Abstract
There is a wide interest in designing peptides able to bind to a specific region of a protein with the aim of interfering with a known interaction or as starting point for the design of inhibitors. Here we describe PepComposer, a new pipeline for the computational design of peptides binding to a given protein surface. PepComposer only requires the target protein structure and an approximate definition of the binding site as input. We first retrieve a set of peptide backbone scaffolds from monomeric proteins that harbor the same backbone arrangement as the binding site of the protein of interest. Next, we design optimal sequences for the identified peptide scaffolds. The method is fully automatic and available as a web server at http://biocomputing.it/pepcomposer/webserver.
INTRODUCTION
Protein–peptide interactions are important mediators of many cellular processes, constituting a major component of protein–protein interaction (PPI) networks where they are estimated to account for about 40% of the interaction events (1). Hence they are critical elements in our understanding of biological systems, giving insights on how protein complexes and networks operate and how we can modulate them (2).
Comprehensive structural studies and collections of protein–peptide complexes have highlighted relevant features of their architecture and mode of binding (3–6). Compared to other type of protein interactions, i.e. PPI, protein–peptide interactions show some peculiarities: their interface is better packed and enriched of main-chain hydrogen bonds (5). Few peptide residues, ‘hot-spots’, contribute most of the binding energy and, in addition to the high frequency of aromatic residues as observed in PPI ‘hot-spots’, the peptide ones tend to be enriched in leucines and isoleucines. Furthermore, it has been shown that protein–peptide interactions often adopt the same structural arrangement observed between different regions of monomeric proteins (3).
These evidences suggest that rational design strategies, which have been successfully applied to specific protein families or domains (7–10), can be generally applicable. At present, however, automated computational methods able to perform all the necessary steps to design peptides binding to a given protein without requiring either the knowledge of the structure of a protein complex or some information about the peptide to be designed are still lacking (11–13).
Structure-based strategies often use peptide fragments or interaction motifs derived from either protein–protein or protein–peptide complexes (11). A recent implementation of this approach is the PiPreD software (13). The method relies on the availability of a protein complex where the anchor residues, defined as those mediating the interaction with the protein of interest, are used to guide the sampling and modeling of peptides derived from a database of complex fragments.
De novo design methods attempt to design the peptide without any prior information about its sequence or structure. The FlexPepDock ab initio protocol (12) of the Rosetta modeling suite (14), given an initial model of the peptide–protein complex, performs a Monte-Carlo simulation for de novo folding and docking of the peptide to the protein surface. The pepspec method (15), another example of de novo design method, does not use information on the complex structure but requires at least an approximate position of one peptide residue in the binding pocket. The ‘in silico panning’ method (16) uses the target protein structure to generate peptides that are subsequently evolved using the docking energy as fitness function. VitAL (17) first identifies the binding site using a coarse grained Gaussian Network Model and subsequently generates all possible amino acid sequences and calculates the binding energies between these pairs and the specific location on the protein. Another recently developed strategy (18) performs simultaneous sampling of peptide sequences and conformational space to estimate the relative free energies of the designed peptides.
By and large, methods using information from known protein–protein or protein–peptide complex structures have been shown so far to be the most successful, leading in some cases to the development of potent inhibitors and drugs (19–21). However, if no information is available about a complex of the target protein with a protein or a peptide, one has to recur to de novo design methods and therefore needs to select an appropriate backbone and optimize its relative orientation with respect to the target protein and its sequence (11).
To simplify and streamline this latter process, we developed PepComposer, a computational pipeline for the design of protein-binding peptides that requires as input only the target protein structure and an approximate definition of the binding site.
As mentioned before, protein–peptide interactions often adopt similar relative arrangements as those found between interacting fragments of monomeric proteins (11). Indeed, it has been shown that this is true in about 80% of known protein–peptide interactions. Notably, the backbone similarity is not influenced by the side chain similarity. We use motifs found in monomeric proteins as backbone scaffolds. This has a clear advantage with respect to pure de novo methods since the identified backbone fragments are stereo chemically plausible and already suitable for binding to a protein region (11). After deriving a set of peptide backbones (scaffolds), we use a Monte Carlo procedure as implemented in PyRosetta (22) to design a set of sequences for the identified peptide scaffolds, and select the best ones according to the predicted binding energy. The method is fully automatic, available as a web server and can effectively reproduce known protein peptide interactions.
MATERIALS AND METHODS
Our approach, described in more detail in the following sections, consists in (i) defining the query region, i.e. the region of the input target protein containing the desired binding site; (ii) searching for regions structurally similar to the query region in a non redundant database of experimentally solved monomeric proteins; (iii) retrieving continuous backbone fragments in contact with the query region and merging them in the appropriate relative position with the target protein and (iv) designing the peptide sequence using repeated cycles of structure diversification and sequence design. These steps are described below.
Define the query region: the query region can be defined in two ways, i.e. by listing a specific set of residues or by selecting a single residue res and a query region radius r. In the latter case, all residues having at least one atom within rÅ from any atom of res are included in the query.
Identify regions structurally similar to the query region: a structure similarity search of the query region as defined above is performed against a non-redundant database of solved monomeric proteins (filtered at the level of 70% sequence identity). We use Triangle Match (23,24) with default parameters to select backbone regions (hit regions) structurally similar to the query region in an amino acid sequence and order independent fashion.
-
Retrieve the appropriate backbones: we retrieve backbone scaffolds by considering regions of the hit proteins in contact with the hit region. This is achieved analyzing contact graphs. Contacts are pre-calculated for all proteins in our database using Almost Delaunay tessellation (25) as implemented in the ADCGAL program (26) and used to construct graphs using the NetworkX package for Python (27). The conformations of the top 2000 longer hit regions are analyzed further. Fragments in contact with the hit regions (that we call scaffolds) are retrieved. Those that do not have an extended conformation are excluded since regions with helical or turn conformations in the native structure are unlikely to preserve their conformation in isolation. Extended backbone scaffolds are defined as the fragments for which ne/(L - 2) ≥ 0.5 where ne is the number of residues that have ϕ angles in the range −185° to −35° and ψ angles in the range from 85° to 160° and L is the length of the fragment defined as the number of its residues. Regions shorter than four residues are not considered.
The contact density (defined as the number of contacts per residue) between the remaining potential scaffolds and the corresponding hit region is used to re-sort the list and retain the top 500 scaffolds (or more if there are regions with the same contact density at position 500). This is based on the assumptions that higher contact density backbone scaffolds are more likely to support sequences leading to high affinity binding peptides in the subsequent sequence design step. Selected backbone scaffolds are then merged with the query protein to build putative protein–peptide complexes on the basis of the superposition between the query and the hit regions. The superposition is performed using Triangle Match (23,24).
Sequence design: sequence design is performed using PyRosetta (22), a Python-based interface to the Rosetta molecular modeling package (28) and the Rosetta full atoms energy function with Talaris2013 energy term weights (29). The design consists of two different stages, both including structure diversification and sequence design.
First, a relaxation step is performed using PackRotamersMover to allow changes in side chain rotamers of both the protein and the peptide. Next, small rigid-body movements of the peptide and small local flexible backbone moves of the protein–peptide complex structure are performed using RigidBodyPerturbMover with translation and rotation steps of 0.08 Å and 0.3° respectively followed by five rounds of BackrubMover on the whole complex (22). Energy minimization of the protein–peptide complex is performed using MinMover DFP minimization with 0.01 Å tolerance. Next, the amino acid sequence of the peptide is optimized using a standard simulated annealing Monte Carlo method, using the PackRotamersMover function, where rotamers are changed in both the protein and the peptide while amino acids are mutated only in the peptide. For every backbone scaffold, 10 different peptide sequences are calculated and the resulting complexes scored and ranked using FoldX (30). The 100 backbone scaffolds corresponding to top protein peptide complexes in terms of lowest FoldX binding energy are selected for the subsequent exhaustive sequence design step.
In the next stage of design, each peptide is subjected to three iterations of backrub movements, energy minimization and sequence design and 100 peptides are generated from every backbone scaffold selected in the pre-design stage. A further structure refinement step is performed on the protein–peptide complex following the Rosetta Classic Relax protocol (31). In this final step the backbone of the protein is kept fixed and all residues within 20 Å from any atoms of the query region are considered in the calculation. Resulting models differing by more than 1Å in terms of Cα RMSD from the initial protein–peptide structure are filtered out in order to avoid both significantly distorted structures and peptide conformations that deviate too much from the starting backbone scaffold. The remaining peptides corresponding to the same backbone are grouped by sequence identity using CLUSEQ (32) and each group is assigned the average FoldX binding energy of its members.
The parameters used in the pipeline described above have been selected on the basis of their ability to retrieve peptides similar in both structure and sequence to experimentally known cases. A few selected examples of the results are illustrated later; more are available in the ‘Example’ section of the web server.
THE PEPCOMPOSER WEB SERVER
The procedure described above is implemented as an automatic pipeline and is freely available on the web at http://biocomputing.it/pepcomposer/webserver. The website interface is built using standard HTML/CSS code. Responsive layouts are implemented using the Bootstrap CSS framework, JavaScript and JQuery. Back-end operations are implemented in PHP, PERL and Python.
In the home page, the required input is a PDB protein structure of the target protein and the definition of the region of interest. Once the PDB is uploaded, the structure of the target protein is displayed in a Jsmol window. Upon inputting and confirming the query region, selected residues and chain(s) are highlighted in the graphics window. Each run requires on average 12 h (depending on the size of the input), therefore users can retrieve their results via a provided Job Id, a link to a page that can be bookmarked or, optionally, via email.
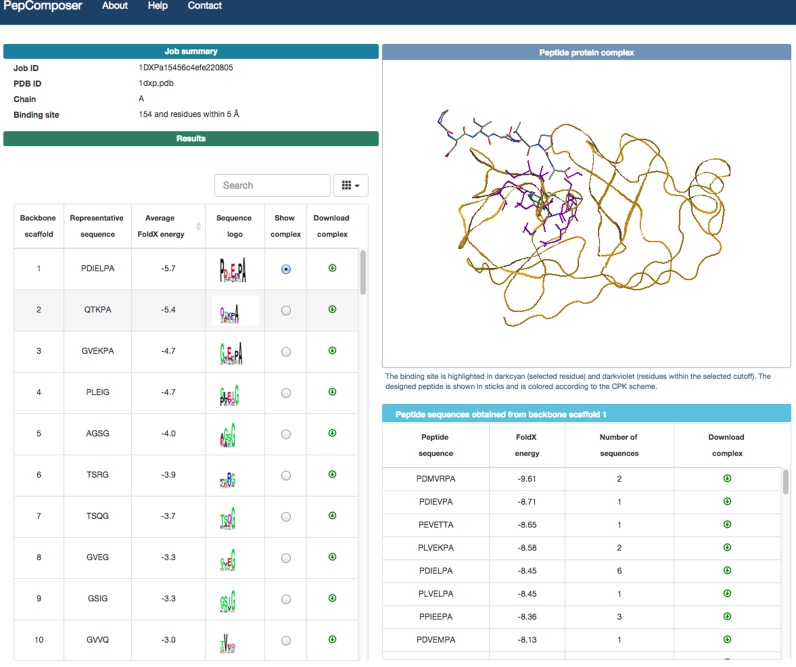
The result page (Figure 1) includes a table showing for each backbone scaffold: (i) the peptide sequence with the lowest FoldX energy (ii) the average FoldX binding energy of the generated complexes for that backbone (iii) a sequence logo obtained using WebLogo (33); (iv) a radio button to display the list of peptides obtained from the same backbone and the complex structure of the representative one in the jsmol window (http://www.jmol.org/) (the representative peptide is the one with the lowest energy from the most populated group) and (v) a button to the download the coordinates of the latter complex in PDB format. The sequence logo is obtained using all sequences for that backbone. The list of peptides includes the number of times each sequence has been obtained and the corresponding representative peptide.
Figure 1.
Example of the output returned by the PepComposer web server.
RESULTS
Validation of the method
PepComposer is meant to provide suggestions for peptides putatively binding to a given region of a protein without using any information on the nature of the peptide. An extensive validation of the method would require the synthesis of several peptides, the experimental validation of their interaction with the target protein and the identification of the binding site. It follows that classical metrics for evaluating the method on established benchmarks cannot be applied to our case (nor to design methods in general).
A possible strategy to validate the method is to use it on sets of proteins for which the structure of their complex with a binding peptide is known. We selected to use the test set of 53 peptide–protein complexes described in (34) and used by the authors to assess the performance of different docking methods. Although we are aware that if a designed peptide does not match the native one, it can still be a possible binder, the native peptide is certainly able to bind the protein and can therefore be used as a true positive for comparison.
We report in Supplementary Table S1 the results of PepComposer applied to this benchmark. They can also be inspected form the web site under the tab ‘LEADS- PEP Benchmark’. Although a docking method uses the knowledge of the peptide sequence to be docked, while our method only uses the target structure, in about 50% of the cases (23 out of 53), the median backbone RMSD between the designed peptide ranking first and the native one is 1.9 Å. The value decreases to 1.1 Å if the best out of the first 10 ranked peptides is considered, accounting for 25 cases out of 53.
This compares rather favorably with the docking results reported in (34), although this comparison has to be taken with much care. While on one side we do not use the sequence of the docked peptide, which clearly puts us at a disadvantage, on the other our designed peptides are usually shorter than the native ones (see Supplementary Table S1) and therefore expected to have a higher probability to achieve a lower RMSD. Furthermore in our case one residue belonging to the binding site is used as input. Given all these caveats, it is worth mentioning that, according to the analysis performed in (34) only the Surflex method (see Table 2 of (34)) retrieves near native peptides in 20 out of the 53 tested cases (for all other methods the figure ranges from 8 to 16) and, in the Surflex case, the median RMSD achieved for the near native peptides is around 1.1 Å, and 4.8 Å if all docked peptides are considered.
In 5 out of the 53 cases, PepComposer did not find any backbone scaffold satisfying the selected thresholds. The remaining 23 cases include 16 peptides that assume a helical or beta strand structure in the complex and that are excluded from our design strategy as described above. In these cases, however, it cannot be discounted that the designed peptides mimic the interactions of the native peptide in the context of different backbone geometries. Indeed, as shown in Supplementary Table S1, in 65% of cases (15 out of 23) the designed peptides interact with at least a third of the residues in contact with the native peptide and only in two cases there is no overlap between the sites contacted by the native peptide and by the best scoring designed one. On average 44% of the residues involved in the known protein binding site are contacted by the best scoring designed peptides.
We believe that these data show that the accuracy of the method is appropriate for the purpose it has been developed.
Below, we describe in detail a few biomedically relevant examples of application of PepComposer. Further examples of the results of PepComposer are available under the ‘Examples’ tab of the server.
Example 1: The FimG protein
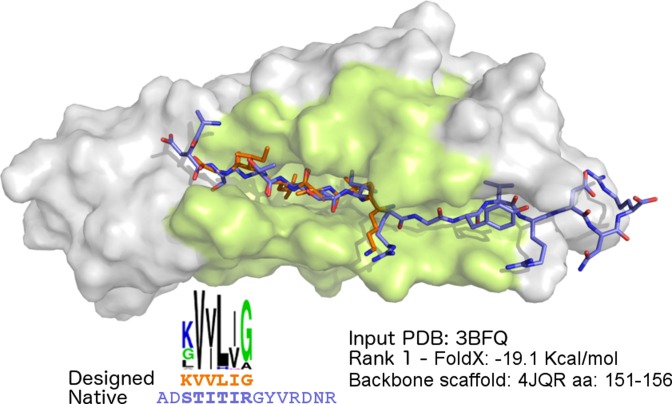
The FimG protein belongs to the fimbrial protein family and together with FimF and FimH plays a major role in the fimbrial morphology, acting as longitudinal modulator. A crystal structure (PDB ID: 3BFQ) of FimG from Escherichia coli is available in complex with the donor strand peptide DSF (sequence: ADSTITIRGYVRDNR) from its FimF partner (35). We used this structure as input to PepComposer after removing the peptide from the PDB file. The query region was defined by selecting a single amino acid approximately at the center of the binding region, namely residue 25, and a query region radius of 10 Å. The method produced many significant predictions, with top ranked peptides displaying a near-native conformation (backbone RMSD < 1Å) compared to the N-terminal portion of the native peptide. The complex including the best peptide in the first cluster (sequence: KVVLIG, FoldX binding energy −19.1 Kcal/mol) is shown in Figure 2. The peptide superimposes to residues 3–8 of the native peptide with an RMSD of 0.97Å. As it can be appreciated from the sequence logo, several peptides obtained from the same backbone scaffold, including the peptide with the lowest predicted binding energy (−20.8 Kcal/mol, sequence: KVILIA), show two conserved isoleucine residues in positions 3 and 5, the same amino acid side chains present in the corresponding positions (5 and 7) of the native peptide.
Figure 2.
PepComposer results for the FimG protein. The top of the figure shows the FimG target protein (PDB ID: 3BFQ) shown as a gray surface with the query region highlighted, the native peptide (with the carbon atoms shown in violet) and the top ranked designed peptide (carbon atoms in orange). The lower part shows the sequence logo of all the peptide sequences obtained from the scaffold, the amino acid sequence of the designed peptide ranking first (in orange) and of the native peptide (in violet), the PDB code of the hit protein, the FoldX binding energy of the predicted complex and the region of the selected backbone scaffold retrieved from the hit protein.
Example 2: The HCV protease
The second example we discuss here concerns the Hepatitis C virus (HCV) NS3 protease. The enzyme is active as non-covalent heterodimer consisting of a catalytic subunit (the N-terminal of NS3) and an activating cofactor (NS4A), in the presence of which the catalytic triad is in the characteristic position expected for a chymotrypsin-like activity (36). Several inhibitors/drugs have been developed against the enzyme; nearly all of them derive from modifications of the hexapeptide Asp-Asp-Ile-Val-Pro-Cys (DDIVPC) (37), a potent competitive inhibitor derived from the N-terminal cleavage product of HCV.
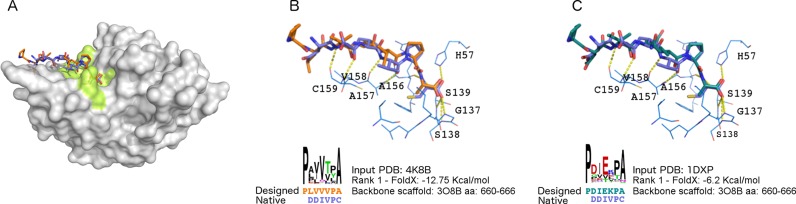
We used the structure of the protease solved in complex with a peptide-like compound (PDB ID: 4K8B). We selected residue F154, known as the main determinant of NS3 substrate specificity (38,39) and a query region radius of 5 Å. As it can be observed in Figure 3A and B, the top ranked peptide (sequence PLVVVPA, FoldX binding energy −12.75 kcal/mol) displays a similar binding mode (backbone RMSD 1.3 Å) as that observed for the known hexapeptide in complex with the protein (PDB ID: 4JMY). Sequence similarity is apparent at the C-terminal, which is known to contribute the most to the binding of DDIVPC (37). Positions corresponding to P3 (Valine) and P2 (Proline) in the native peptide show identical amino acids in the designed one (residue positions are labeled according to the nomenclature described in (40)). This is relevant, since these two residues are responsible for the inhibition activity, as shown by alanine and D-amino acid scanning respectively (40). Several hydrogen bonds are present between the main chain atoms of the peptide and of the protease (P1-NH and Arg155, P3-NH, P3-carbonyl and Ala157, P5-carbonyl and Cys159). The terminal carbonyl of P1-Alanine is within the hydrogen-bond distance of the backbone of Gly137, Ser138 and Ser139 (the oxyanion hole). These interactions are consistent with the typical pattern observed between peptide ligands and serine protease active sites, commonly referred to as the ‘canonical’ binding mode.
Figure 3.
PepComposer results for the Hepatitis C virus NS3 protease. (A) The HCV NS3 target protein (PDB ID: 4K8B) in complex with a native peptide (PDB ID: 4JMY) and with the designed peptide. The input structure of NS3 used here (PDB ID: 4K8B) has been solved in complex with a native-like peptide. (B) Details of the superposition of the native and designed peptide shown in (A). (C) Superposition of a designed peptide obtained using the structure of the NS3 protein in its apo state (PDB ID: 1DXP) superimposed to the native peptide found in complex with the protease in the 4JMY PDB structure. The lower parts of the panels show the sequence logo of all the peptide sequences obtained from the scaffold, the amino acid sequence of the designed peptide ranking first and of the native, the PDB code of the hit protein, the FoldX binding energy of the predicted complex and the region of the selected backbone scaffold retrieved from the hit protein. The color code is the same as in Figure 2.
We also used the method in a more challenging case, i.e. when the structure of the HCV NS3 protein is solved in its apo state (PDB ID: 1DXP), i.e. in absence of binders or inhibitors. Also in this case the highest ranked peptide (sequence PDIEKPA) shows a similar binding pose with respect to the native one (backbone RMSD 1.0 Å), and most of the main chain interactions observed in the known complex are preserved as well (Figure 3C). The important proline in P2 is present in the designed sequence, while a Lysine replaces the Valine in P3.
These results show that PepComposer is able to design a peptide that closely resemble a known binder, and correctly rank this and similar solutions among the top ones.
DISCUSSION
The identification of peptides able to bind to a given protein region is among the most common requests of cell and molecular biologists to computational structural biologists. Although in some cases this can be used as a starting point for the development of non peptidic binders or even inhibitors, more often it is required to test hypotheses about the effect of the inhibition of a native protein-protein interaction on a cellular process (2,10,41).
The use of peptide libraries is often the starting point for the development of peptidic binders, however it is difficult to target a specific region in such experiments (42,43).
The large amount of structural data nowadays available can, in our view, be effectively used to at least provide a starting point for library design and, in some cases, to directly synthesize a specific peptide.
When the aim is to interfere with an interaction and there is information about the complex, the strategy usually consists in analyzing the interacting region trying and identifying a contiguous ‘peptide like’ region of the partner to be used as starting point (7,11). When this information is not available, the first hurdle is the selection of an appropriate backbone likely to support a sequence able to bind to the desired region. Next, one has to design the appropriate sequence and there exist very effective tools to this end (22).
Here, we tried to streamline the process by identifying one or more plausible backbones to be used on the basis of previous observations (3) and to simplify the subsequent operations and the analysis of the results by implementing the various steps as a single web server requiring minimal effort from the user.
An extensive experimental verification of the method in unknown cases would be desirable of course, but rather difficult to perform in a reasonable time frame. We hope that the availability of our server will foster such targeted experiments. Here we decided to verify the correct operation of the pipeline by trying and reconstructing known protein–peptide interactions. We used a dataset including 53 protein structures solved in complex with a peptide reported in (34) and described in detail a few examples in this paper, while more are available on the server web site. In general, either no peptide is retrieved (about 10% of the cases tested so far) or we find backbones that closely resemble the known ones.
We believe that the tool, given its easiness of use and the clarity of the result representation, will be of help as a guide for our experimental colleagues in designing single peptides or peptide libraries.
Supplementary Material
Acknowledgments
The authors are grateful to the other members of the Biocomputing Unit for useful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Epigenomics Flagship Project (EPIGEN); KAUST [KUK-I1-012-43]. Funding for open access charge: Epigenomics Flagship Project (EPIGEN).
Conflict of interest statement. None declared.
REFERENCES
- 1.Neduva V., Linding R., Su-Angrand I., Stark A., de Masi F., Gibson T.J., Lewis J., Serrano L., Russell R.B. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neduva V., Russell R.B. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 2006;17:465–471. doi: 10.1016/j.copbio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Vanhee P., Stricher F., Baeten L., Verschueren E., Lenaerts T., Serrano L., Rousseau F., Schymkowitz J. Protein-peptide interactions adopt the same structural motifs as monomeric protein folds. Structure. 2009;17:1128–1136. doi: 10.1016/j.str.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Vanhee P., Reumers J., Stricher F., Baeten L., Serrano L., Schymkowitz J., Rousseau F. PepX: a structural database of non-redundant protein-peptide complexes. Nucleic Acids Res. 2010;38:D545–D551. doi: 10.1093/nar/gkp893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.London N., Movshovitz-Attias D., Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Singh S., Chaudhary K., Dhanda S.K., Bhalla S., Usmani S.S., Gautam A., Tuknait A., Agrawal P., Mathur D., Raghava G.P. SATPdb: a database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2016;44:D1119–D1126. doi: 10.1093/nar/gkv1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin H., Slusky J.S., Berger B.W., Walters R.S., Vilaire G., Litvinov R.I., Lear J.D., Caputo G.A., Bennett J.S., DeGrado W.F. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 8.Grigoryan G., Reinke A.W., Keating A.E. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moellering R.E., Cornejo M., Davis T.N., Del Bianco C., Aster J.C., Blacklow S.C., Kung A.L., Gilliland D.G., Verdine G.L., Bradner J.E. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart M.L., Fire E., Keating A.E., Walensky L.D. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhee P., van der Sloot A.M., Verschueren E., Serrano L., Rousseau F., Schymkowitz J. Computational design of peptide ligands. Trends Biotechnol. 2011;29:231–239. doi: 10.1016/j.tibtech.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Raveh B., London N., Zimmerman L., Schueler-Furman O. Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One. 2011;6:e18934. doi: 10.1371/journal.pone.0018934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliva B., Fernandez-Fuentes N. Knowledge-based modeling of peptides at protein interfaces: PiPreD. Bioinformatics. 2015;31:1405–1410. doi: 10.1093/bioinformatics/btu838. [DOI] [PubMed] [Google Scholar]
- 14.Das R., Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 15.King C.A., Bradley P. Structure-based prediction of protein-peptide specificity in Rosetta. Proteins. 2010;78:3437–3449. doi: 10.1002/prot.22851. [DOI] [PubMed] [Google Scholar]
- 16.Yagi Y., Terada K., Noma T., Ikebukuro K., Sode K. In silico panning for a non-competitive peptide inhibitor. BMC Bioinformatics. 2007;8:1–11. doi: 10.1186/1471-2105-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unal E.B., Gursoy A., Erman B. VitAL: Viterbi algorithm for de novo peptide design. PLoS One. 2010;5:e10926. doi: 10.1371/journal.pone.0010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacherjee A., Wallin S. Exploring protein-peptide binding specificity through computational peptide screening. PLoS Comput. Biol. 2013;9:e1003277. doi: 10.1371/journal.pcbi.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watt P.M. Screening for peptide drugs from the natural repertoire of biodiverse protein folds. Nat. Biotechnol. 2006;24:177–183. doi: 10.1038/nbt1190. [DOI] [PubMed] [Google Scholar]
- 20.Naider F., Anglister J. Peptides in the treatment of AIDS. Curr. Opin. Struct. Biol. 2009;19:473–482. doi: 10.1016/j.sbi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crunkhorn S. Anticancer drugs: Stapled peptide reactivates p53. Nat. Rev. Drug Discov. 2013;12:741. doi: 10.1038/nrd4133. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhury S., Lyskov S., Gray J.J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 2010;26:689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfson H.J., Rigoutsos I. Geometric hashing: an overview. Comput. Sci. Eng. 1997;4:10–21. [Google Scholar]
- 24.Nussinov R., Wolfson H.J. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc. Natl. Acad. Sci. U.S.A. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandyopadhyay D., Snoeyink J. Proceedings of the fifteenth annual ACM-SIAM symposium on Discrete algorithms. Soc. Ind. Appl. Math. 2004:410–419. [Google Scholar]
- 26.Bandyopadhyay D., Snoeyink J. Almost-Delaunay simplices: robust neighbor relations for imprecise 3D points using CGAL. Comput. Geom. 2007;38:4–15. [Google Scholar]
- 27.Hagberg A., Swart P., Schult D. Exploring network structure, dynamics, and function using NetworkX. In: Varoquaux G, Vaught T, Millman J, editors. Proceedings of the 7th Python in Science Conference. 2008. pp. 11–15. [Google Scholar]
- 28.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leaver-Fay A., O'Meara M.J., Tyka M., Jacak R., Song Y., Kellogg E.H., Thompson J., Davis I.W., Pache R.A., Lyskov S., et al. Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradley P., Misura K.M., Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 32.Yang J., Wang W. Proceedings of the 19th IEEE International Conference on Data Engineering (ICDE) 2003. CLUSEQ: Efficient and effective sequence clustering; pp. 101–112. [Google Scholar]
- 33.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauser A.S., Windshugel B. LEADS-PEP: a benchmark data set for assessment of peptide docking performance. J. Chem. Inf. Model. 2016;56:188–200. doi: 10.1021/acs.jcim.5b00234. [DOI] [PubMed] [Google Scholar]
- 35.Puorger C., Eidam O., Capitani G., Erilov D., Grutter M.G., Glockshuber R. Infinite kinetic stability against dissociation of supramolecular protein complexes through donor strand complementation. Structure. 2008;16:631–642. doi: 10.1016/j.str.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Kim J.L., Morgenstern K.A., Lin C., Fox T., Dwyer M.D., Landro J.A., Chambers S.P., Markland W., Lepre C.A., O'Malley E.T., et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 37.LaPlante S.R., Nar H., Lemke C.T., Jakalian A., Aubry N., Kawai S.H. Ligand bioactive conformation plays a critical role in the design of drugs that target the hepatitis C virus NS3 protease. J. Med. Chem. 2014;57:1777–1789. doi: 10.1021/jm401338c. [DOI] [PubMed] [Google Scholar]
- 38.Koch J.O., Bartenschlager R. Determinants of substrate specificity in the NS3 serine proteinase of the hepatitis C virus. Virology. 1997;237:78–88. doi: 10.1006/viro.1997.8760. [DOI] [PubMed] [Google Scholar]
- 39.Pizzi E., Tramontano A., Tomei L., La Monica N., Failla C., Sardana M., Wood T., De Francesco R. Molecular model of the specificity pocket of the hepatitis C virus protease: implications for substrate recognition. Proc. Natl. Acad. Sci. U.S.A. 1994;91:888–892. doi: 10.1073/pnas.91.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llinas-Brunet M., Bailey M., Fazal G., Goulet S., Halmos T., Laplante S., Maurice R., Poirier M., Poupart M.A., Thibeault D., et al. Peptide-based inhibitors of the hepatitis C virus serine protease. Bioorg. Med. Chem. Lett. 1998;8:1713–1718. doi: 10.1016/s0960-894x(98)00299-6. [DOI] [PubMed] [Google Scholar]
- 41.Sackett D.L., Sept D. Protein-protein interactions: making drug design second nature. Nat. Chem. 2009;1:596–597. doi: 10.1038/nchem.427. [DOI] [PubMed] [Google Scholar]
- 42.Tornatore L., Sandomenico A., Raimondo D., Low C., Rocci A., Tralau-Stewart C., Capece D., D'Andrea D., Bua M., Boyle E., et al. Cancer-selective targeting of the NF-kappaB survival pathway with GADD45beta/MKK7 inhibitors. Cancer Cell. 2014;26:495–508. doi: 10.1016/j.ccr.2014.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wells J.A., McClendon C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.