Abstract
The MPI Bioinformatics Toolkit (http://toolkit.tuebingen.mpg.de) is an open, interactive web service for comprehensive and collaborative protein bioinformatic analysis. It offers a wide array of interconnected, state-of-the-art bioinformatics tools to experts and non-experts alike, developed both externally (e.g. BLAST+, HMMER3, MUSCLE) and internally (e.g. HHpred, HHblits, PCOILS). While a beta version of the Toolkit was released 10 years ago, the current production-level release has been available since 2008 and has serviced more than 1.6 million external user queries. The usage of the Toolkit has continued to increase linearly over the years, reaching more than 400 000 queries in 2015. In fact, through the breadth of its tools and their tight interconnection, the Toolkit has become an excellent platform for experimental scientists as well as a useful resource for teaching bioinformatic inquiry to students in the life sciences. In this article, we report on the evolution of the Toolkit over the last ten years, focusing on the expansion of the tool repertoire (e.g. CS-BLAST, HHblits) and on infrastructural work needed to remain operative in a changing web environment.
INTRODUCTION
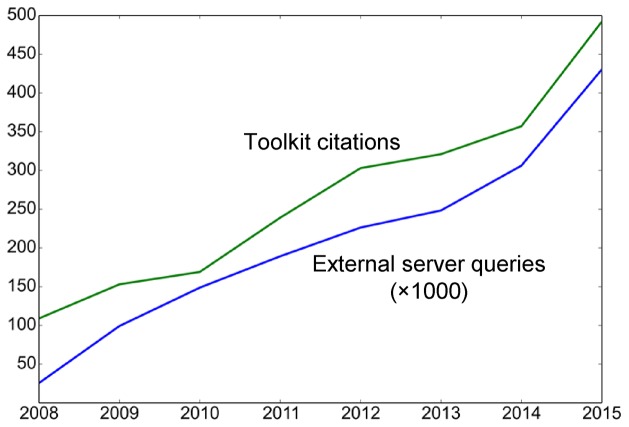
Over the last two decades, bioinformatic analysis of proteins has become central to molecular biology research. In fact, several new stand-alone tools as well as specialized web services are developed and published every month. However, the large majority are hard to install and/or use, and therefore remain out of reach for most bench biologists. To alleviate this problem, a number of easy-to-use web services have been developed, including the NCBI Resources (http://www.ncbi.nlm.nih.gov/guide/all), the SIB Bioinformatics Resource Portal (ExPASy; http://www.expasy.org), the EMBL-EBI Bioinformatics Web Services [(1); http://www.ebi.ac.uk/services], the Protein Analysis Toolkit [PAT (2); http://pat.cbs.cnrs.fr], the PredictProtein server [(3); https://www.predictprotein.org] and the CBS Prediction Servers (http://www.cbs.dtu.dk/services). Motivated by the work of our department in both computational and experimental biology, we wished to provide our colleagues at the bench with access to cutting-edge bioinformatics tools; we therefore developed a simple web-based system that combined the most useful external and internal tools. This evolved into the MPI Bioinformatics Toolkit, which we made public in a beta version in 2005 with 30 tools and published in the 2006 Web Server issue of Nucleic Acids Research (4). The current production-level release was launched in 2008 with 37 tools and has been up and running reliably ever since, servicing over 1.6 million external queries. Over this time period, the number of queries, as well as of citations of the Toolkit and the tools developed by us, have increased fairly linearly from year to year (Figure 1).
Figure 1.
Toolkit usage and citations. The number of queries from external IP addresses in thousands is shown by the blue line and the citations of the Toolkit framework and the tools developed by us, as indicated by Google Scholar, is shown by the green line.
Although the Toolkit has become an essential resource to experimental scientists mainly through its state-of-the-art remote homology detection tool HHpred (5), which is currently accessed more than 1000 times a day, its tools are in fact used broadly, with 18 of 53 called up more than five times a day on average (Tables 1 and 2). These highly accessed tools include both internal developments, such as CS-BLAST (6), HHblits (7) and PCOILS (8), and external tools, e.g. BLASTClust (9), Modeller (10) and MARCOIL (11) (Table 2). Interestingly, our 6FrameTranslation tool, which translates a given nucleotide sequence into the six possible frames, is also among the most accessed tools, suggesting that the Toolkit is not just a platform for advanced bioinformatic analyses, e.g. with HHpred, but that it has also grown into a general bioinformatics resource.
Table 1. An overview of tools available in the Toolkit.
| Category | Tools |
|---|---|
| Search | CS-BLAST (6), HHblits (7), HHpred (5), HHsenser (45), HMMER3 (46), PatternSearch, ProtBLAST (9), ProtBLAST+ (15), PSI-BLAST (14), PSI-BLAST+ (15), SimShiftDB (47), PDBalert (37) |
| Alignment | AlignmentViewer, Blammer, Clustal Omega (38), GLProbs (42), HHalign, Kalign (39), MAFFT (48), MSAProbs (40), MUSCLE (49), ProbCons (50), TCoffee (41) |
| Sequence Analysis | Aln2Plot, COILS/PCOILS (8), FRpred (51), HHrep (25), HHrepID (13), MARCOIL (11), REPPER (52), TPRpred (53) |
| Secondary Structure | Ali2D, HHomp (30), Quick2D |
| Tertiary Structure | bFit (54), HHfrag (55), HHpred (5), Modeller (10), SamCC (31) |
| Classification | ANCESCON (12), BLASTClust (9), CLANS (56), ClubSub-P (57), daTAA (28), GCView (58), HHcluster (27), PHYLIP-NEIGHBOR (59) |
| Utilities | 6FrameTranslation, Backtranslator, Extract GIs, GI2Promoter, HHfilter, Reformat, RetrieveSeq |
The categories listed here correspond to the section tabs in the menu bar located at the top of the Toolkit page. Section-specific tools are listed in a submenu within each tab. All tools can also be accessed through the tool index displayed on the homepage of the Toolkit. Tools developed by us are shown in boldface and tools added since our last publication on the Toolkit in 2006 are underlined.
Table 2. Tools used more than five times a day on average: usage in 2008 and 2015.
| Tool | 2008 | 2015 |
|---|---|---|
| Ali2D | 136 | 2620 |
| AlignmentViewer | 339 | 4997 |
| BLASTClust | 980 | 6531 |
| Clustal Omega | 575 | 3416 |
| CS-BLAST | 67 | 2063 |
| HHalign | 205 | 1906 |
| HHblits | – | 4353 |
| HHpred | 12562 | 328973 |
| MAFFT | 203 | 3637 |
| MARCOIL | – | 4289 |
| Modeller | 2401 | 20134 |
| MUSCLE | 414 | 2706 |
| PCOILS | 584 | 5542 |
| ProbCons | 245 | 2343 |
| PSI-BLAST | 1302 | 3049 |
| Quick2D | 811 | 5783 |
| 6FrameTranslation | 328 | 5298 |
| TPRpred | 364 | 4350 |
| Total (all 53 tools) | 25 611 | 430 296 |
Tools developed in our group are shown in boldface and tools added since our last publication on the Toolkit in 2006 are underlined.
KEY FEATURES
Our primary motivation behind developing and maintaining the Toolkit has always been the desire to provide experimental biologists (starting with our colleagues in our own department) with a simple web-based, one-stop platform that integrates a limited number of highly useful bioinformatic tools for the analysis of protein sequences and structures. In our opinion, the following features make our Toolkit useful to experts and non-experts alike:
Ease of use
We offer easy, web-based access to a number of tools that are otherwise only accessible from the command line and are often hard to install and get to work for a non-expert user [e.g. BLASTClust (9), ANCESCON (12), HHpred (5) or HHrepID (13)]. For many external tools, we also offer enhanced functionalities; for instance, our BLAST tools allow searches against nonstandard databases, such as the nonredundant (nr) protein sequence database clustered down to a pairwise sequence identity of 90% (nr90) or 70% (nr70), or personal databases uploaded by the user. Further, our implementations of PSI-BLAST (14) and PSI-BLAST+ (15) allow users to change the database between iterations. Using this feature, users can, for instance, train a profile on a large database (e.g. nr70) and then search for all homologs of the query protein in a specific genome or in a database uploaded by the user, with a much higher sensitivity than would be available if the profile had been trained only on the genome or the user database in question.
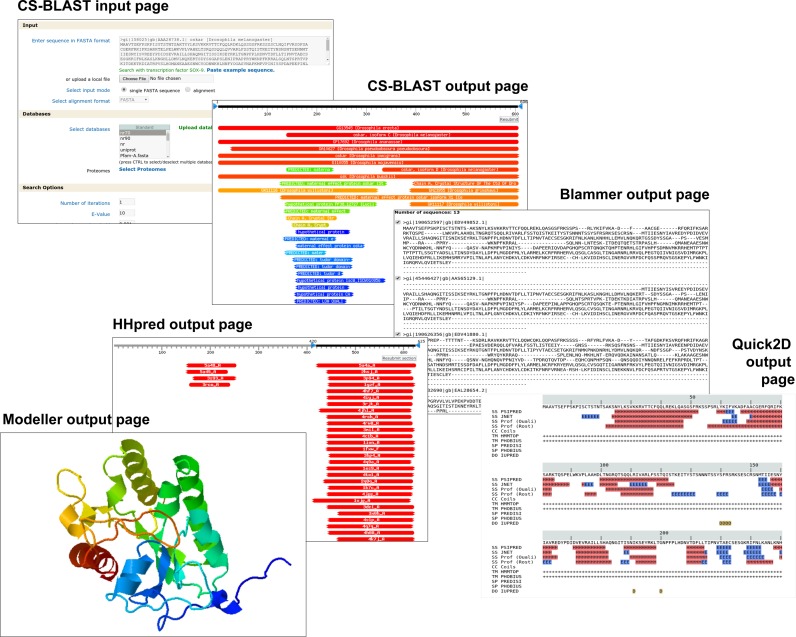
A further important feature of our Toolkit is the tight interconnection between most tools on offer, allowing the results of one tool to be forwarded as input to several others. The users could, for instance, start a sequence search with a protein of interest against a database of choice, using the sensitive search tool CS-BLAST, and then forward the results to Blammer to parse out a multiple alignment of the obtained sequence hits (Figure 2). This multiple alignment could then be forwarded further to Quick2D, to obtain an overview of secondary structure features such as α-helices, β-strands, coiled coils, transmembrane helices and intrinsically disordered regions. In parallel, the alignment could be forwarded to HHpred in order to detect distant homologs of known structure. Subsequently, the hits obtained from HHpred could be forwarded to Modeller (10) as templates for comparative modelling, resulting in a structural model for the protein of interest. This tight interconnection of the tools thus allows for complex bioinformatic analyses in a simple and straightforward manner, starting with just a single protein sequence.
Figure 2.
Interconnection of the tools in the Toolkit. The output of most tools can be forwarded as input to many other tools. One such possible forwarding pipeline is shown, wherein the output of the sensitive search method CS-BLAST is forwarded to Blammer to parse out a multiple alignment, which is subsequently forwarded to Quick2D for secondary structure prediction and HHpred for the identification of remote homologs. The output of HHpred is then forwarded to Modeller in order to obtain a structural model.
High quality, up-to-date tools and databases
Since we rely heavily on the Toolkit for our own research into the structure, function and evolution of proteins, we have been able to maintain its tools and databases at a high level and to detect and fix bugs rapidly. We update the databases regularly and ensure that the various internal and external tools are up-to-date as well. In addition to standard databases, such as the nonredundant protein sequence database (nr), the Protein Data Bank [PDB; (16)], the Pfam database (17) and the UniProt database (18) (including their variants, such as nr70 or nr90), we provide the databases of profile HMMs needed for HHblits [UniProt20] and HHpred [e.g. PDB70, SCOPe95 (19), Pfam (17), CDD (20), representative proteomes] and also allow users to upload their own sequence databases. We strive to react to bug reports and update/feature requests sent to us by our users in a timely manner and are currently engaged in revising and expanding our help pages.
Job management and personal workspace
One of the main design goals during the development of the Toolkit was to provide users with easy access to their jobs and the possibility to share them, in order to facilitate collaborative research. In line with this, every job submitted to the Toolkit is assigned either an automatic or a user-specified identifier, which upon submission of the job is displayed along with its current status in a sidebar on the left side of the browser. Users can click on previously submitted jobs to check their current status, get to the results page or return to an earlier job. Furthermore, users can share these job identifiers with their collaborators, allowing them to see the same output page as the user. For uploading custom sequence databases and to preserve jobs for a longer period of time, we offer users the possibility to create a personal account; their jobs are then stored for two months, rather than for two weeks, as without a log-in. Jobs in a personal account are private and cannot be viewed by other users. For a more detailed description of job submission and management, please refer to our previous paper on the Toolkit (4).
Links to external resources
Complementary to the tools offered in the Toolkit, we collate links to external tools that we think are particularly useful; these links are found on the front pages of the individual sections. For example, in the ‘Sequence Analysis’ section, we provide links to the function prediction servers The Seed (21) and String (22), and to the de-novo repeat detections servers RADAR (23) and TRUST (24). In this we emulate other highly used bioinformatic platforms such as ExPASy.
NEW TOOLS
Since our previous article in 2006, the Toolkit has grown from 30 to 53 tools (Table 1), more than half of which were developed internally. New developments concern sensitive sequence searching, address the classification of domains or are structure-based tools.
Sensitive sequence comparison tools
We have included two new sequence comparisons tools, CS-BLAST (6) and HHblits (7). CS-BLAST is a BLAST-like tool that gains sensitivity by including context-specific pseudocounts and finds twice as many homologs as BLAST at the same error rate and a comparable runtime (6). This tool can also be used iteratively and two iterations of it are typically more sensitive than five iterations of PSI-BLAST (6). HHblits is a remote homology detection tool based on iterative HMM–HMM comparison. In the first step, it converts the input sequence or multiple sequence alignment (MSA) to a profile HMM, which it then uses to iteratively search through profile HMMs in the UniProt20 database, employing an algorithm similar to the one used by HHsearch. Target sequences found to be significantly similar in each iteration are added to the query profile HMM for the next iteration. Compared to PSI-BLAST, HHblits is twice as sensitive, faster and produces alignments that are more accurate (7). We therefore now use HHblits as the preferred method for the MSA generation steps of HHpred (5), HHrep (25) and HHrepID (13). The latter is also a new addition to the Toolkit, built for the de novo detection of highly divergent repeats based on profile HMM comparison. We have previously used this tool to detect evidence for the homology of structural repeats in outer membrane proteins [OMPs; (26)] and TIM barrels (13).
Domain annotation/classification tools
Over the last years, we have developed and included further classification tools into the Toolkit. One, HHcluster, allows users to explore homologous connections between superfamilies with different structures in our galaxy of folds, which is a two-dimensional map of sequence relationships in protein fold space (27). We constructed this map by performing pairwise HMM-HMM comparisons for all domains in the SCOP database filtered to a maximum of 20% sequence identity and subsequently clustering them by a force-directed procedure using the statistical significance of these comparisons.
Two other tools address the detection of domains belonging to specific superfamilies. daTAA (28) provides a platform for the annotation of trimeric autotransporter adhesins (TAAs), an important family of pathogenic determinants in Gram-negative bacteria. TAAs present special challenges for automated domain annotation due to their high sequence diversity, mosaic-like arrangement of constituent domains, fuzzy domain boundaries and the frequent presence of extended regions of low sequence complexity, some of which we recognized as compositionally unusual coiled coils (29). daTAA meets these challenges through a combination of knowledge-based rules and HMM-based sequence analyses against manually curated alignments. The second, HHomp (30), is a tool for the prediction and classification of outer membrane proteins (OMPs), which are a major component of the outer membranes of Gram-negative bacteria, mitochondria and plastids. The transmembrane domains of OMPs comprise 4–12 β-hairpins that organize themselves around a central pore to form a β-barrel. We have previously shown that the β-barrels of all bacterial OMPs share a common ancestry and that they may have evolved by amplification of a single, ancestral β-hairpin (26). HHomp exploits this evolutionary observation; for a given input sequence, it builds a profile HMM and compares it with a database containing profile HMMs for ∼20 000 OMPs, in order to detect and classify new members.
Structure-based tools
We have also extended the repertoire of structure-based tools: (i) SamCC (31) measures the local structural parameters of parallel and antiparallel four-helical bundles, and compares these with the ideal values of four-helical coiled coils. We developed it in order to quantify departures from the ideal state and thus make variants of one domain comparable to each other in a quantitative way. Based on SamCC analyses of HAMP domains, we proposed a model for transmembrane signal transduction in TCST receptors (32–34). (ii) Ali2D is a tool that annotates multiple sequence alignments. It accepts MSAs as input, predicts the secondary structure of the constituent sequences with PSIPRED (35) and their membrane propensity with MEMSAT2 (36), and maps the results onto the MSA. This gives a consensus overview of secondary structure and membrane insertion in a given protein family, and alerts the user to potentially misaligned regions. Ali2D has become quite popular and was among the top third most accessed tools last year (Table 2). (iii) PDBalert (37), finally, is a tool that notifies users of the availability of PDB structures (released or on hold) with homology to a given protein of interest. This tool is only accessible from a personal user account (see above).
External tools
Of the external tools added to the Toolkit in recent years, most are multiple protein sequence alignment methods and include Clustal Omega (38), Kalign (39), MSAProbs (40), TCoffee (41) and GLProbs (42). Other newly incorporated external tools are the NCBI tools BLAST+ and PSI-BLAST+ (15) and the coiled coil detection tool MARCOIL (11).
TEACHING
In addition to establishing itself as a resource for protein bioinformatic analysis, the Toolkit has also become a useful platform for teaching bioinformatic enquiry to students in the life sciences. Due to its broad array of tools and their tight interconnection, its simple web interface, and its intuitive job management features that allow to pre-compute and share jobs, the Toolkit empowers students to efficiently progress to the scientific aspects of bioinformatic analysis, without the need to install programs and learn how to connect these with scripts. We ourselves use it as a primary resource to teach the ‘Bioinformatics for Biochemists’ practical course at the University of Tübingen and the graduate students of our institute. We are currently striving to make it more attractive for teaching purposes by including more detailed help pages and tutorials.
OUTLOOK
The growing use of the Toolkit gives us confidence that providing easy access to state-of-the-art bioinformatic tools will remain an important endeavor. In order to continue this and meet the software challenges of the next decade, our current focus is on replacing Java Applets with JavaScript-based solutions, to ensure the usability of our Toolkit on all different browsers. For instance, we now use JSmol (43), a JavaScript-based molecular viewer, to display protein structures, the BioJS MSA viewer (44) to display multiple sequence alignments, and the BioJS Tree viewer (44) to display phylogenetic trees. We are currently making the transition to accession.version identifiers for tools that use sequence GI identifiers, because NCBI is phasing out GIs this September. As mentioned earlier, Toolkit usage has grown linearly over the years, passing the 400 000 mark last year. This year we expect to cross the 500 000 mark and in anticipation of this and further growth in the future, we are upgrading our computational resources and are migrating to a more scalable architecture.
Acknowledgments
We would like to thank Andreas Biegert, Alexander Diemand, Klaus Faidt, Klaus O. Kopec, Jörn Marialke, Christian Mayer, Markus Meier, Andre Noll, Michael Remmert, Tina Streich, Christina Wassermann and Johannes Wörner for their contributions to the development and maintenance of the Toolkit over the years, as well as our current undergraduate students working on the Toolkit: Andrew Stephens, Jonas Kübler and Lukas Zimmermann. We would also like to thank all our users and members of our department for helping us to improve the Toolkit through their bug reports and feature requests. AL gratefully acknowledges Kristin K. Brown (GlaxoSmithKline) for many discussions, particularly in the early stages of the Toolkit.
FUNDING
Institutional funds of the Max Planck Society; German Federal Ministry of Education and Research (BMBF) (to J.S.) within the framework of e:Med [e:AtheroSysMed, 01ZX1313A-2014] and e:bio [SysCore]. Funding for open access charge: Institutional funds of the Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gracy J., Chiche L. PAT: a protein analysis toolkit for integrated biocomputing on the web. Nucleic Acids Res. 2005;33:W65–W71. doi: 10.1093/nar/gki455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T., Honigschmid P., Schafferhans A., Roos M., Bernhofer M., et al. PredictProtein–an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014;42:W337–W343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biegert A., Mayer C., Remmert M., Soding J., Lupas A.N. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biegert A., Soding J. Sequence context-specific profiles for homology searching. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3770–3775. doi: 10.1073/pnas.0810767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remmert M., Biegert A., Hauser A., Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 8.Gruber M., Soding J., Lupas A.N. Comparative analysis of coiled-coil prediction methods. J. Struct. Biol. 2006;155:140–145. doi: 10.1016/j.jsb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 11.Delorenzi M., Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- 12.Cai W., Pei J., Grishin N.V. Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 2004;4:33. doi: 10.1186/1471-2148-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biegert A., Soding J. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 2008;24:807–814. doi: 10.1093/bioinformatics/btn039. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox N.K., Brenner S.E., Chandonia J.M. SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heger A., Holm L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins. 2000;41:224–237. doi: 10.1002/1097-0134(20001101)41:2<224::aid-prot70>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.Szklarczyk R., Heringa J. Tracking repeats using significance and transitivity. Bioinformatics. 2004;20(Suppl. 1):i311–i317. doi: 10.1093/bioinformatics/bth911. [DOI] [PubMed] [Google Scholar]
- 25.Soding J., Remmert M., Biegert A. HHrep: de novo protein repeat detection and the origin of TIM barrels. Nucleic Acids Res. 2006;34:W137–W142. doi: 10.1093/nar/gkl130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remmert M., Biegert A., Linke D., Lupas A.N., Soding J. Evolution of outer membrane beta-barrels from an ancestral beta beta hairpin. Mol. Biol. Evol. 2010;27:1348–1358. doi: 10.1093/molbev/msq017. [DOI] [PubMed] [Google Scholar]
- 27.Alva V., Remmert M., Biegert A., Lupas A.N., Soding J. A galaxy of folds. Protein Sci. 2010;19:124–130. doi: 10.1002/pro.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szczesny P., Lupas A. Domain annotation of trimeric autotransporter adhesins–daTAA. Bioinformatics. 2008;24:1251–1256. doi: 10.1093/bioinformatics/btn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassler J., Hernandez Alvarez B., Hartmann M.D., Lupas A.N. A domain dictionary of trimeric autotransporter adhesins. Int. J. Med. Microbiol. 2015;305:265–275. doi: 10.1016/j.ijmm.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Remmert M., Linke D., Lupas A.N., Soding J. HHomp–prediction and classification of outer membrane proteins. Nucleic Acids Res. 2009;37:W446–W451. doi: 10.1093/nar/gkp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunin-Horkawicz S., Lupas A.N. Measuring the conformational space of square four-helical bundles with the program samCC. J. Struct. Biol. 2010;170:226–235. doi: 10.1016/j.jsb.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 32.Ferris H.U., Zeth K., Hulko M., Dunin-Horkawicz S., Lupas A.N. Axial helix rotation as a mechanism for signal regulation inferred from the crystallographic analysis of the E. coli serine chemoreceptor. J. Struct. Biol. 2014;186:349–356. doi: 10.1016/j.jsb.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Ferris H.U., Dunin-Horkawicz S., Hornig N., Hulko M., Martin J., Schultz J.E., Zeth K., Lupas A.N., Coles M. Mechanism of regulation of receptor histidine kinases. Structure. 2012;20:56–66. doi: 10.1016/j.str.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Ferris H.U., Dunin-Horkawicz S., Mondejar L.G., Hulko M., Hantke K., Martin J., Schultz J.E., Zeth K., Lupas A.N., Coles M. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure. 2011;19:378–385. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 36.Nugent T., Jones D.T. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal V., Remmert M., Biegert A., Soding J. PDBalert: automatic, recurrent remote homology tracking and protein structure prediction. BMC Struct. Biol. 2008;8:51. doi: 10.1186/1472-6807-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lassmann T., Sonnhammer E.L. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Schmidt B. Multiple protein sequence alignment with MSAProbs. Methods Mol. Biol. 2014;1079:211–218. doi: 10.1007/978-1-62703-646-7_14. [DOI] [PubMed] [Google Scholar]
- 41.Notredame C., Higgins D.G., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 42.Ye Y., Cheung D.W., Wang Y., Yiu S.M., Zhan Q., Lam T.W., Ting H.F. GLProbs: aligning multiple sequences adaptively. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;12:67–78. doi: 10.1109/TCBB.2014.2316820. [DOI] [PubMed] [Google Scholar]
- 43.Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L. JSmol and the next-generation web-based representation of 3D molecular structure as applied to proteopedia. Isr. J. Chem. 2013;53:207–216. [Google Scholar]
- 44.Yachdav G., Goldberg T., Wilzbach S., Dao D., Shih I., Choudhary S., Crouch S., Franz M., Garcia A., Garcia L.J., et al. Anatomy of BioJS, an open source community for the life sciences. Elife. 2015;4 doi: 10.7554/eLife.07009. doi:10.7554/eLife.07009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soding J., Remmert M., Biegert A., Lupas A.N. HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 2006;34:W374–W378. doi: 10.1093/nar/gkl195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginzinger S.W., Coles M. SimShiftDB; local conformational restraints derived from chemical shift similarity searches on a large synthetic database. J. Biomol. NMR. 2009;43:179–185. doi: 10.1007/s10858-009-9301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Do C.B., Mahabhashyam M.S., Brudno M., Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer J.D., Mayer C.E., Soding J. Prediction of protein functional residues from sequence by probability density estimation. Bioinformatics. 2008;24:613–620. doi: 10.1093/bioinformatics/btm626. [DOI] [PubMed] [Google Scholar]
- 52.Gruber M., Soding J., Lupas A.N. REPPER–repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 2005;33:W239–W243. doi: 10.1093/nar/gki405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karpenahalli M.R., Lupas A.N., Soding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mechelke M., Habeck M. Robust probabilistic superposition and comparison of protein structures. BMC Bioinformatics. 2010;11:363. doi: 10.1186/1471-2105-11-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalev I., Habeck M. HHfrag: HMM-based fragment detection using HHpred. Bioinformatics. 2011;27:3110–3116. doi: 10.1093/bioinformatics/btr541. [DOI] [PubMed] [Google Scholar]
- 56.Frickey T., Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 57.Paramasivam N., Linke D. ClubSub-P: cluster-based subcellular localization prediction for Gram-negative bacteria and archaea. Front. Microbiol. 2011;2:218. doi: 10.3389/fmicb.2011.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grin I., Linke D. GCView: the genomic context viewer for protein homology searches. Nucleic Acids Res. 2011;39:W353–W356. doi: 10.1093/nar/gkr364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]