, very close to ZDOCK3.0.2 performances (84.1%) (24). All decoys are clustered using frodockcluster method implementing an explicit comprehensive algorithm (25) with a ligand RMSD threshold of 4.0 Å and ranked with respect to their FRODOCK scores (Figure 1).
Figure 1.
Workflow of the InterEvDock pipeline. Three major steps are performed in InterEvDock: (i) the exhaustive sampling using the rigid-body method FRODOCK; (ii) the scoring by three scores, FRODOCK itself, SOAP-PP and InterEvScore; (iii) the clustering and selection of the best InterEvDock consensus.
Scoring
The best 10 000 FRODOCK cluster representatives are re-scored using InterEvScore (18) and SOAP-PP (20) potentials. InterEvScore is a scoring function using a coarse-grained statistical potential including two- and three-body interactions combined with evolutionary information. InterEvDock runs the calculation mode in which evolutionary information is scored only for residues belonging to apolar patches which was shown to give the best performance (18). SOAP-PP is a statistically optimized atomic potential (SOAP) designed so as to, for instance, capture orientation-dependent interactions such as hydrogen bonds (20). For each of the three scores, the top1000 models are clustered using the Fraction of Common Contacts (FCC) clustering method which groups together structural models according to their fraction of common contacts at the interface (for InterEvDock, the fraction of common contacts threshold was set to 0.5) (26) (Figure 1). FCC was shown to run very fast with specific advantages when dealing with symmetrical assemblies and large macromolecular complexes. Clusters are then ranked according to the mean score of the 30% models with best scores as in (18). The best model in each of the 10 best clusters is returned on the webpage (top10 predictions for every score) (Table 1) and the best models of the 50 best clusters are provided in the zip file. The scoring methods in InterEvDock server were all developed paying attention to the potential risks of overfitting. For instance, the structural data set used for the statistical development of InterEvScore was deprived of any structure related with the complexes of Weng's benchmarks and as for SOAP-PP, distinct test and validation data sets were considered. Criteria used to define a solution as acceptable follow those established by the Critical Assessment of Predicted Interactions (CAPRI) consortium (27).
Table 1. Performances of the predictions of the InterEvDock server for three levels of difficulty categories: rigid-body, medium and difficult.
| All | Rigid-body | Medium | Difficult | ||
|---|---|---|---|---|---|
| Number of cases | 85 | 43 | 23 | 19 | |
| Top 10 success rate | InterEvScore | 21 (25%) | 19 (44%) | 2 (9%) | 0 (0%) |
| SOAP_PP | 17 (20%) | 14 (33%) | 2 (9%) | 1 (5%) | |
| FRODOCK v1 | 13 (15%) | 10 (23%) | 2 (9%) | 1 (5%) | |
| InterEvDock consensus | 25 (29%) | 21 (49%) | 4 (17%) | 0 (0%) | |
| SwarmDock server 2013 | 25 (29%) | 18 (42%) | 6 (26%) | 1 (5%) | |
| Zdock 3.0.2 | 17 (20%) | 12 (28%) | 3 (13%) | 2 (11%) | |
| Residue interface prediction | InterEvDock ≥1 correct in top 10 (5 receptor + 5 ligand) | 77 (91%) | 40 (93%) | 20 (87%) | 17 (89%) |
| InterEvDock ≥1 correct in top 5 receptor AND in top 5 ligand | 46 (54%) | 24 (56%) | 12 (52%) | 10 (53%) | |
| Zdock3.0.2 ≥1 correct in top 10 (5 receptor + 5 ligand) | 75 (88%) | 39 (90%) | 20 (87%) | 16 (84%) | |
| Zdock3.0.2 ≥1 correct in top 5 receptor AND in top 5 ligand | 37 (44%) | 19 (44%) | 11 (48%) | 7 (38%) |
The benchmark is made of the 85 targets of the Weng benchmark v4 (176 cases) (21) for which pairs of co-evolved multiple sequence alignments with more than 10 sequences could be obtained. In the top lines, top10 success rates of six methods report the number of cases for which at least one model out of 10 is an acceptable or better solution (27). The methods are InterEvScore (18), SOAP-PP (20), FRODOCK (19), InterEvDock consensus (this work), Zdock3.0.2 (11) and SwarmDock (10). In the bottom lines, the number and percentage of cases for which at least one residue out of 10 could be predicted correctly as present in the complex interface are assessed for InterEvDock and Zdock3.0.2. The best result for each category is highlighted in bold.
Multiple sequence alignments for co-evolved partners
The InterEvScore potential is calculated based on MSAs of co-evolved homologs of both input partners. The server runs a fast and automatic protocol as used in (28) to retrieve homologs of both partners. For every input protein, homologous sequences are searched using a single BLASTp search (29) against the Uniprot-KB database (The UniProt Consortium, 2014) with the threshold sequence identity > 30%, coverage > 75% and E-value < 10–4. In case several sequences were found for one species, only the sequence with the highest sequence identity (and highest coverage if sequence identities are identical) is chosen. Pairs of sequences in every MSA belonging to the same species are collected. Redundant paired sequences with sequence identity higher than 90% are removed and MSAs are re-aligned by MAFFT program (30). In the end, a set of two MSAs is recovered containing exactly the same number of sequences in the same species order. When less than 10 sequences are retrieved, a warning message in the log indicates that little information will be gained from the MSAs and that models selected by InterEvScore may be less reliable.
Consensus models selection
From the top10 models of each of the three scoring methods, InterEvDock server extracts a consensus set of 10 models. Models similar between the three scoring methods are clustered together (two models are considered similar if they have a ligand-RMSD below 10 Å). The first selection step is then to take the best representative models for every score (4, 3 and 3 from InterEvScore, SOAP-PP and FRODOCK, respectively). Such a selection does not account for the fact that models well predicted by at least two different scoring methods have more chances to be correct. To account for this property, each of the 3*top10 decoys is assigned a score reporting the number of similar decoys selected in the top50 of the other two scores. Decoys connected with at least two other decoys are ranked and inserted in the top10 list of decoys. In case of equality, priority is given to InterEvDock decoys, next to SOAP-PP and then FRODOCK.
Selection of five residues on each partner
From the top10 models of each of the three scoring methods, the InterEvDock server extracts a consensus set of five residues predicted as the most likely interface residues on each of the two partners. This selection is performed based on the number of decoys displaying contacts involving those residues among the 30 decoys. Contacts are defined as in CAPRI (27), i.e. two residues are assumed to be in contact if any non-hydrogen atom in the first residue is within 5 Å of any atom in the second residue. In the event of a tie, residues are selected on the basis of the number of decoys with contacts involving this residue among the 10 best InterEvScore decoys; if this does not resolve the tie, a second step of selection is performed based on the 10 best SOAP-PP decoys.
RESULTS
Benchmarking
InterEvDock server predictions were assessed using the subset of the Weng BM4 (21) for which sufficient co-evolutionary information could be retrieved. It corresponds to the 85 complexes previously described (18) which do not for instance include either antibody-antigen or pathogen-host complexes (http://bioserv.rpbs.univ-paris-diderot.fr/services/InterEvDock/table.html). Targets of the Weng BM4 are classified into three difficulty levels depending on the interface RMSD (iRMSD) and the fraction of non-native residue contacts (F-non-nat) obtained after superimposition of the unbound structures with the reference complex (21). ‘Rigid-body’ category corresponds to cases with iRMSD < 1.5 Å and F-non-nat < 40%, ‘difficult’ category has iRMSD > 2.2 Å and the remaining cases belong to the ‘medium’ class. The subset of 85 targets is enriched in ‘medium’ and ‘difficult’ targets with respect to ‘rigid-body’ ones. 70% of the complexes in the entire Weng BM4 (176 complexes) belong to the ‘rigid-body’ category while they represent only 51% of the 85 complexes subset. For the ‘rigid-body’ category (43 cases, see Table 1), InterEvScore, SOAP-PP and FRODOCK individual scores identified an acceptable or better solution in 44%, 33% and 23% of the cases, respectively. The consensus model selection of InterEvDock server increases the rate of success up to 49% of the cases in the top10, emphasizing the interest of combining together three scores reporting for different interface properties. Detailed results about the consensus models are reported in Supplementary Table S4.
The results of InterEvDock server were compared to those published for the Zdock (11) and SwarmDock (10) servers on the Weng benchmark v4 (21) (Table 1). Zdock3.0.2 is a widely used and efficient rigid-body docking method with a large number of successes in the international protein–protein docking experiment (CAPRI). The SwarmDock web service (10) is a reference server for the flexible modeling of protein–protein complexes based on algorithm which incorporates a normal mode approach. Overall, InterEvDock and SwarmDock performed similarly. For the rigid-body category, InterEvDock consensus performance was higher than SwarmDock (10), but the latter performed better on medium and difficult cases. These results are consistent with the advantages of SwarmDock running flexible docking steps, yet at the cost of longer simulations. As for SwarmDock, a typical docking run was estimated to take up to 36 h (10). Tested on 60 complexes from the Weng BM4, it took on average 45 min for InterEvDock server to return results with 20 targets submitted at a time. When the 15 largest structures of heterodimeric complexes were selected (500–1000 residues in total), the average running time was 1 h 30 min. On average the multiple sequence co-alignment step lasted 20 min. Interestingly, about 30% of the targets predicted by InterEvDock were not predicted by SwarmDock and vice-versa (Supplementary Table S1) (as the example in Figure 2B 2G77, see below). When comparing Zdock and InterEvDock, more than 40% of the targets were correctly predicted by InterEvDock but not by Zdock, while only for 17% of cases, good solutions were obtained by Zdock but not by InterEvDock (Supplementary Table S1). Altogether this benchmark emphasizes the complementarity of the different approaches tested and the interest in combining their results to increase the coverage of complexes which can be successfully predicted.
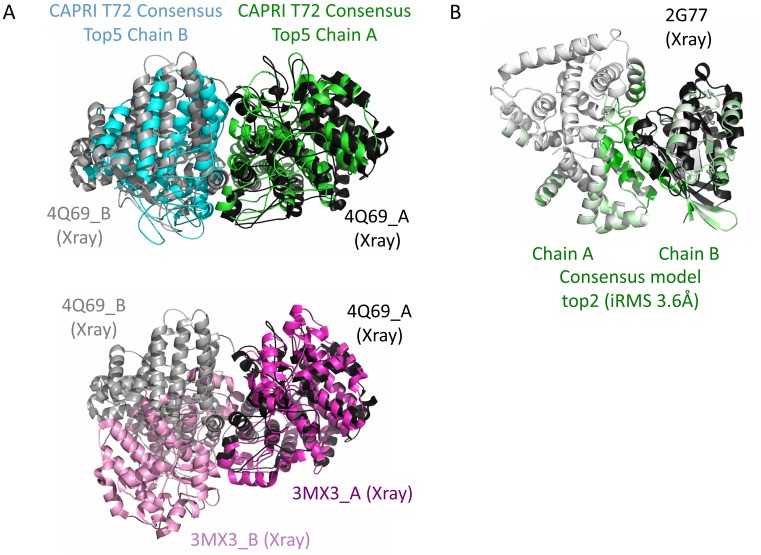
Figure 2.
Successful examples from a CAPRI challenge and from Weng benchmark v4. (A) The challenge for CAPRI target T72 (round 30) was to predict the homodimeric assembly of a protein whose monomeric structure could be modeled at 20% sequence identity using the template 3MX3 (pink structure). Modeling the homodimer using template-based approach based on 3MX3 dimeric assembly (pink) would have led to incorrect prediction since the experimental structure of T72 (4Q69 shown in black and gray) was eventually found to assemble in a different manner. Still, during CAPRI round 30 our group obtained the lowest iRMSD of 3.5Å for this prediction which was only well predicted by two other methods (Haddock and SwarmDock). InterEvDock server successfully identified in its top10 consensus an acceptable model rank as top5 (cyan and green model). (B) 2G77 example from Weng benchmark v4 (21). The model is shown as white and green cartoon while the X-ray structure is in black with only the B chain displayed. Green color reports for the likelihood of a residue to be at the interface calculated from the consensus method over the 3xtop10 decoys. It supports the prediction of the 10 most likely residues at interface provided by the server which is very precise for 2G77 with all 10 predicted residues at interface. InterEvDock consensus detected an acceptable model ranked top2 although it was missed by SwarmDock. This figure illustrates that the residues predicted to lie in the consensus interface provide very good hints to guide the mutagenesis.
Table 1 also reports the capacity of InterEvDock server to predict residues making contacts at the interface of a complex based on the analysis of all the interfaces of the top10 decoys for all three scores (30 models). In up to 91% of the cases, at least one residue out of 10 could be correctly predicted as present at the interface. There is almost no decrease in precision from the rigid body to the difficult cases (93% to 89%, respectively) (detailed statistics for receptor and ligand independently are provided in Supplementary Table S2). In the perspective of providing ambiguous docking restraints, we can note that in 54% of the cases, at least one correct residue is predicted on both sides of the interface, with very close performances for both rigid-body and difficult targets. Zdock reached a lower performance with 44% of the cases in which at least one correct residue is predicted on both sides of the interface. Performance was also compared with the EVcomplex server (16) on a subset of 13 bacterial complexes of Weng BM4 (Supplementary Table S3). In most cases, InterEvDock performed better with 6 versus 2 targets out of 13 for which residues could be correctly predicted on both sides of the interface. Altogether, predictions of InterEvDock server can thus be used for several types of applications. For bioinformaticians, these predictions can provide useful priors to help focusing docking simulations and refinement on specific regions. They can provide a good starting point for methods that integrate more flexibility in the structures of the binding partners. For experimentalists, they may help in designing interface mutants helpful to dissect the functional role of an interaction.
Examples from CAPRI
The consensus predictions provided by the InterEvDock methods were previously used to guide functional analysis of complexes (31) and also challenged through our participation in all the CAPRI rounds since 2013 (from round 28 to round 35). So far, only the results of round 30 which was coupled to CASP11 were officially communicated. In round 30 (targets T68–T94), our group performed among the top3 best CAPRI predictors (Lensink et al., in revision, see http://www.ebi.ac.uk/msd-srv/capri/round30/CAPRI_R30_v20141224.SW.pdf). In particular, relying on the scoring methods used in the InterEvDock pipeline, for target T72, one of our models was ranked the best according to the interface RMSD (3.5 Å) while only two other predictors managed to identify an acceptable solution. For this target, InterEvDock server selects as top5 of its consensus selection, an acceptable model with respect to the crystal structure of the complex (Figure 2A). For other recent CAPRI rounds, partial results are already available. For Target T59 proposed in round 28, we were the only group proposing a model reaching the medium accuracy level (https://www.ebi.ac.uk/msd-srv/capri/round28/R28_T59/, our group was predictor P07, unpublished work). In InterEvDock server, InterEvScore ranks an acceptable solution for T59 in the top1, and the consensus selection between all three scores returns an acceptable solution in the top2 position. These examples illustrate the robustness of the InterEvDock server in identifying original solutions that are not easily detected by other methods.
Step-by-step description of a running case
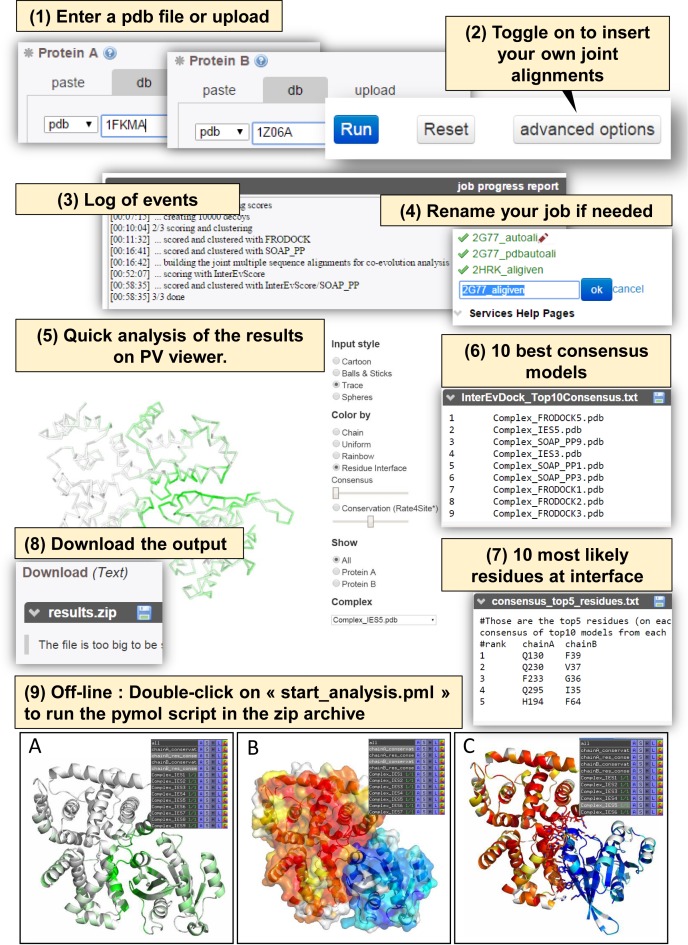
To illustrate how the server runs and how results can be exploited, let us consider an example from the Weng benchmark v4 of the complex 2G77 to be predicted from the unbound proteins the GTPase-activating protein GYP1 (PDB:1FKM chain A) and the Ras-related protein Rab-33B (PDB:1Z06 chain A). The InterEvDock consensus ranks an acceptable solution in the top2 (Figure 2B). PDB files of the unbound form can be uploaded from the input files distributed in Weng BM4 (http://bioserv.rpbs.univ-paris-diderot.fr/services/InterEvDock/table.html) (or directly retrieved using the PDB codes and the chain label (Figure 3 step 1)). As shown in Figure 3 step 2, users can also upload their own MSAs by toggling on the advanced options. As the server runs, a log of events is provided indexed by the time required for each step to be completed (Figure 3 step 3). At any time, the name of the job can be modified for the sake of clarity as illustrated in Figure 3 step 4. As output, the server returns first a graphical panel from which top10 models for each score can be visualized (Figure 3 step 5). In this example, the models Complex_IES2 and Complex_IES5 are acceptable and Complex_FRODOCK9 is medium according to CAPRI criteria (Figure 3 step 6). Several display options allows mapping information useful for the analysis. The first useful display mode is the residue interface consensus scheme in which residues are colored from green to white, from most to least likely to be involved in interface of the 30 decoys (Figure 3 step 5 & step 9). The second coloring scheme allows the visualization of the conservation index as calculated by the rate4site algorithm (32,33) (Figure 3 step 9 for pymol representation). The other important information concerns residues that are predicted as part of the interface (Figure 3 step 7). In the case of 2G77, the prediction works very efficiently since all ten residues are part of the correct interface. Decoy structures can be downloaded from the html table or from the zip file attached below (Figure 3 step 8). All these results can also be viewed easily thanks to the pymol script ‘start_analysis.pml’, included in the zip archive (Figure 3 step 9).
Figure 3.
Example of input and output in the InterEvDock server. InterEvDock integrates a number of important features that are indexed from 1 to 9. (1) Input pdbs can be either uploaded or retrieved automatically by specifying the pdb code and the chain. (2) Multiple sequence alignments can be either automatically generated on the server or uploaded by clicking on this option. (3) A log allows following the time required for the different steps of the docking. (4) Jobs can be easily renamed in the left panel at any time. (5) At the end of the docking process, the top10 decoys for every scoring methods can be analyzed in the PV viewer. Several coloring options are proposed to map either the probability of a residue to be at interface (from green to white) or the conservation index as calculated by rate4site (32,33) (from red to white through yellow). (6) List of the 10 best consensus models predicted by InterEvDock server. (7) List of the 10 most likely residues at interface predicted from the consensus analysis. (8) Zip archive containing all the top50 models selected by InterEvScore, SOAP-PP and FRODOCK. (9) A ‘start_analysis.pml’ pymol script is distributed in every zip archive which can be opened in a single click to generate pre-processed views of the 3xtop10 models, including color mapping schemes as in (5).
CONCLUSION
InterEvDock is the first rigid-body docking server accounting for co-evolutionary information. It relies on the integration of various components for decoy generation and scoring. Particularly, the combination of the SOAP-PP, FRODOCK and InterEvScore makes it very efficient for the identification of complex conformations not undergoing large conformational changes. The server has many advantages: a user-specific workspace for easy job management, fast evaluation of several tens of thousands of models, high success rates of the consensus method and a user-friendly graphical interface. Owing to its modular architecture, InterEvDock could in the future benefit from the contributions of new scores. Another interesting direction could be to consider the modeling of complexes of higher order, which seems feasible owing to the reasonably low calculation time, but which remains an open challenge. In its present form, InterEvDock should be a useful tool for biologists looking for hypotheses about mechanisms and residues involved in protein–protein interactions.
Supplementary Material
Acknowledgments
We are very grateful to Pablo Chacon (FRODOCK), Andrej Sali (SOAP-PP), João Rodrigues and Alexandre Bonvin (FCC), Tal Pupko (Rate4Site) and Marco Biasini (ProteinViewer) for making their methods available for implementation in the InterEvDock server.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Infrastructure for Integrated Structural Biology (FRISBI) [ANR-10-INSB-05-01]; ANR-IAB-2011-BIP:BIP [ANR-10-BINF-0003]; IFB [ANR-11-INBS-0013].
Conflict of interest statement. None declared.
REFERENCES
- 1.Rodrigues J.P., Bonvin A.M. Integrative computational modeling of protein interactions. FEBS J. 2014;281:1988–2003. doi: 10.1111/febs.12771. [DOI] [PubMed] [Google Scholar]
- 2.Mosca R., Tenorio-Laranga J., Olivella R., Alcalde V., Ceol A., Soler-Lopez M., Aloy P. dSysMap: exploring the edgetic role of disease mutations. Nat. Methods. 2015;12:167–168. doi: 10.1038/nmeth.3289. [DOI] [PubMed] [Google Scholar]
- 3.Vreven T., Hwang H., Pierce B.G., Weng Z. Evaluating template-based and template-free protein-protein complex structure prediction. Brief. Bioinform. 2014;15:169–176. doi: 10.1093/bib/bbt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tovchigrechko A., Vakser I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyskov S., Gray J.J. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macindoe G., Mavridis L., Venkatraman V., Devignes M.D., Ritchie D.W. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:W445–W449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimenez-Garcia B., Pons C., Fernandez-Recio J. pyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics. 2013;29:1698–1699. doi: 10.1093/bioinformatics/btt262. [DOI] [PubMed] [Google Scholar]
- 10.Torchala M., Moal I.H., Chaleil R.A., Fernandez-Recio J., Bates P.A. SwarmDock: a server for flexible protein-protein docking. Bioinformatics. 2013;29:807–809. doi: 10.1093/bioinformatics/btt038. [DOI] [PubMed] [Google Scholar]
- 11.Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries S.J., Schindler C.E., Chauvot de Beauchene I., Zacharias M. A web interface for easy flexible protein-protein docking with ATTRACT. Biophys. J. 2015;108:462–465. doi: 10.1016/j.bpj.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries S.J., van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez-Garcia B., Pons C., Svergun D.I., Bernado P., Fernandez-Recio J. pyDockSAXS: protein-protein complex structure by SAXS and computational docking. Nucleic Acids Res. 2015;43:W356–W361. doi: 10.1093/nar/gkv368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopf T.A., Scharfe C.P., Rodrigues J.P., Green A.G., Kohlbacher O., Sander C., Bonvin A.M., Marks D.S. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014;3:e03430. doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ovchinnikov S., Kamisetty H., Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreani J., Faure G., Guerois R. InterEvScore: a novel coarse-grained interface scoring function using a multi-body statistical potential coupled to evolution. Bioinformatics. 2013;29:1742–1749. doi: 10.1093/bioinformatics/btt260. [DOI] [PubMed] [Google Scholar]
- 19.Garzon J.I., Lopez-Blanco J.R., Pons C., Kovacs J., Abagyan R., Fernandez-Recio J., Chacon P. FRODOCK: a new approach for fast rotational protein-protein docking. Bioinformatics. 2009;25:2544–2551. doi: 10.1093/bioinformatics/btp447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong G.Q., Fan H., Schneidman-Duhovny D., Webb B., Sali A. Optimized atomic statistical potentials: assessment of protein interfaces and loops. Bioinformatics. 2013;29:3158–3166. doi: 10.1093/bioinformatics/btt560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang H., Vreven T., Janin J., Weng Z. Protein-protein docking benchmark version 4.0. Proteins. 2010;78:3111–3114. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faure G., Andreani J., Guerois R. InterEvol database: exploring the structure and evolution of protein complex interfaces. Nucleic Acids Res. 2012;40:D847–D856. doi: 10.1093/nar/gkr845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S.Y. Exploring the potential of global protein-protein docking: an overview and critical assessment of current programs for automatic ab initio docking. Drug Discov. Today. 2015;20:969–977. doi: 10.1016/j.drudis.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Pierce B.G., Hourai Y., Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozakov D., Clodfelter K.H., Vajda S., Camacho C.J. Optimal clustering for detecting near-native conformations in protein docking. Biophys. J. 2005;89:867–875. doi: 10.1529/biophysj.104.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues J.P., Trellet M., Schmitz C., Kastritis P., Karaca E., Melquiond A.S., Bonvin A.M. Clustering biomolecular complexes by residue contacts similarity. Proteins. 2012;80:1810–1817. doi: 10.1002/prot.24078. [DOI] [PubMed] [Google Scholar]
- 27.Mendez R., Leplae R., De Maria L., Wodak S.J. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 28.Iserte J., Simonetti F.L., Zea D.J., Teppa E., Marino-Buslje C. I-COMS: Interprotein-COrrelated Mutations Server. Nucleic Acids Res. 2015;43:W320–W325. doi: 10.1093/nar/gkv572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisboa J., Andreani J., Sanchez D., Boudes M., Collinet B., Liger D., van Tilbeurgh H., Guerois R., Quevillon-Cheruel S. Molecular determinants of the DprA-RecA interaction for nucleation on ssDNA. Nucleic Acids Res. 2014;42:7395–7408. doi: 10.1093/nar/gku349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pupko T., Bell R.E., Mayrose I., Glaser F., Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl. 1):S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 33.Glaser F., Pupko T., Paz I., Bell R.E., Bechor-Shental D., Martz E., Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.