Abstract
Several high-throughput technologies have been developed to probe RNA base pairs and loops at the transcriptome level in multiple species. However, to obtain the final RNA secondary structure, extensive effort and considerable expertise is required to statistically process the probing data and combine them with free energy models. Therefore, we developed an RNA secondary structure prediction server that is enhanced by experimental data (RNAex). RNAex is a web interface that enables non-specialists to easily access cutting-edge structure-probing data and predict RNA secondary structures enhanced by in vivo and in vitro data. RNAex annotates the RNA editing, RNA modification and SNP sites on the predicted structures. It provides four structure-folding methods, restrained MaxExpect, SeqFold, RNAstructure (Fold) and RNAfold that can be selected by the user. The performance of these four folding methods has been verified by previous publications on known structures. We re-mapped the raw sequencing data of the probing experiments to the whole genome for each species. RNAex thus enables users to predict secondary structures for both known and novel RNA transcripts in human, mouse, yeast and Arabidopsis. The RNAex web server is available at http://RNAex.ncrnalab.org/.
INTRODUCTION
Several high-throughput technologies have been developed recently to probe RNA secondary structures at the transcriptome level in human, mouse, yeast and Arabidopsis (1–5). These technologies have been based on enzyme cleavage or chemical modification of nucleotides with specific structural states (e.g. loop regions or double-stranded regions), which can be detected by high-throughput sequencing via the stops they cause during reverse transcription (RT stops). For example, parallel analysis of RNA structure (PARS) utilizes RNase V1 and nuclease S1 simultaneously to probe RNA structures (2,5). DMS-seq or Structure-seq uses the small molecule dimethyl sulfate (DMS) to modify adenines and cytosines in single-stranded status both in vivo and in intro (3,4). The recently improved icSHAPE protocol uses NAI-N3 to modify the backbone of all four nucleotides in single-stranded states, which also enables the analysis of both in vivo and in vitro RNA structures (1). These protocols generate data that are quite different in both signal distribution and control experiment design. Therefore, deriving the structural reactivity from different structure-probing data in a way that fully accounts for background noise and local bias is rather challenging (6). Furthermore, these data alone can only reveal the structural state of individual nucleotides, but fail to reflect the pairing relationships between different nucleotides. To determine the final secondary structure, the probing data need to be correctly incorporated into an energy model with the proper folding algorithm (7). The use of probing data with proper energy models is currently limited to experts in the relevant field.
Traditionally, RNA secondary structure (involving canonical AU, GC and GU base pairs) was commonly predicted by computational methods based on the nearest neighbor model (a free energy model) (8,9). Three major types of algorithms have been developed to predict an optimal secondary structure for a given RNA sequence: maximizing expected accuracy (MEA) (10–12), sampling (13,14) and minimizing free energy (MFE) (15–17). Later, these prediction methods were improved by incorporating the probing data as restraints (18,19). For example, restrained MaxExpect (RME) (7) used a posterior probabilistic model to transform various types of probing data into pairing probabilities. Then, these probabilities were used to restrain the partition function and predict RNA secondary structure with the MEA algorithm. A method based on the sampling algorithm, SeqFold (20), was specifically designed for PARS data. It first transformed the sequencing read counts based on Fisher's exact test. Next, the structure centroid with minimal distance to the PARS data was selected from the sampling results. A method based on MFE algorithm, RNAstructure (Fold) (21), was improved to incorporate SHAPE or DMS restraints as extra pseudo-energy terms to predict RNA secondary structure. Moreover, starting from version 2.2.0, ViennaRNA (19) supported soft-constraints and provided three different approaches for converting probing data into pseudo energy contributions (21–23). All these methods require sophisticated analytical or statistical processing of the raw probing data (18).
Four major steps are generally required to process the raw probing data and predict the data-enhanced RNA secondary structure. The first step is to map the reads, which is time-consuming and largely variable as the adaptor sequences specific to each probing experiment need to be trimmed. The second step is to calculate the RT stop read counts. Because enzyme cleavage truncates the RNA transcripts and chemical modification halts reverse transcription before the modification sites (24), each mapped read only gives information for the base immediately 5′ of the first mapping position (25). The third step is to derive the structural reactivity from the RT stop counts. Usually, there are two libraries in each probing experiment for controlling the background noise (1–5). A commonly used method is to control the effect of transcript abundance and length in each library, and then subtract the normalized counts in the control library from the normalized counts in the treatment library (4,7,24,25). The fourth step is to incorporate the experimental restraints into the final structure prediction. The structural reactivity derived from the third step needs to be transformed into the proper inputs (e.g. probabilities based on a statistical model) for a given structure-folding algorithm.
Several programs and platforms have been developed to accomplish the four steps that are required to use the latest structure-probing data. StructureFold provides an integrated solution to analyze the structure-probing data via the Galaxy platform (24). RSF provides a Perl framework for data processing and structure inference based on structure-probing data (25). Although these packages are incredibly helpful, intensive computational efforts and expertise in analyzing the probing data are required to use them, which generate large bottlenecks for wet lab biologists. The SAVoR web server was developed for structure prediction with experimental restraints and data visualization along the predicted structures (26), but users need to create and input the read alignment files. SAVoR does not provide optional folding methods other than RNAfold (19).
We designed the novel web server RNAex to bridge the gap between accumulating structure-probing data and improved RNA secondary structure prediction. RNAex enables non-specialists to easily access state-of-art high-throughput probing data in multiple species (human, mouse, yeast and Arabidopsis),and predict RNA secondary structures enhanced by in vivo and in vitro experimental data. We have re-mapped the raw sequencing reads of published structure-probing experiments and transformed the read counts into corresponding inputs for four representative structure-folding methods [RME (7), SeqFold (20), RNAstructure (Fold) (21) and RNAfold (19)]. RNAex predicts RNA secondary structure with many control options and provides an interactive visualization interface for users to explore the probing data, the processed structure profile and the predicted secondary structures. RNAex also displays the post-transcriptional regulation and mutation information for each predicted RNA secondary structure, including RNA modification sites (human and mouse) (27), RNA editing (28,29) and SNP sites (30) (human). This enables the predicted RNA structure to be effectively annotated and linked with function, disease and post-transcriptional regulation events.
DATASETS
We first collected various representative high-throughput structure-probing data in human, mouse, yeast and Arabidopsis (Table 1). We analyzed the raw sequencing data and mapped the short reads to the whole genome for each species (see detailed mapping methods in Supplementary File 1). The following genomes were used: hg19 for human, mm10 for mouse, SacCer2 for yeast and TAIR10 for Arabidopsis. Then, we calculated the RT stop counts for each individual nucleotide following the same procedures that were described in the original papers (1–5).
Table 1. The structure-probing data used in the RNAex web server.
| Species | Build | Annotationb | Data type | Sample | Condition | Raw data | Citation |
|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | TAIR10 | TAIR10 | Structure-seq (DMS) | Control | in vitro | SRP027216 | Ding et al., 2014 Nature (4) |
| DMS | in vivo | ||||||
| Saccharomyces cerevisiae | SacCer2 | SacCer2 | PARS | V1 | in vitro | GSE22393 | Kertesz et al., 2010 Nature (5) |
| S1 | in vitro | ||||||
| DMS-seq | Control | in vitro | GSE45803 | Rouskin et al., 2014 Nature (3) | |||
| DMS_vitro | in vitro | ||||||
| DMS_vivo | in vivo | ||||||
| Homo sapiens | hg19 | GENCODE_v19 | DMS-seq | Control_K562 | in vitro | GSE45803 | Rouskin et al., 2014 Nature (3) |
| Vitro_K562 | in vitro | ||||||
| Vivo_K562 | in vivo | ||||||
| Control_Fibroblast | in vitro | ||||||
| Vitro_Fibroblast | in vitro | ||||||
| Vivo_Fibroblast | in vivo | ||||||
| hg19/ | GENCODE_v19/ | PARS | V1_Mother | in vitro | GSE50676 | Wan et al., 2014 Nature (2) | |
| Transcriptomea | GENCODE_v12 | V1_Father | in vitro | ||||
| V1_Child | in vitro | ||||||
| S1_Mother | in vitro | ||||||
| S1_Father | in vitro | ||||||
| S1_Child | in vitro | ||||||
| Mus musculus | mm10 | GENCODE_v2 | icSHAPE | DMSO | in vitro | GSE64169 | Spitale et al., 2015 Nature (1) |
| NAI_vitro | in vitro | ||||||
| NAI_vivo | in vivo | ||||||
| Frag-seq | Control_ESC | in vitro | GSE24622 | Underwood et al., 2010 Nature Methods (38) | |||
| Control_NPC | in vitro | ||||||
| P1_ESC | in vitro | ||||||
| P1_NPC | in vitro | ||||||
| CIRS-seq | NT | in vivo | GSE54106 | Incarnoto et al., 2014 Genome Biology (39) | |||
| DMS | in vivo | ||||||
| CMCT | in vivo |
aWe provide the whole genome data (hg19) by re-mapping the raw reads, and transcriptome only data (transcriptome) mapped in the original paper.
bThe version of genome annotation is selected based on the original paper.
Next, we collected the post-transcriptional regulation and mutation information for human and mouse to display in the predicted structures. The RNA modification sites for human and mouse were downloaded from the RMBase database (27). The RNA editing sites for human were obtained from the RADAR and DARNED databases (28,29). The human SNP information was downloaded from NCBI dbSNP (30).
ALGORITHMS
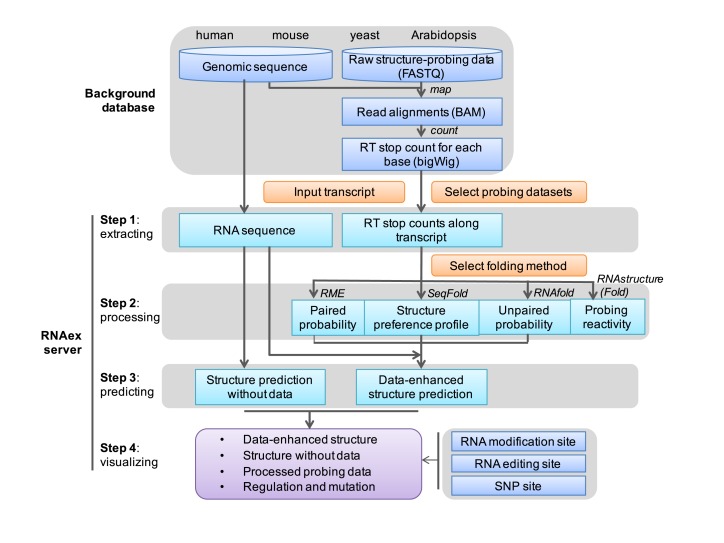
The RNAex server embedded the structure-probing data and the regulation and mutation information in a background database and implemented all computation steps through backward scripts. Users input their transcripts of interest (IDs or genomic locations) and RNAex performs four major steps. (i) RNAex extracts the sequence and structure-probing data for the given transcripts. (ii) RNAex processes the structure-probing datasets selected by the user. (iii) RNAex predicts the data-enhanced RNA secondary structures using any of the four well known folding methods [RME; Seqfold; RNAstructure (Fold) and RNAfold], which are selected by the user. (iv) RNAex visualizes the predicted structures, the processed structure-probing data and the post-transcriptional regulation and mutation information (Figure 1).
Figure 1.
Schematic of the RNAex web server. RNAex embeds the structure-probing data, post-transcriptional regulation data and mutation information. Users select specific probing datasets and a folding method and input their selected transcripts of interest. Then, RNAex performs four major steps in the background: extracting the sequence and structure-probing data, processing the structure-probing data, predicting the structures and visualizing the results.
The first step is to extract the RNA sequence and the RT stop counts of the structure-probing experiments for the selected transcripts. RNAex displays the sequences in FASTA format as a check page for users to confirm. Then, it extracts the probing read counts along the transcript from the user-selected structure-probing datasets. RNAex checks if sufficient probing data were mapped on the selected transcripts. RNAex only proceeds to fold the data-enhanced structure for transcripts that have been mapped with sufficient probing data. Otherwise, RNAex predicts the structure without the enhanced data, which generates a structure prediction that is no different from those of other sequence-based RNA secondary structure prediction servers (15).
The next step is to process the structure-probing data, which differs according to the four different folding methods available [RME, SeqFold, RNAstructure (Fold) and RNAfold]. Usually, as mentioned in the above introduction section, all methods need a paired control data as the background to compare and normalize the signal data (Supplementary Table S1). For instance, we used a denatured sample as the control for the signal data of DMS-seq, as did by the original paper (3). The denatured RNA molecules were considered to be unstructured and served as a control to capture the intrinsic variability in reactivity, which was much higher in structured RNAs. If the user selects RME as the folding method, RNAex will calculate a posterior pairing probability for each nucleotide based on a posterior probabilistic model (7). If the user selects SeqFold, RNAex will calculate a structure preference profile vector using Fisher's exact test followed by multiple test correction (20). For each nucleotide, the RT stop counts from the treatment and control samples are compared by Fisher's test. If there is a significant difference, then a binary value is given to the nucleotide to denote its structure status. If the user selects RNAstructure (Fold), RNAex will transform the RT stop counts to a probing reactivity (4,24). Briefly, the RT stop read counts are normalized to the RNA transcript abundance and length in each sample. Then, the normalized counts in the two samples are subtracted to obtain the final probing reactivity. If the user selects RNAfold (19), RNAex will provide three approaches/algorithms in the advanced options: RNAfold (D) (21), RNAfold (Z) (22) and RNAfold (W) (23). We pre-calculated the unpaired probabilities based on the probing data using RME's scripts (7) and gave them to the three algorithms of RNAfold. Moreover, we optimized the parameters (Supplementary Table S1 and File 2) of RNAfold based on our training RNAs (Supplementary Table S2). These parameters can also be changed in the advanced options.
After the first two steps, the sequence and processed data for each selected input transcript are available for the prediction step. Subsequently, RNAex will continue to perform the data-enhanced structure prediction by one of the four folding methods. The parameters (Supplementary Table S1) of all methods were learned for each probing experiment based on RNAs with known structures (Supplementary Table S2). The performances of different methods on each dataset were reported for the training RNAs (Supplementary File 2). RNAex also provides advanced options that allow users to change each method's parameters in the submit page. Detailed usage information is provided in the manual on the RNAex server and in the README files of each method [i.e. RME, SeqFold, RNAstructure (Fold) and RNAfold].
The last step is visualizing the predicted structures and all relevant information. For each selected input transcript, RNAex provides a module for structure folding without incorporating probing data, which is convenient for users to compare and evaluate the contributions of the structure-probing data. If users are interested in the raw data, RNAex provides a genome browser page that directly displays the RT stop counts. RNAex also provides post-transcriptional regulation and mutation information on the predicted structures for human and mouse RNAs, which can be searched and viewed. The interactive controls on the visualization page are described in the next section.
RNAex WEB SERVER
The RNAex web server is free to access and is unrestricted (without a login procedure). We adapted several JavaScript libraries and the HTML framework Bootstrap to build the RNAex server. The genome browser is based on JBrowse (31). The server is accessible from http://RNAex.ncrnalab.org (redirected to http://lulab.life.tsinghua.edu.cn/RNAex) and is compatible with most web browsers (Mozilla Firefox 15+, Google Chrome version 38+, Internet Explorer 9+ and Safari 8+). The processing and predicting functions of the RNAex server also are available through the command line version of RME, SeqFold, RNAstructure (Fold) and RNAfold, which are open source and available for downloading.
Inputs
To use RNAex, users are required to first select two matched probing datasets and a folding method. Then, users need to input their transcripts of interest for structure prediction, by providing the transcript IDs or the genomic locations in GTF format. RNAex can run multiple transcripts in parallel.
RNAex accepts the following transcript IDs for known transcripts: TAIR (v10) ID (32) for Arabidopsis transcripts, SGD (SacCer2) ID (33) for yeast transcripts, GENCODE (v19 for DMS-seq and v12 for PARS data) ID (34) for human transcripts and GENCODE (v2) ID (35) for mouse transcripts. RNAex also accepts genomic locations in GTF format for novel transcripts.
Users are free to adjust various parameters by selecting ‘Advanced options’. For example, users can define the criteria of sufficient probing data for transcripts by adjusting the options for ‘percentage of nucleotides with sufficient probing reads’ and ‘minimum number of reads to define sufficiency’. Users also can easily choose whether to compare the data-enhanced structure prediction with the structure prediction without data. Users also can adjust the contribution of probing data in the structure prediction by changing the parameters for each folding method, including the weight (m), gamma1 and gamma2 for RME (7), the P-value cutoff for SeqFold (20), the slope and intercept for RNAstructure (Fold) (21) and the algorithm, slope, intercept for RNAfold(19). Detailed descriptions of these parameters and the available options are provided in the manual on the RNAex server.
Selecting the ‘Submit’ button triggers RNAex to run the job. A user-provided E-mail address is optional but advisable when submitting computationally intensive jobs. A message containing the URL for the result page will be sent to the E-mail address when the job is finished.
Computation
After submitting the job, RNAex provides a check page that users can browse to confirm the job. The check page displays the extracted RNA sequence for each input transcript in FASTA format. Users can check job accuracy in this page, and then go back to revise the job if they find input errors. RNAex also shows a warning or error panel if it detects errors; the warning message contains a brief description that helps users identify potential problems. Users should note that structure prediction for long transcripts is quite time-consuming and is not as accurate as predictions for short transcripts (36). RNAex allows the users to fold long transcript by dividing the sequence into small chunks (600 nt each). Then, the server will merge the chunks into one structure file (ct file) for the users to download. In the visualization page, considering the visualization effect might be largely affected if we incorporate too long sequence into one viewer, the server still makes these chunks being visualized and interacted separately. Moreover, because most of the transcripts have less than 3000 nt, we still have a limitation on a maximum length as 3000 nt. Therefore, we also provide download links for the command line version of each folding method, which can be used if users choose to fold an entire long transcript locally.
After submitting and checking the input transcripts, the folding jobs will run on the server. Structure-folding analyses are usually time consuming, and the results may not be available immediately. A waiting page is provided that displays a URL to the result page. Users can retrieve their results later using the URL. Users also can bookmark the waiting page, which will be directed to the same URL. Results remain on the server for up to one month. The real-time log information is provided on the waiting page to make the server more user-friendly; the log actively reports the current processing steps and the job status.
Outputs
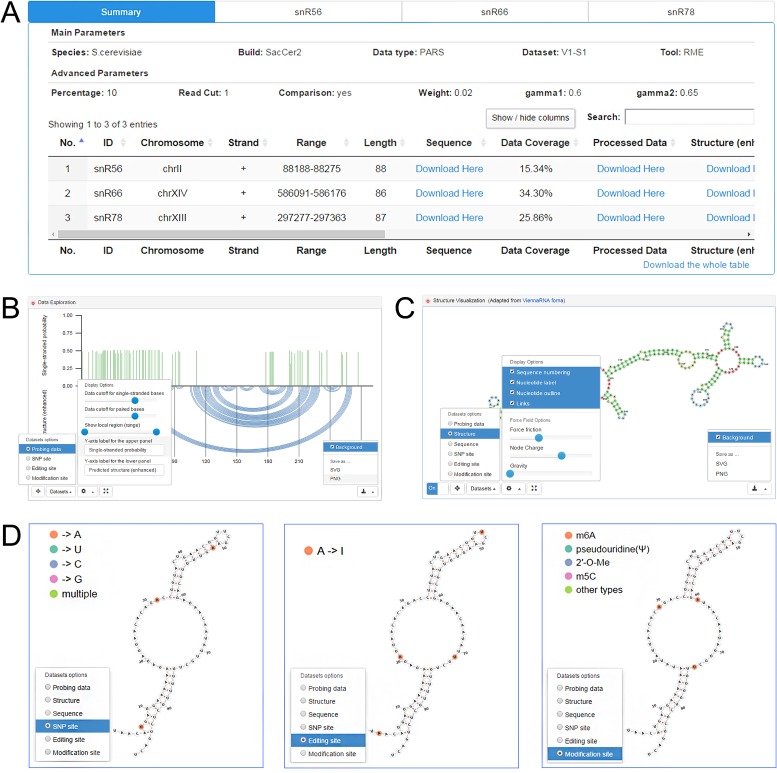
The final output of RNAex contains a summary page showing all the result files and the parameters used for the run (Figure 2A). The output displays four sections in the following order.
Main parameters. General information is shown for the structure-probing data (species, build, data type and dataset) and the folding method [RME, SeqFold, RNAstructure (Fold) or RNAfold] selected by the user.
Advanced parameters. Three types of advanced parameters are listed in order. The first type is the user-defined or default criteria to determine whether there are sufficient experimental probing data mapped onto the selected transcripts, including percentage of nucleotides with sufficient probing reads and read cut (the minimum number of reads to define sufficiency). The second type of advance parameter is the option to perform comparisons between the data-enhanced structure prediction and the prediction without data enhancement. The default selection is ‘yes’. The third type of advance parameter includes the weighting parameters for each folding method, which are explained in the manual on the RNAex server.
- Summary table of results (Figure 2A). Each row of the table lists various types of information for one selected transcript, including basic description, structure-probing data, predicted structures, post-transcriptional regulation and mutation information.
- The basic description includes the transcript ID, the genomic coordinate (chromosome, strand, range and length) and the downloadable sequence file in FASTA format for each transcript. The ‘range’ column records the detailed starts and ends for all exons of the transcript, which are separated by commas.
- The structure-probing data information includes a statistic and a downloadable file. The statistic represents the average ‘data coverage’, which is the percent of nucleotides associated with structure-probing data for the selected transcript in two different samples. The downloadable file is provided in the ‘processed data’ column, which has different meaning for each probing method. For RME and RNAfold, the raw data are processed into single-stranded probabilities on every nucleotide. For SeqFold, the raw data are processed into single-stranded probabilities, but the values are discrete (either 1 or 0). For RNAstructure (Fold), the processed data represent the probing reactivity. A nucleotide with higher reactivity value has higher probability to be single-stranded. These descriptions are included in the data file as comment lines.
- Structure information includes the predicted secondary structure enhanced by the probing data [structure (enhanced)], and the predicted secondary structure without restrained data [structure (no data)]. These can be downloaded in CT format (17). The predicted folding free energies for these two kinds of structures also are provided, and their difference (structure difference) is calculated by dividing the number of unique base pairs by the number of all base pairs in the two structures.
- Post-transcriptional regulation and mutation sites are displayed on the RNA structures, including RNA modification sites (human and mouse) and RNA editing and SNP sites (human). If these data are missing, an ‘NA’ value is displayed.
- The link to visualize corresponding transcript's genomic location in jbrowse is also provided.
Bulk download button. In addition to the summary table where files for each transcript can be downloaded separately, a bulk download button also is provided. An Excel file containing all information and the URL for each downloadable file can be obtained by clicking the button.
Figure 2.
Visualization of structure-probing data, the predicted structures, regulation and mutation information. (A) The summary page contains the main parameters, advanced parameters, a summary table of results and a bulk download button. (B) The first visualization module displays various datasets in bar plots and the predicted structure in arc diagrams. The control options in the bottom-left corner are unfolded for illustration. (C) The second visualization module displays the secondary structures with control options unfolded. (D) Example visualization of post-transcriptional regulation and mutation information by selecting the ‘SNP site’, ‘editing site’ and ‘modification site’ options in the ‘datasets’ control panel.
Visualization
RNAex also implements interactive visualization tools for users to view the probing data and the predicted structures. The result page for each run contains n + 1 tabs for an easy visualization, where n is the number of input transcripts or long transcript’ fragments (each fragment is <600 nt and counts as one tab). Users are free to navigate these results through the tabs at the top (Figure 2A). The first tab shown as default is the summary page. The other n tabs are visualization pages for each selected transcript. In each visualization page, RNAex provides two modules to view the probing data and predicted secondary structure. The first module is illustrated in Figure 2B, which is a combination of two plots. The upper plot shows the probing data as a bar plot, where nucleotides with higher values have higher probability to be single-stranded. Users can see the exact value for each base by hovering the mouse over the base. The lower plot shows the predicted secondary structure in arc diagrams. RNAex provides another interactive module adapted from ViennaRNA forna (37), which allows users to view the secondary structure (Figure 2C). The structure can be fine-tuned and color-coded according to various datasets, which can be controlled by users.
These two visualization modules can be configured in many ways using the buttons available in the bottom-left corner of the screen. The star at the left of the screen is used to reset the figure to default parameters. The ‘datasets’ button is used to color-code the plots according to various datasets. For example, clicking the ‘SNP site’ button prompts the upper panel of the first module to display several red bars, with each one showing a nucleotide with SNP annotation (Figure 2D). If the transcript does not contain any post-transcriptional regulation information, then only one choice is provided (i.e. ‘probing data’). The next button shows many configuration options. For example, users can edit the y-axis labels through the input boxes for ‘y-axis label for the upper panel’ and ‘y-axis label for the lower panel’. If the transcripts are too long, users can view only a segment of the transcript by sliding the two icons in the ‘show local region (range)’ option. Users also can control the color of the bars shown in the upper panel by sliding the icons appearing in the ‘data cutoff for single-stranded bases’ and ‘data cutoff for paired bases’ options. Finally, a full-screen button is provided to enlarge the figure to the entire display region.
By default, two kinds of secondary structures are predicted for each transcript. One is restrained by the structure-probing data (data-enhanced), whereas the other is not restrained by any probing data (without data). Thus, four visualization panels are available for each input transcript. A fold/unfold option (the ‘+’ button before the panel title) is provided for users to conveniently compare the four different plots. All these figures can be downloaded in SVG or PNG format by clicking the download button in the bottom-right corner. The background color can be set to be transparent or white by changing the option of ‘background’.
DISCUSSION
We present RNAex as a web-based server that enables non-specialists to easily predict RNA secondary structures using cutting-edge high-throughput structure-probing data. Users can easily access and incorporate these data into the RNAex server to enhance the RNA secondary structure prediction using four different folding methods. RNAex results allow users to clearly view, identify and understand SNP and post-transcriptional regulation sites on the RNA structures. Overall, the server is an early web-based platform for the emerging new field of high-throughput RNA structure-probing analysis. We will continue enriching RNAex as additional data become available, by including more species, diverse types of structure-probing data and other types of information related to RNA structure dynamics. RNAex only uses the folding tools to predict a single secondary structure (e.g. the structure with the lowest folding free energy change). However, RNA secondary structure at non-zero temperatures is an ensemble of configurations, not a single structure. We will improve this in the future. Moreover, we also will continue to improve the user experience of the website.
Supplementary Material
Acknowledgments
We thank Kui Xu and Boqin Hu for help with server implementation and Yu-Cheng Yang for the collection of post-transcriptional regulation and mutation information.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Basic Research Program of China [2012CB316503]; National High-Tech Research and Development Program of China [2014AA021103]; National Natural Science Foundation of China [31271402, 31522030]; Tsinghua University Initiative Scientific Research Program [2014z21045]; Computing Platform of National Protein Facilities (Tsinghua University). Funding for open access charge: National Natural Science Foundation of China [31271402].
Conflict of interest statement. None declared.
REFERENCES
- 1.Spitale R.C., Flynn R.A., Zhang Q.C., Crisalli P., Lee B., Jung J.W., Kuchelmeister H.Y., Batista P.J., Torre E.A., Kool E.T., et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan Y., Qu K., Zhang Q.C., Flynn R.A., Manor O., Ouyang Z., Zhang J., Spitale R.C., Snyder M.P., Segal E., et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 5.Kertesz M., Wan Y., Mazor E., Rinn J.L., Nutter R.C., Chang H.Y., Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwok C.K., Tang Y., Assmann S.M., Bevilacqua P.C. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 2015;40:221–232. doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y., Shi B., Ding X., Liu T., Hu X., Yip K.Y., Yang Z.R., Mathews D.H., Lu Z.J. Improved prediction of RNA secondary structure by integrating the free energy model with restraints derived from experimental probing data. Nucleic Acids Res. 2015;43:7247–7259. doi: 10.1093/nar/gkv706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z.J., Turner D.H., Mathews D.H. A set of nearest neighbor parameters for predicting the enthalpy change of RNA secondary structure formation. Nucleic Acids Res. 2006;34:4912–4924. doi: 10.1093/nar/gkl472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 10.Do C.B., Woods D.A., Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:e90–e98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 11.Knudsen B., Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z.J., Gloor J.W., Mathews D.H. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009;15:1805–1813. doi: 10.1261/rna.1643609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Y., Chan C.Y., Lawrence C.E. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Y., Lawrence C.E. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013;41:W471–W474. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X., Wu Y., Lu Z.J., Yip K.Y. Analysis of sequencing data for probing RNA secondary structures and protein-RNA binding in studying posttranscriptional regulations. Brief. Bioinform. 2015 doi: 10.1093/bib/bbv106. doi:10.1093/bib/bbv106. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz R., Luntzer D., Hofacker I.L., Stadler P.F., Wolfinger M.T. SHAPE directed RNA folding. Bioinformatics. 2016;32:145–147. doi: 10.1093/bioinformatics/btv523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouyang Z., Snyder M.P., Chang H.Y. SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. 2013;23:377–387. doi: 10.1101/gr.138545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deigan K.E., Li T.W., Mathews D.H., Weeks K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U.S.A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarringhalam K., Meyer M.M., Dotu I., Chuang J.H., Clote P. Integrating chemical footprinting data into RNA secondary structure prediction. PLoS One. 2012;7:e45160. doi: 10.1371/journal.pone.0045160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washietl S., Hofacker I.L., Stadler P.F., Kellis M. RNA folding with soft constraints: reconciliation of probing data and thermodynamic secondary structure prediction. Nucleic Acids Res. 2012;40:4261–4272. doi: 10.1093/nar/gks009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Y., Bouvier E., Kwok C.K., Ding Y., Nekrutenko A., Bevilacqua P.C., Assmann S.M. StructureFold: genome-wide RNA secondary structure mapping and reconstruction in vivo. Bioinformatics. 2015;31:2668–2675. doi: 10.1093/bioinformatics/btv213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Incarnato D., Neri F., Anselmi F., Oliviero S. RNA structure framework: automated transcriptome-wide reconstruction of RNA secondary structures from high-throughput structure probing data. Bioinformatics. 2016;32:459–461. doi: 10.1093/bioinformatics/btv571. [DOI] [PubMed] [Google Scholar]
- 26.Li F., Ryvkin P., Childress D.M., Valladares O., Gregory B.D., Wang L.S. SAVoR: a server for sequencing annotation and visualization of RNA structures. Nucleic Acids Res. 2012;40:W59–W64. doi: 10.1093/nar/gks310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun W.J., Li J.H., Liu S., Wu J., Zhou H., Qu L.H., Yang J.H. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016;44:D259–D265. doi: 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramaswami G., Li J.B. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiran A., Baranov P.V. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 30.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudge J.M., Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathews D.H. Revolutions in RNA secondary structure prediction. J. Mol. Biol. 2006;359:526–532. doi: 10.1016/j.jmb.2006.01.067. [DOI] [PubMed] [Google Scholar]
- 37.Kerpedjiev P., Hammer S., Hofacker I.L. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31:3377–3379. doi: 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Underwood J.G., Uzilov A.V., Katzman S., Onodera C.S., J.E. Mainzer., Mathews D.H., Lowe T.M., Salama S.R., Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Incarnato D., Neri F., Anselmi F., Oliviero S. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 2014;15:491. doi: 10.1186/s13059-014-0491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.