Abstract
The Dali server (http://ekhidna2.biocenter.helsinki.fi/dali) is a network service for comparing protein structures in 3D. In favourable cases, comparing 3D structures may reveal biologically interesting similarities that are not detectable by comparing sequences. The Dali server has been running in various places for over 20 years and is used routinely by crystallographers on newly solved structures. The latest update of the server provides enhanced analytics for the study of sequence and structure conservation. The server performs three types of structure comparisons: (i) Protein Data Bank (PDB) search compares one query structure against those in the PDB and returns a list of similar structures; (ii) pairwise comparison compares one query structure against a list of structures specified by the user; and (iii) all against all structure comparison returns a structural similarity matrix, a dendrogram and a multidimensional scaling projection of a set of structures specified by the user. Structural superimpositions are visualized using the Java-free WebGL viewer PV. The structural alignment view is enhanced by sequence similarity searches against Uniprot. The combined structure-sequence alignment information is compressed to a stack of aligned sequence logos. In the stack, each structure is structurally aligned to the query protein and represented by a sequence logo.
INTRODUCTION
Comparative analyses of protein sequences and structures play a fundamental role in understanding proteins and their functions. Assuming an evolutionary continuity of structure and function, describing the structural similarity relationships between protein structures allows scientists to infer the functions of newly discovered proteins.
Until the early 1990s it was possible for one scientist to hold the shapes of all protein structures solved to date in memory. Structural classifications ((1,2) http://kinemage.biochem.duke.edu/teaching/anatax/) used to be done manually and visually. With the steadily growing size of the Protein Data Bank (PDB), automated protein structure comparison programs started to produce a string of discoveries, where an interesting fold similarity had been missed in the original publication of a structure (e.g. (3–10)). From the middle 1990s automated structure comparison programs started to gain acceptance and their wider use was propelled by their availability as network services ((11,12) and references therein). The current PDB contains over 110 000 structure entries and over 40 000 distinct structures (chains) with less than 90% sequence identity, making automated search and comparison tools necessary.
The Dali server (not to be confused with a similarly named server for Internet-Enabled Remote Control, http://www.diamondsystems.com/dali/) has processed over 175 000 PDB searches in the last 8 years (according to usage statistics at http://ekhidna.biocenter.helsinki.fi/dali_server/.results/stat.html). The Dali program optimizes a structural alignment, that is, a sequential set of one-to-one correspondences between C-alpha atoms (13,14). A large variety of scoring functions have been proposed (15). The most important categories of scoring functions are (i) those based on the root mean square deviation of rigid-body superimposition and (ii) those allowing flexible superimposition or plastic deformations. Early works based on visual analysis of folds stressed the importance of plastic deformations in the evolution of protein structure. Dali's scoring function belongs to the latter category, and it has been shown to yield structural dendrograms that agree well with expert classifications (13,16–18).
The major changes since the previous publication on the Dali server (12) have been made to the user interface. (i) Jmol has been replaced by PV (Marco Biasini. (2015). pv: v1.8.1. Zenodo. 10.5281/zenodo.20980) as protein viewer. Jmol is a Java applet. Unfortunately, Java has become very cumbersome to use due to security checks. A JavaScript successor of Jmol, JSmol is too slow on large proteins. PV is a JavaScript viewer built on WebGL to visualize protein structures directly in modern browsers. It is very fast and does not require plugins. (ii) The primary output from a PDB search is an ordered list of structural neighbours. What are the relationships between those structures? To answer this type of question, we added an option for all-against-all comparisons of a selected subset of structures. The structural relationships are visualized as a dendrogram (19) generated by agglomerative clustering. Correspondence analysis generates an alternative view of ‘structure space’ which does not enforce a strictly hierarchical bifurcating model. (iii) Structural information can be leveraged to homologous proteins based on sequence comparison. The Basic Local Alignment Search Tool (BLAST) program has dominated the field for many years but is too slow for interactive visualization purposes or multiple simultaneous searches. The advent of a new generation of very fast sequence search tools ((20) and references therein) has made it possible to generate sequence profiles on the fly. We introduce stacked sequence logos as a way to condense huge multiple sequence alignments that result from augmenting structural alignments with homologous sequences aligned to each of the participating structures. Instead of hundreds or thousands of sequence rows, we display just one logo per structure. Gaps are inserted in the logos so that conserved sequence motifs at structurally equivalent positions line up according to the structural alignment.
MATERIALS AND METHODS
The PDB database is mirrored from RCSB (http://www.rcsb.org/) and updated weekly.
There have been no major changes to the algorithms for structural alignment ((12,21,22); a full bibliography of methods is available from the Dali server web site). The server is hosted on a new computer cluster. Load balancing was improved by master/slave parallelization. Whilst this does not increase throughput, per se, users experience faster turnaround when the load is low.
The Dali server performs three types of structure comparisons: PDB search, pairwise comparison and all against all structure comparison. We have dropped the database option of the old server (12), which returned results from a pre-computed database. Usage statistics showed that at most a quarter of the pre-computed results would ever be looked at.
The all against all structure comparison is a new option. The user inputs a set of N structures and the server computes the N × N matrix of pairwise similarities (Dali Z-scores). Dali uses various heuristics to optimize the alignment score. Although Dali has been shown to generate close to optimal solutions on a benchmark of small proteins (23), we observed some gaps and inconsistencies in the matrix after direct pairwise comparison. The inconsistencies can be caused by poorly defined secondary structure, inconsistent definition of domain boundaries or too greedy optimization. The program therefore performs a few rounds of transitive alignment (involving triplets to improve the score of the weakest link) followed by refinement as long as the sum of Z-scores over the matrix increases. From the similarity matrix, a dendrogram is derived using average linkage clustering. An algorithm for correspondence analysis was recycled from code already included in the DaliLite package (21).
Sequence logos are computed for an input sequence. First, the SANSparallel server (20) is called to collect and align 100 sequence neighbours from the UniRef50 database. The alignment is converted to an HMMer profile and visualized by a Skylign server (24). Skyalign options are frag = frag and letter_height = info_content_above. One logo is generated in about 8 s. The slow step is the generation of the HMMer profile whilst the SANSparallel search only takes a fraction of a second.
RESULTS AND DISCUSSION
User interface
Inputs
The input to the server is one or more protein structures in PDB format. The query structure can be specified as a PDB identifier plus chain identifier, or a PDB file uploaded by the user. All backbone atoms (N, CA, C, O) are required and the minimum chain length is 30 amino acids. If only the amino sequence is known, it can be mapped to the closest known structure by a sequence similarity search against PDB using e.g. the SANSparallel server (20).
Outputs
PDB search, pairwise comparison and all-against-all comparison produce summaries of structural neighbours in a common output format and share interactive analysis tools. The all-against-all comparison additionally generates a dendrogram and correspondence analysis plot of the similarity matrix (Figure 1). The summaries consist of (i) a list of structural neighbours, ranked by Z-score, and (ii) the alignment data. The results are presented as plain text for downloading by downstream applications, and as hypertext for interactive analysis. A subset of matches to PDB90, filtered at 90% sequence identity, is provided for convenience. Selected subsets of matching structures can be (i) visualized as stacked alignments, (ii) visualized in 3D superimposition (Figure 2), or their amino acid sequences can be sent to (iii) the SANSparallel server for comparison against Uniprot, UniRef50 or PDB sequences or to (iv) the PANNZER2 server for automated function assignment (25). The stacked alignment view (i) shows the amino acid sequences and secondary structures of the selected structures. A stacked alignment is equivalent to progressive alignment based on a tree with star topology where the query protein is the hub. The sequences can be replaced by sequence logos of the selected structures so that the logos are displayed in stacked alignment as in Figure 3. A traditional multiple sequence alignment can be retrieved through the SANSparallel server and displayed in Jalview.
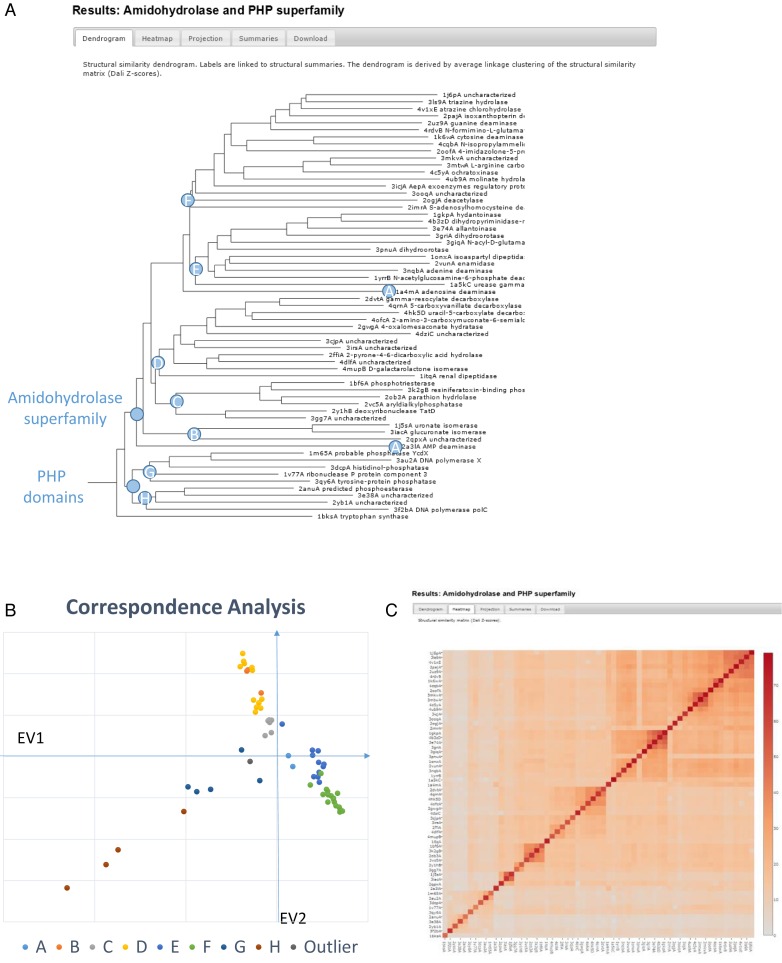
Figure 1.
Outputs from the newly added all-against-all comparison option of the Dali server (manual annotation of internal nodes). Structural dendrogram (A) of 61 selected structures from the PHP and amidohydrolase superfamilies and tryptophan synthase as outgroup. The leaves are linked to structural alignment pages. Branches A–H of the dendrogram are labelled in the correspondence analysis plot (B) of the structural similarity matrix (C). Graphics generated with JSPhyloSVG (19), Microsoft Excel and Plotly (Plotly Technologies Inc. Collaborative data science. Montréal, QC, 2015. https://plot.ly).
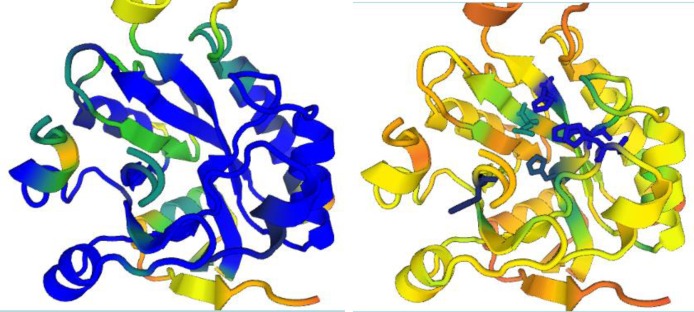
Figure 2.
Onthe left, view down the (beta-alpha)-barrel of phosphoesterase 2anuA. Colouring is by structural conservation amongst the members of the PHP superfamily (from blue for the highest through green to red for the lowest conservation). The green barrel strand at the middle left runs in reverse direction compared to the phosphatase and polymerase X members of the PHP superfamily, which have a parallel (beta–alpha) barrel. Onthe right, the same view but this time coloured by sequence conservation. The most highly conserved side chains are shown and reveal the location of the active site. Graphics generated with PV (Protein Viewer).
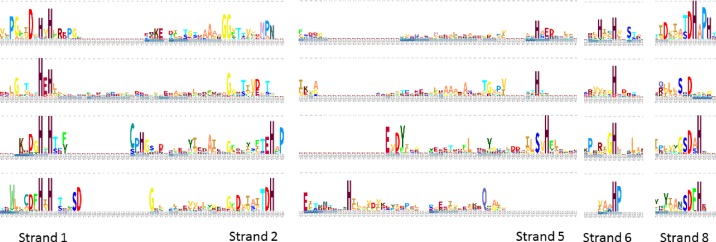
Figure 3.
Comparison between the sequence motifs of the amidohydrolase and PHP superfamilies. The top two rows represent distant members of the amidohydrolase superfamily (dihydroorotase 3griA and phosphotriesterase 1hzyA (27)). The bottom two rows represent distant members of the PHP superfamily (phosphatase 3dcpA and esterase 2anuA). All segments having two identically conserved amino acids in one or both superfamilies are shown. The structural alignment is anchored on 3griA. There is partial overlap between the sequence motifs but overall the active site is constructed differently. Logos are generated with Skylign from sequence alignments by SANSparallel.
Example
As an example, we revisit the amidohydrolase and PHP superfamilies by generating a structural overview of them. The amidohydrolase superfamily unifies a large set of metal-dependent hydrolases. The superfamily was initially defined by urease, adenosine deaminase and phosphotriesterase, and 13 other enzymes sharing a sharp sequence signature at the active site and the same predicted (beta–alpha)8-barrel fold (9). The superfamily was further extended by sequence-based predictions of novel members of this superfamily (our unpublished observations). Many of these predictions have been later verified by structure determination. Representative structures from this superfamily (with less than 40% sequence identity and different functions) were collected from structural neighbour lists generated using the PDB search option. Representatives of the PHP superfamily (26) were added. The fold and sequence signatures of the PHP domain partly resemble those of the amidohydrolase superfamily. Tryptophan synthase has a prototypical (beta–alpha)8-barrel fold and was added as an outlier. The selected set consisted of 61 structures.
The all-against-all comparison option returned a structural dendrogram that clearly separates the amidohydrolases and PHP superfamily as distinct groups (Figure 1). The PHP superfamily has a deep dichotomy. Visual inspection of the superimposed structures revealed that one strand of the (beta–alpha) barrel cannot be aligned (sequentially) because it has reversed direction (Figure 2). The set of proteins that we study here is highly divergent. Therefore sequence logos yield a convenient summary of conserved features in a protein family. The stacked sequence logo view indicates which features are conserved across different families, which are so distant that they can only be reliably aligned based on structure comparison (illustrated with PDB structures 3griA, 1hzyA (27), 3dcpA and 2anuA in Figure 3).
The amidohydrolase superfamily supports an exceptionally wide spectrum of enzyme functions, including recently evolved novel activities to break down environmental toxins. Nevertheless, enzyme classes tend to be grouped together. This can be interpreted as reflecting divergent evolution from a common ancestral enzyme activity. As a corollary, structural neighbours can suggest possible functions to uncharacterized proteins. On the other hand, if we assume evolutionary continuity of structure and function, then incongruently placed functions alert to possible misannotation. Indeed, the structure 2ogj is annotated as a dihydroorotase in the header of the PDB entry, although the protein has been shown experimentally to be a deacetylase lacking dihydroorotase activity (28). Sequence searches by SANSparallel show that the correct function has not propagated to its homologues in Uniprot, which remain misannotated as dihydroorotases.
CONCLUSION
Genomics and structural genomics are pushing out vast quantities of data which can feel overwhelming. The Dali server provides tools to navigate, integrate and organize some of these data into a form that is easier to comprehend.
Acknowledgments
We thank Heli Koskimaki and Alan Medlar for technical assistance.
FUNDING
Biocenter Finland. Funding for open access charge: Biocenter Finland.
Conflict of interest statement. None declared.
REFERENCES
- 1.Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976;261:5552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- 2.Richardson J. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 3.Holm L., Sander C. Globin fold in a bacterial toxin. Nature. 1993;361:309. doi: 10.1038/361309a0. [DOI] [PubMed] [Google Scholar]
- 4.Holm L., Sander C. Structural similarity between plant endochitinase and lysozymes from animals and phage: an evolutionary connection. FEBS Lett. 1994;340:129–132. doi: 10.1016/0014-5793(94)80187-8. [DOI] [PubMed] [Google Scholar]
- 5.Holm L., Murzin A., Sander C. Three sisters, different names: 3alpha,20beta-hydroxysteroid dehydrogenase, dihydropteridine reductase and UDP-galactose 4-epimerase. Nat. Struct. Biol. 1994;1:146–147. doi: 10.1038/nsb0394-146. [DOI] [PubMed] [Google Scholar]
- 6.Holm L., Sander C., Ruterjans H., Schnarr M., Fogh R., Boelens R., Kaptein R. LexA repressor and iron-uptake regulator from E. coli: new members of the CAP-like DNA-binding domain superfamily. Protein Eng. 1994;7:1449–1453. doi: 10.1093/protein/7.12.1449. [DOI] [PubMed] [Google Scholar]
- 7.Holm L., Sander C. Evolutionary link between glycogen phosphorylase and a DNA modifying enzyme. EMBO J. 1995;14:1287–1293. doi: 10.1002/j.1460-2075.1995.tb07114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm L., Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 9.Holm L., Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 10.Holm L., Sander C. Enzyme HIT. Trends Biochem. 1997;22:116–117. doi: 10.1016/s0968-0004(97)01021-9. [DOI] [PubMed] [Google Scholar]
- 11.Holm L., Sander C. Dali: a network tool for protein structure comparison. TiBS. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 12.Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 14.Holm L., Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa H., Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:381–389. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Dietmann S., Holm L. Identification of homology in protein structure classification. Nat. Struct. Biol. 2001;8:953–957. doi: 10.1038/nsb1101-953. [DOI] [PubMed] [Google Scholar]
- 17.Andreeva A., Howorth D., Chandonia J.-M., Brenner S.E., Hubbard T.J.P., Chothia C., Murzin A.G. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2007;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox N.K., Brenner S.E., Chandonia J.M. SCOPE: Structural Classification of Proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits S.A., Ouverney C.C. jsPhyloSVG: a javascript library for visualizing interactive and vector-based phylogenetic trees on the web. PLoS ONE. 2010;5:e12267. doi: 10.1371/journal.pone.0012267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somervuo P., Holm L. SANSparallel: interactive homology search against Uniprot. Nucleic Acids Res. 2015;43:W24–W29. doi: 10.1093/nar/gkv317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holm L., Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 22.Holm L., Kääriäinen S., Rosenström P., Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wohlers I., Andonov R., Klau G.W. DALIX: optimal DALI protein structure alignment. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013;10:26–36. doi: 10.1109/TCBB.2012.143. [DOI] [PubMed] [Google Scholar]
- 24.Wheeler T.J., Clements J., Finn R.D. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics. 2014;15:7. doi: 10.1186/1471-2105-15-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koskinen P., Törönen P., Nokso-Koivisto J., Holm L. PANNZER—high-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics. 2015;31:1544–1552. doi: 10.1093/bioinformatics/btu851. [DOI] [PubMed] [Google Scholar]
- 26.Aravind L., Koonin E.V. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holden H.M., Benning M.M., Raushel F.M., Shim H. High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta. Biochemistry. 2001;40:2712–2722. doi: 10.1021/bi002661e. [DOI] [PubMed] [Google Scholar]
- 28.Ornelas A., Korczynska M., Ragumani S., Kumaran D., Narindoshvili T., Shoichet B.K., Swaminathan S., Raushel F.M. Functional annotation and three-dimensional structure of an incorrectly annotated dihydroorotase from cog3964 in the amidohydrolase superfamily. Biochemistry. 2013;52:228–238. doi: 10.1021/bi301483z. [DOI] [PMC free article] [PubMed] [Google Scholar]