Abstract
G-protein-coupled receptors (GPCRs) play important physiological roles related to signal transduction and form a major group of drug targets. Prediction of GPCR–ligand complex structures has therefore important implications to drug discovery. With previously available servers, it was only possible to first predict GPCR structures by homology modeling and then perform ligand docking on the model structures. However, model structures generated without explicit consideration of specific ligands of interest can be inaccurate because GPCR structures can be affected by ligand binding. The Galaxy7TM server, freely accessible at http://galaxy.seoklab.org/7TM, improves an input GPCR structure by simultaneous ligand docking and flexible structure refinement using GALAXY methods. The server shows better performance in both ligand docking and GPCR structure refinement than commonly used programs AutoDock Vina and Rosetta MPrelax, respectively.
INTRODUCTION
G-protein-coupled receptors are involved in various signal transduction pathways in the cell, mediating response to different types of extracellular signals ranging from photons and small compounds to peptides and proteins (1,2). Signal transduction by GPCRs underlies a variety of physiological processes such as neurological, cardiovascular, endocrine and reproductive functions, making GPCR a major group of drug targets (3). Since ligand binding controls the activation of GPCRs, GPCR–ligand complex structures can provide invaluable information for understanding and regulating GPCR functions. Due to the advances in experimental techniques related to GPCR structure determination since 2000, structures for about 30 different GPCRs have been revealed (4). Hence, the number of GPCR sequences for which homology modeling can produce reasonable model structures is also on the rise (5).
In this context, a community-wide blind prediction experiment called GPCR Dock has been held three times since 2008 (6–8). During the GPCR Dock experiments, the community was challenged to predict GPCR–ligand complex structures from GPCR sequences and ligand 2D structures. According to the GPCR Dock assessment, high-accuracy predictions were still limited to targets with closely related template structures available in the structure database, e.g. with sequence identity >35%. In addition, the best predictions of GPCR Dock required human intervention, and it seems not easy to achieve as good performance with an automatic server yet. Two server methods, GOMoDo (9) and GPCRautomodel (10), are currently available for GPCR modeling and docking, to the best of our knowledge. These servers perform ligand docking using AutoDock Vina (11) after GPCR modeling using MODELLER (12). However, in their implementation MODELLER generates a protein structure without explicit consideration of ligand binding. Autodock Vina then tries to dock a ligand to the protein structure that isn't structurally prepared for the ligand. GOMoDo provides another docking option that uses HADDOCK (13,14). This option allows flexibility in the assigned protein residues by performing molecular dynamics (MD) simulation at the final step of docking.
Different methods have been employed to relax and refine the complex structures after docking to a homology-generated protein structure. MD simulation with explicit lipid molecules has often been used (15,16). However, MD simulations are computationally expensive. Cavasotto et al. (17) and Katritch et al. (18) reported useful methods that perform ligand-guided receptor optimization, but they are not available as a server.
The Galaxy7TM server performs flexible GPCR–ligand docking from an input GPCR structure and ligand by applying an efficient refinement method called GalaxyRefine (19,20) after initial docking with GalaxyDock (21). GalaxyRefine can consistently refine protein model structures, as evaluated in CASP (Critical Assessment of techniques for protein Structure Prediction) experiments (22). Receptor flexibility is also considered in the initial docking stage by generating an ensemble of receptor structures perturbed in normal mode directions. In this way, the server accounts for full receptor flexibility upon ligand binding. The method has been extensively tested on a test set of 125 GPCR model structure–ligand inputs, where input GPCR model structures were generated by GalaxyTBM (23), MODELLER (12) and GPCR-I-TASSER (24). Galaxy7TM showed higher docking success rates (20.8 and 24.8%) than AutoDock Vina (16.0 and 15.2%) in terms of ligand RMSD from the crystal structure (≤2.0 Å) and ratio of predicted contacts to native contacts (≥30%), respectively. The server also improved receptor structure as well as docking pose with higher performance than Rosetta MPrelax (25).
THE GALAXY7TM METHOD
Galaxy7TM docks an input ligand to an input receptor in two stages, by initial docking and subsequent refinement docking. Structural flexibility of a receptor is considered in both stages, as described below.
In the initial docking stage, receptor flexibility is taken into account by docking a ligand to an ensemble of receptor structures. An ensemble of 30 receptor structures is generated by perturbing the initial receptor structure in the normal mode directions of an anisotropic network model (26). More specifically, 200 perturbations are first generated within an RMSD range of 0.5–2.5 Å from the initial structure using randomly selected low-frequency modes, and sidechain structures are optimized with SCWRL4 (27). The structures are then clustered into 30 representative structures using RMSD of binding pocket residues as a distance measure. Binding pocket residues are predicted from the experimentally determined ligand-bound GPCR structure closest to the input receptor structure as detected by TM-align (28). If ligand-binding residues are provided as an optional input, residues <4 Å from the average positions of the provided residues are assigned as binding residues. Ligand docking is performed to each member of the receptor ensemble structures using GalaxyDock (21). The docking grid box is centerd at the average position of the predicted binding pocket residues. For each receptor structure, four ligand conformations with the lowest docking energies are selected for further refinement.
In the refinement docking stage, each of the 120 GPCR–ligand complex structures generated by initial docking is refined using a method based on GalaxyRefine (20,29). GalaxyRefine is a protein structure refinement method that applies iterative sidechain repacking and overall structure relaxation. In the current application of GalaxyRefine to a GPCR–ligand complex, sidechains of the receptor are repacked and then both receptor and ligand are allowed to relax. To treat membrane proteins properly, the FACTS implicit solvation free energy term (30) of the GalaxyRefine energy was substituted by the FACTSMEM solvation free energy for membrane proteins (31). Harmonic restraints to the input structure are added as in GalaxyRefine. After refinement, the ligand structure is minimized using the GalaxyDock energy. Among the 120 refined complex structures, the final 10 structures are selected by the sum of the rank by the refinement energy and half of the rank by the docking energy.
The server was tested on a set of 125 GPCR structure–ligand inputs, as listed in Supplementary Table S1. The input GPCR structures were constructed as follows. For each of the 23 GPCR sequences for which crystal structures non-covalently bound to small organic compounds were determined, two GPCR structures were obtained by homology modeling with GalaxyTBM (23) and MODELLER (12). An additional structure built by GPCR-I-TASSER deposited in the GPCR-HGmod database (24) was included if available for the sequence. For modeling with GalaxyTBM and MODELLER, a single template structure was used by selecting the closest structure to the crystal structure in terms of RMSD. Global sequence alignment was generated using the MAC algorithm of HHalign (32). The target-template sequence identity range was 9.2–59.9%.
Performance of the method
The Galaxy7TM server has been tested on a set of 125 GPCR and ligand structure inputs which are combinations of structures made for 23 GPCR sequences and 47 ligands. Supplementary Tables S1–3 provide detailed information on the prediction results. Performance of the server is compared with that of AutoDock Vina (11) for docking and that of Rosetta MPrelax (25) for membrane protein structure refinement in Table 1. Galaxy7TM shows a higher percentage of successful cases in all docking and receptor structure accuracy measures than compared methods.
Table 1. Comparison of Galaxy7TM with AutoDock Vina applied to input GPCR structures (Input-Vina) and to GPCR structures refined by MPrelax (MPrelax-Vina), respectively, in terms of docking accuracy and comparison with GalaxyRefine and MPrelax in terms of improvement in receptor structure quality for full structure (and for binding pocket residues in parentheses) on a test set of 125 GPCR structure-ligand inputs.
| Percentage of successful cases for the best of 10 predictions | |||
|---|---|---|---|
| Docking accuracy | Galaxy7TM | Input-Vina | MPrelax-Vina |
| Ligand RMSD (≤2.0Å) | 20.8 | 16.0 | 6.4 |
| Contact ratio (≥30.0%) | 24.8 | 15.2 | 12.0 |
| Improvement in receptor structure quality | Galaxy7TM | GalaxyRefine | MPrelax |
| ΔGDT-HA (>0.0) | 78.4 (68.0) | 77.6 (55.2) | 46.4 (52.8) |
| ΔGDC-SC (>0.0) | 93.6 (88.8) | 84.0 (52.8) | 84.8 (80.0) |
| ΔCα-RMSD (<0.0) | 74.4 (75.2) | 71.2 (64.8) | 62.4 (60.8) |
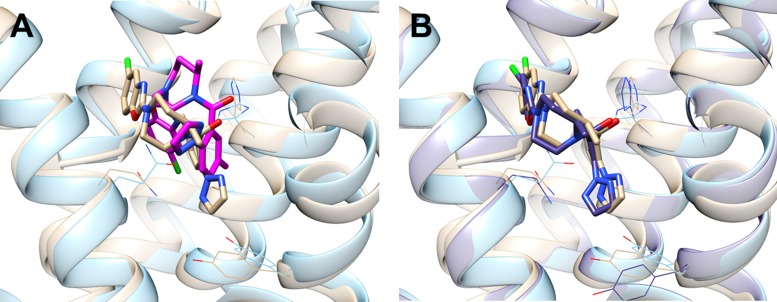
Galaxy7TM predicted ligand poses within 2 Å RMSD from the native poses in 20.8% of the cases, compared to 16.0% with AutoDock Vina when the best of ten predictions were considered for each target (Table 1). When the binding conformations were analyzed in terms of correctly predicted receptor–ligand contacts, the server returned conformations with ≥30% of native contacts (defined as two atoms <4 Å in the crystal structure) in 24.8% of the cases, compared to 15.2% with AutoDock Vina. Interestingly, when AutoDock Vina was applied to GPCR structures refined by Rosetta MPrelax (25), the success rates became worse, implying that receptor structure refinement without considering ligand can be limited in accuracy. AutoDock Vina can be run allowing receptor side chain flexibility, but flexible side chains have to be assigned manually. The success rates with flexible AutoDock Vina were 16.0 and 16.0% in terms of RMSD and contacts, respectively. A successful example in which Galaxy7TM produced a high-accuracy prediction due to sidechain improvement is presented in Figure 1. More examples and explanations are provided in Supplementary Figures S1 and 2.
Figure 1.
Result of applying (A) AutoDock Vina and (B) Galaxy7TM to a GPCR model structure built by MODELLER for human orexin receptor type 2 and a ligand, suvorexant. AutoDock Vina gave contact ratio of 22.0% (magenta in A), and Galaxy7TM 41.0% (purple in B). Input GPCR model and the crystal structure (PDB ID: 4S0V) are shown in sky blue and brown, respectively.
Improvement in receptor structure accuracy is also reported in Table 1 in terms of the CASP measures GDT-HA for backbone accuracy and GDC-SC for sidechain accuracy and Cα-RMSD. The percentage of the improved cases (ΔGDT-HA > 0, ΔGDC-SC < 0 and ΔCα-RMSD < 0) by Galaxy7TM were 78.4, 93.6 and 74.4%, respectively (22,33). The success rates are higher than those of GalaxyRefine (19,20) developed for soluble proteins (77.6, 84.0 and 71.2%, respectively) and Rosetta MPrelax (25) developed for membrane proteins (46.4, 84.8 and 62.4%, respectively). Similar trends were observed when structure accuracy was measured for ligand-binding residues. Although the percentage of improvement is rather high, the magnitude of improvement is limited. For example, it is challenging to predict large conformational changes of receptor such as helix rearrangements. It is also challenging to discriminate difference between agonists and antagonists/inverse agonists with the current server.
THE GALAXY7TM SERVER
Hardware and software
The Galaxy7TM server runs on a cluster of Linux servers, which utilizes 60 2.33 GHz Intel Xeon processors. The web application is constructed by Python and MySQL database. The whole Galaxy7TM calculation pipeline is implemented by using the Python language. The protein-ligand docking and refinement methods are implemented as part of the GALAXY program package (34,35) written in Fortran 90. The final models are visualized using the JavaScript Protein Viewer (http://biasmv.github.io/pv). Interactions between the ligand and GPCR are visualized by LIGPLOT (36).
Input and output
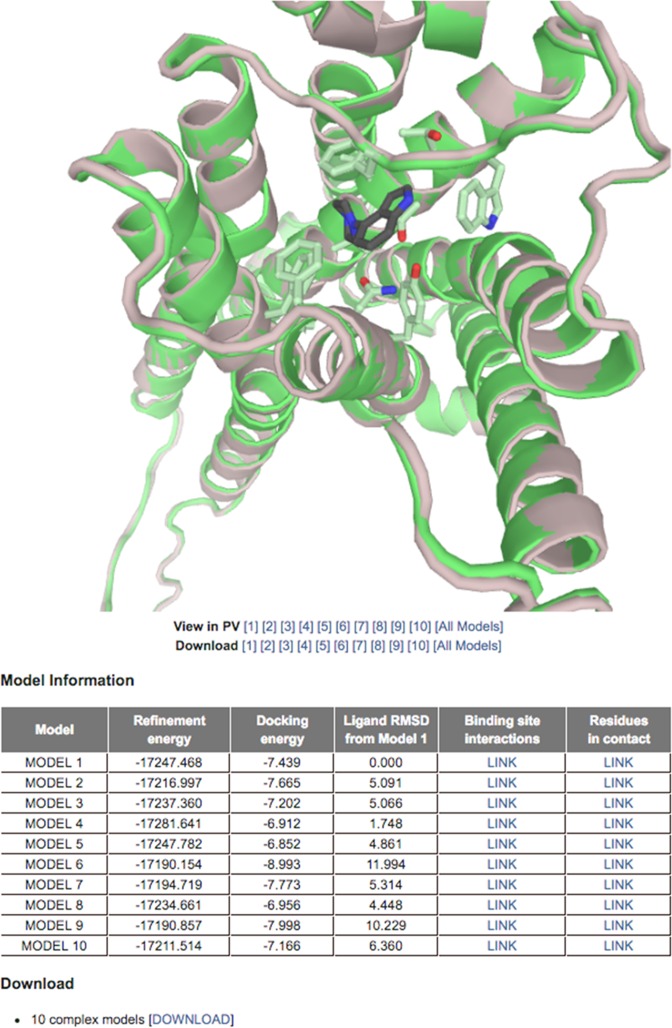
Two required inputs of the server are a GPCR structure file in PDB format and a ligand structure file in PDB, MOL2 or XYZ format. The server can deal with a GPCR structure with up to five gaps if its full sequence is provided. In addtion, up to 10 GPCR residues expected to interact with the ligand can be submitted as input. Average run time is 2–3 h. Ten GPCR–ligand complex conformations are visualized and available for download in PDB format (Figure 2). Detailed information for each prediction such as refinement energy, docking energy, ligand RMSD from Model 1 and interactions between GPCR and ligand is provided in the results table.
Figure 2.
An example output page of Galaxy7TM. Ten selected models are visualized using the JavaScript Protein Viewer. The models can be downloaded in PDB format. Additional information such as refinement energy, docking energy and a link to the LIGPLOT image showing the interactions between GPCR and ligand is provided in a table.
CONCLUSION
The Galaxy7TM server predicts GPCR–ligand complex structures by flexible docking and refinement. When tested on a set of GPCR models built by different homology modeling methods, the server could predict GPCR–ligand complex structures with higher accuracy than AutoDock Vina and Rosetta MPrelax. Galaxy7TM was especially successful in predicting contacts between GPCR and ligand and may potentially be applicable to practical problems related to drug discovery.
Supplementary Material
Acknowledgments
We would like to thank Dr. Michael Feig, Martín Carballo-Pacheco and Dr. Birgit Strodel for providing data related to FACTSMEM. We are also grateful to Dr. Lim Heo for his help with the server construction, Minkyung Baek for her advice on running GalaxyDock and Andrew H. Beaven for his comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning [2013R1A2A1A09012229, 2012M3C1A6035362]; Korea Institute of Science and Technology Information Supercomputing Center [KSC-2015-C2-001]. Funding for open access charge: Seoul National University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lappano R., Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 2.Katritch V., Cherezov V., Stevens R.C. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagerstrom M.C., Schioth H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 4.Isberg V., Mordalski S., Munk C., Rataj K., Harpsoe K., Hauser A.S., Vroling B., Bojarski A.J., Vriend G., Gloriam D.E. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016;44:D356–D364. doi: 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavasotto C.N., Palomba D. Expanding the horizons of G protein-coupled receptor structure-based ligand discovery and optimization using homology models. Chem. Commun. (Camb) 2015;51:13576–13594. doi: 10.1039/c5cc05050b. [DOI] [PubMed] [Google Scholar]
- 6.Michino M., Abola E., Brooks C.L. 3rd, Dixon J.S., Moult J., Stevens R.C. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat. Rev. Drug Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kufareva I., Rueda M., Katritch V., Stevens R.C., Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19:1108–1126. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kufareva I., Katritch V., Stevens R.C., Abagyan R. Advances in GPCR modeling evaluated by the GPCR Dock 2013 assessment: meeting new challenges. Structure. 2014;22:1120–1139. doi: 10.1016/j.str.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandal M., Duy T.P., Cona M., Zung H., Carloni P., Musiani F., Giorgetti A. GOMoDo: a GPCRs online modeling and docking webserver. PLoS One. 2013;8:e74092. doi: 10.1371/journal.pone.0074092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Launay G., Teletchea S., Wade F., Pajot-Augy E., Gibrat J.F., Sanz G. Automatic modeling of mammalian olfactory receptors and docking of odorants. Protein Eng. Des. Sel. 2012;25:377–386. doi: 10.1093/protein/gzs037. [DOI] [PubMed] [Google Scholar]
- 11.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 14.de Vries S.J., van Dijk A.D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A.M. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 15.Wolf S., Bockmann M., Howeler U., Schlitter J., Gerwert K. Simulations of a G protein-coupled receptor homology model predict dynamic features and a ligand binding site. FEBS Lett. 2008;582:3335–3342. doi: 10.1016/j.febslet.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 16.Mortier J., Rakers C., Bermudez M., Murgueitio M.S., Riniker S., Wolber G. The impact of molecular dynamics on drug design: applications for the characterization of ligand-macromolecule complexes. Drug Discov. Today. 2015;20:686–702. doi: 10.1016/j.drudis.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Cavasotto C.N., Orry A.J., Murgolo N.J., Czarniecki M.F., Kocsi S.A., Hawes B.E., O'Neill K.A., Hine H., Burton M.S., Voigt J.H., et al. Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J. Med. Chem. 2008;51:581–588. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 18.Katritch V., Rueda M., Lam P.C., Yeager M., Abagyan R. GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex. Proteins. 2010;78:197–211. doi: 10.1002/prot.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heo L., Park H., Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41:W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee G. R., Heo L., Seok C. Effective protein model structure refinement by loop modeling and overall relaxation. Proteins. 2015 doi: 10.1002/prot.24858. doi:10.1002/prot.24858. [DOI] [PubMed] [Google Scholar]
- 21.Shin W.H., Kim J.K., Kim D.S., Seok C. GalaxyDock2: protein-ligand docking using beta-complex and global optimization. J. Comput. Chem. 2013;34:2647–2656. doi: 10.1002/jcc.23438. [DOI] [PubMed] [Google Scholar]
- 22.Nugent T., Cozzetto D., Jones D.T. Evaluation of predictions in the CASP10 model refinement category. Proteins. 2014;82(Suppl. 2):98–111. doi: 10.1002/prot.24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko J., Park H., Seok C. GalaxyTBM: template-based modeling by building a reliable core and refining unreliable local regions. BMC Bioinformatics. 2012;13:198. doi: 10.1186/1471-2105-13-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Yang J., Jang R., Zhang Y. GPCR-I-TASSER: a hybrid approach to G protein-coupled receptor structure modeling and the application to the human genome. Structure. 2015;23:1538–1549. doi: 10.1016/j.str.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alford R.F., Koehler Leman J., Weitzner B.D., Duran A.M., Tilley D.C., Elazar A., Gray J.J. An integrated framework advancing membrane protein modeling and design. PLoS Comput. Biol. 2015;11:e1004398. doi: 10.1371/journal.pcbi.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakan A., Meireles L.M., Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krivov G.G., Shapovalov M.V., Dunbrack R.L., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin W.H., Lee G.R., Seok C. Evaluation of GalaxyDock Based on the Community Structure-Activity Resource 2013 and 2014 Benchmark Studies. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00309. doi:10.1021/acs.jcim.5b00309. [DOI] [PubMed] [Google Scholar]
- 30.Haberthur U., Caflisch A. FACTS: fast analytical continuum treatment of solvation. J. Comput. Chem. 2008;29:701–715. doi: 10.1002/jcc.20832. [DOI] [PubMed] [Google Scholar]
- 31.Carballo-Pacheco M., Vancea I., Strodel B. Extension of the FACTS implicit solvation model to membranes. J. Chem. Theory Comput. 2014;10:3163–3176. doi: 10.1021/ct500084y. [DOI] [PubMed] [Google Scholar]
- 32.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 33.Kopp J., Bordoli L., Battey J.N., Kiefer F., Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl. 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 34.Ko J., Park H., Heo L., Seok C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40:W294–W297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin W.H., Lee G.R., Heo L., Lee H., Seok C. Prediction of protein structure and interaction by GALAXY protein modeling programs. Bio Des. 2014;2:1–11. [Google Scholar]
- 36.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.