Abstract
Enrichment analysis is a popular method for analyzing gene sets generated by genome-wide experiments. Here we present a significant update to one of the tools in this domain called Enrichr. Enrichr currently contains a large collection of diverse gene set libraries available for analysis and download. In total, Enrichr currently contains 180 184 annotated gene sets from 102 gene set libraries. New features have been added to Enrichr including the ability to submit fuzzy sets, upload BED files, improved application programming interface and visualization of the results as clustergrams. Overall, Enrichr is a comprehensive resource for curated gene sets and a search engine that accumulates biological knowledge for further biological discoveries. Enrichr is freely available at: http://amp.pharm.mssm.edu/Enrichr.
INTRODUCTION
The Gene Ontology (GO), which was first published in the year 2000 (1), introduced the concept of associating a collection of genes with a functional biological term in a systematic way. GO was needed because methods such as cDNA microarrays that measure mRNA expression at a global genome-wide scale produce lists of differentially expressed genes that are difficult to interpret. The creation of GO enabled the analysis of gene lists in the context of prior knowledge. Early tools such as FatiGO (2), BiNGO (3) and TermFinder (4) first realized this concept. Initially, most enrichment analyses of sets of differentially expressed genes, integrated with prior knowledge, were limited to either GO terms, or gene sets were projected onto known protein–protein interaction networks and signaling pathways. These include, for example, membership of genes in pathway databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) (5). Later on, other types of annotated gene sets for enrichment analysis emerged; for example, chromosome location of genes, computationally predicted targets of microRNAs and transcription factors, and gene modules identified computationally from large collections of gene expression data (6). Subsequently, improved enrichment analysis algorithms (7,8) and enrichment analysis tools (9–13) emerged. Here we present a major update to the enrichment analysis tool Enrichr, which was first published in 2013. Since its initial publication, we added many new features and data sets to Enrichr. The new gene set libraries that were added include differentially expressed genes after drug, gene, disease and pathogen perturbations extracted from the national center for biotechnology information (NCBI) gene expression omnibus (GEO) through a crowdsourcing project. Furthermore, we have implemented the ability to submit fuzzy sets, upload BED files, a calendar that shows the number of lists submitted each day, an improved application programming interface (API), an enhanced help documentation, an improved Find a Gene feature, and visualization of the results as clustergrams. In this manuscript, we also provide updated benchmarking results of the different scoring schemes implemented in Enrichr and visualize the overlap between the data sets currently within Enrichr compared with other comparable web-server tools and resources that serve gene set libraries.
ENHANCEMENTS AND UPDATES
New gene set libraries
Since the original publication of Enrichr in 2013 (14), we have systematically added new gene set libraries (Table 1). We created gene set libraries from HumanCyc (15), a metabolic pathway resource stored in BioPAX format (16); gene and small-molecule perturbations from the LINCS L1000 data set; NCI-Nature pathways (17); protein complexes from the NURSA project (18); pathways from the PANTHER resource (19); targets of phosphatases from DEPOD (20); human phenotypes from the Human Phenotype Ontology (HPO) (21); genes associated with grants using NIH RePORTER and GeneRIF (22); transcription factor targets computed from the ChIP-seq data from the ENCODE project (23); differentially expressed genes from the Allen Brain Atlas (24); tissue expression extracted from the Genotype-Tissue Expression (GTEx) project (25); protein expression in tissues and cell types from ProteomicsDB (26) and the Human Proteome Map (HPM) (27); genes associated with cell survival from the Achilles Project (28); and more. More details about constructing these new libraries are available as supporting online materials. These libraries are open source, freely available for download from the libraries page of Enrichr. In the updated version of Enrichr, we added a new category of gene set libraries called ‘Crowd’. These libraries were created by an independent crowdsourcing project where participants extracted gene expression signatures for six specific themes as described below.
Table 1. Details of the new gene set libraries added to Enrichr since its original publication.
| Gene set library | Terms | Gene coverage | Mean genes per term | Source | PMID |
|---|---|---|---|---|---|
| Achilles fitness decrease | 216 | 4271 | 128 | http://www.broadinstitute.org/achilles | 25984343 |
| Achilles fitness increase | 216 | 4320 | 129 | http://www.broadinstitute.org/achilles | 25984343 |
| Allen Brain Atlas down | 2192 | 13 877 | 304 | http://www.brain-map.org/ | 23193282 |
| Allen Brain Atlas up | 2192 | 13 121 | 305 | http://www.brain-map.org/ | 23193282 |
| BioCarta 2015 | 239 | 1678 | 21 | http://pid.nci.nih.gov/download.shtml | |
| ChEA 2015 | 395 | 48 230 | 1429 | http://amp.pharm.mssm.edu/lib/chea.jsp | 20709693 |
| dbGaP | 345 | 5613 | 36 | http://www.ncbi.nlm.nih.gov/gap | 24297256 |
| Disease Perturbations from GEO down | 839 | 23 939 | 293 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| Disease Perturbations from GEO up | 839 | 23 561 | 307 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| Drug Perturbations from GEO down | 906 | 23 877 | 302 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| Drug Perturbations from GEO up | 906 | 24 350 | 299 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| ENCODE Histone Modifications 2015 | 412 | 29 065 | 2123 | http://genome.ucsc.edu/ENCODE | 26527727 |
| ENCODE TF ChIP-seq 2015 | 816 | 26 382 | 1811 | http://genome.ucsc.edu/ENCODE | 26527727 |
| Roadmap Epigenomics HM ChIP-seq | 383 | 22 288 | 4368 | http://www.roadmapepigenomics.org/ | 22690667 |
| Genes Associated with NIH Grants | 32876 | 15 886 | 9 | http://exporter.nih.gov | 25355515 |
| GO Biological Process 2015 | 5192 | 14 264 | 58 | http://www.geneontology.org | 25428369 |
| GO Cellular Component 2015 | 641 | 13 236 | 82 | http://www.geneontology.org | 25428369 |
| GO Molecular Function 2015 | 1136 | 12 753 | 57 | http://www.geneontology.org | 25428369 |
| GTEx Tissue Sample Gene Expression Profiles down | 2918 | 16 725 | 1443 | http://www.gtexportal.org/ | 25954001 |
| GTEx Tissue Sample Gene Expression Profiles up | 2918 | 19 249 | 1443 | http://www.gtexportal.org/ | 25954001 |
| HomoloGene | 12 | 19 129 | 1594 | http://www.ncbi.nlm.nih.gov/homologene | |
| Human Phenotype Ontology | 1779 | 3096 | 31 | http://www.human-phenotype-ontology.org/ | 24217912 |
| HumanCyc | 125 | 756 | 12 | http://humancyc.org/ | 15642094 |
| KEA 2015 | 428 | 3102 | 25 | http://amp.pharm.mssm.edu/lib/kea.jsp | 19176546 |
| KEGG 2015 | 179 | 3800 | 48 | http://www.kegg.jp/kegg/download/ | 24214961 |
| Kinase Perturbations from GEO | 37 | 25 858 | 2081 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| Kinase Perturbations from L1000 | 49 | 12 441 | 4052 | http://www.lincscloud.org/ | |
| LINCS L1000 chem pert down | 33132 | 9448 | 63 | http://www.lincscloud.org/ | |
| LINCS L1000 chem pert up | 33132 | 9559 | 73 | http://www.lincscloud.org/ | |
| MGI Mammalian Phenotype Level 3 | 71 | 10 406 | 715 | http://www.informatics.jax.org/ | 18981050 |
| MGI Mammalian Phenotype Level 4 | 476 | 10 493 | 200 | http://www.informatics.jax.org/ | 18981050 |
| NCI-Nature | 209 | 2541 | 39 | http://pid.nci.nih.gov/ | 18832364 |
| NURSA Human Endogenous Complexome | 1796 | 10 231 | 158 | https://www.nursa.org | 21620140 |
| Panther | 104 | 1918 | 39 | http://www.pantherdb.org/ | 23193289 |
| Phosphatase Substrates from DEPOD | 59 | 280 | 9 | http://www.koehn.embl.de/depod/ | 25332398 |
| Reactome 2015 | 1389 | 6768 | 47 | http://www.reactome.org/download | 24243840 |
| Single Gene Perturbations from GEO down | 2460 | 30 832 | 302 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| Single Gene Perturbations from GEO up | 2460 | 31 132 | 298 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| TargetScan microRNA | 222 | 7504 | 155 | http://www.targetscan.org | 26267216 |
| TF-LOF Expression from GEO | 269 | 34 061 | 641 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| Tissue Protein Expression from Human Proteome Map | 30 | 6454 | 301 | http://www.humanproteomemap.org | 24870542 |
| Tissue Protein Expression from ProteomicsDB | 207 | 13 572 | 301 | https://www.proteomicsdb.org/ | 24870543 |
| Transcription Factor PPIs | 290 | 6002 | 77 | ||
| Virus Perturbations from GEO down | 323 | 17 576 | 300 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| Virus Perturbations from GEO up | 323 | 17 711 | 300 | http://www.ncbi.nlm.nih.gov/geo/ | 23193258 |
| WikiPathways 2015 | 404 | 5863 | 51 | http://www.wikipathways.org | 26481357 |
PMID stands for PubMed identifiers.
Differentially expressed genes after drug, gene, disease, ligand and pathogen perturbations extracted from GEO by the crowd
To extract gene sets from gene expression data deposited in the GEO (29), we established a crowdsourcing microtask project that asks participants to extract gene sets from GEO for the following categories: (1) single-gene perturbations in mammalian cells; (2) comparison of diseased versus normal tissues; (3) single-drug perturbations in mammalian cells; (4) perturbations applied to MCF7 cells; (5) comparison between young and old mammalian tissues; (6) endogenous ligand perturbations of mammalian cells; and (7) comparison of before and after pathogen infection of human cells. Participants of the microtasks were recruited via two Coursera massive online open courses (MOOCs) and worked voluntarily on finding relevant studies from the GEO database. Participants were instructed to identify control and perturbation samples (GSM files), and to add additional metadata such as cell-line/tissue used in each study, as well as IDs for genes, diseases and small molecules. Participants were also instructed to use the browser extension GEO2Enrichr (30) to extract differentially expressed gene sets from GEO. The metadata and gene sets were submitted to our crowdsourcing database and then converted to gene set libraries for Enrichr.
To ensure the quality of these crowd-generated gene set libraries, we performed both automatic and manual sanitizations. We first programmatically re-processed all the entries submitted by the participants to calculate differentially expressed gene sets using the metadata submitted by the participants using the Characteristic Direction method (31). Incorrect entries where samples did not belong to the particular study were automatically filtered. We also automatically filtered out entries with invalid gene symbols and mismatched organisms. Entries from curators who submitted more than 10% invalid entries were removed entirely. Entries that passed these filters were randomly sampled for manual inspection to ensure that the metadata, such as the perturbed genes, were in fact perturbed in the study, and control samples and perturbation samples were correctly selected. As a result, approximately 20% of the submitted entries were removed for each microtask.
In addition, to encourage Enrichr users to contribute their own lists to the crowd category, we added a checkbox on the submission page that enables user-submitted lists to be added to a collection that can then be searched by other users. The default settings of the checkbox are unchecked to avoid users exposing their lists by accident. So far, ∼600 lists were contributed by users of Enrichr. In the future, we plan to make these contributed lists available for search by the community.
Benchmarking enrichment methods
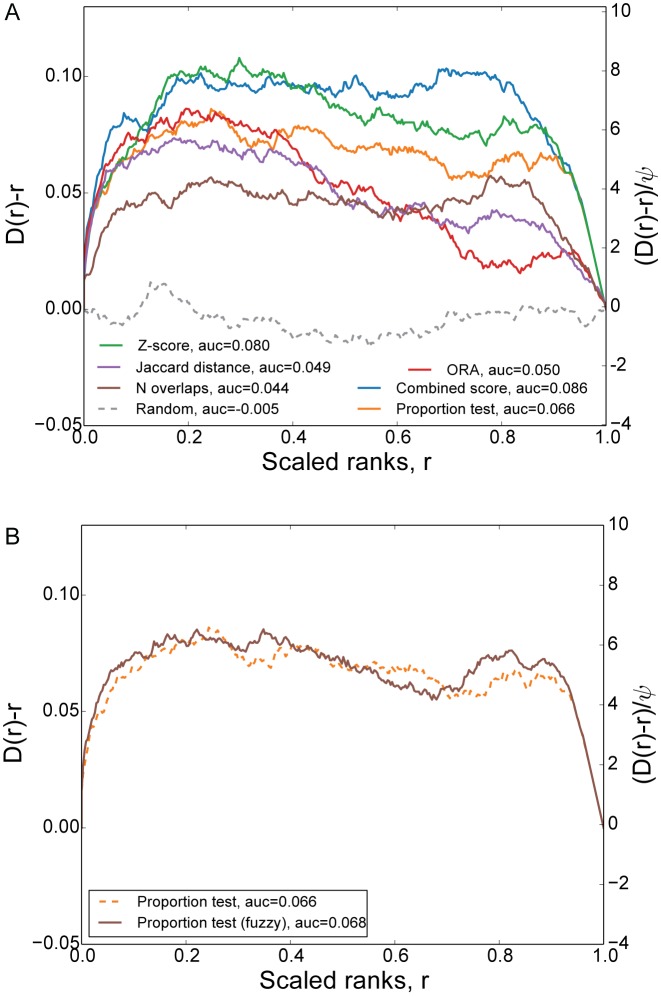
To benchmark the performance of the various enrichment analysis methods implemented within Enrichr, namely, the proportion test, the Z-score and the combined score, as well as other similar published methods, for example, the over representation analysis (ORA) method (11), as well as simple methods such as the Jaccard distance or the number of overlapping genes, we processed 489 experiments that genetically perturbed (knockdown, knockout or overexpression) transcript factors (TFs) from 293 studies available from GEO. We identified the differentially expressed genes from these studies using the Characteristic Direction (CD) method (31). We then performed enrichment analysis against the ChIP-X enrichment analysis (ChEA) gene set library, ranking TFs with the different scoring methods (32). The hypothesis behind this benchmarking idea is that genes that are differentially expressed after genetic perturbations of a TF are enriched for the targets of the TF as determined by ChIP-seq regardless of cell type, mammalian organism or microarray platform. We then find the ranks of the perturbed TFs for each enrichment analysis scoring methods and plot their cumulative distributions. Our results demonstrate that the combined score and the Z-score methods recover more of the ‘correct’ terms compared with the other methods we tested (Figure 1A). This result is consistent with our results from 2013, presented in the original Enrichr publication.
Figure 1.
Benchmarking different enrichment analysis methods. (A) Deviation of the cumulative distribution from uniform of the scaled ranks of TFs derived from different enrichment analysis methods; (B) Comparison between crisp and fuzzy version of the proportion test. The ranking distribution of randomly ordered ChEA terms is plotted in gray dashed line. The area under the curve (AUC) is indicated in the legend as a measure of the degree of deviation from uniform.
Fuzzy enrichment analysis
A fuzzy set is composed of a pair 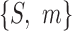 , where S is a set and m is a membership function defined over the members of the set:
, where S is a set and m is a membership function defined over the members of the set: 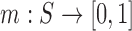 . For each
. For each 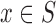 , the value m(x) is the grade of membership of x, such that if
, the value m(x) is the grade of membership of x, such that if 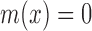 then x is termed ‘not in the set’ and if
then x is termed ‘not in the set’ and if 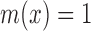 then x is termed ‘completely in the set’, and intermediate values of x are considered to have intermediate fuzzy membership. In these terms, the simple gene sets referred to above are called ‘crisp sets’ because all the genes in these sets have a membership value of 1. Another common representation for fuzzy sets is
then x is termed ‘completely in the set’, and intermediate values of x are considered to have intermediate fuzzy membership. In these terms, the simple gene sets referred to above are called ‘crisp sets’ because all the genes in these sets have a membership value of 1. Another common representation for fuzzy sets is
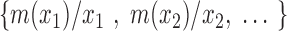 |
To perform enrichment analysis with fuzzy sets, we require the fuzzy equivalent of set intersection. For the fuzzy sets 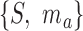 and
and 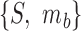 , this is defined such that
, this is defined such that
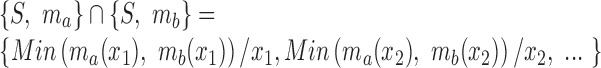 |
In addition, we need the cardinality of a fuzzy set which is defined as the sum of the grades of membership of each element,
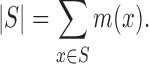 |
The fuzzy P-value enrichment score can be calculated by decomposing the null distribution into two parts; firstly we denote by Z the number of non-zero grades of membership in the fuzzy intersection between two null fuzzy sets: 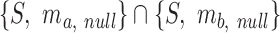 . Then Z is a random variable that is distributed by the hypergeometric distribution:
. Then Z is a random variable that is distributed by the hypergeometric distribution:
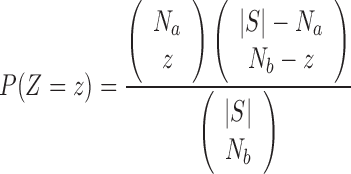 |
While intuitively fuzzy enrichment analysis should be more accurate than ‘crisp’ enrichment analysis, because ‘fuzzy’ enrichment considers the ranks and magnitude of genes in both the input set and the library sets, our initial results so far only show a marginal enhancement, utilizing the same TF-centered benchmark presented above (Figure 1B). In the future, we plan to further explore ways to improve the performance of the fuzzy set enrichment analysis idea. It is also important to note that with the fuzzy set enrichment analysis, the scaling method used to convert typical values that represent, for example, level of differential expression, into membership values between 0 and 1 is important. Overall effective use of fuzzy enrichment analysis requires advanced computational expertise. However, in the near future, we plan to make such transformations easier and more transparent.
Uploading BED files
The introduction of ChIP-seq and ChIP-chip technologies enables the detection of de novo transcription factor binding sites and changes in histone modifications in mammalian genomes. Efforts such as the ENCODE project supply a large compendium of this type of data. To identify the exact location of protein-DNA binding, genomic regions with statistically enriched reads, called peaks, are detected. The final step in such analyses is to associate peaks with genes. The updated version of Enrichr features similar functionality developed for the popular tool Genomic Regions Enrichment of Annotations Tool (GREAT) (33) by allowing users to upload BED files describing genomics region peaks. A Java module in Enrichr maps the chromosome coordinates listed in input BED files to their nearest coding mouse or human genes. User options allow the specification of whether the input is for human or mouse, and the number of genes to return based on distance to the transcription start site (TSS). The identified nearest genes are automatically uploaded to Enrichr for enrichment analysis. Enrichr now has a new button that enables users to view, cut and paste the uploaded lists. This feature can be used to analyze the nearest genes from any input BED file containing peaks using other tools.
Visualization of the results with clustergrams
One of the new features of Enrichr is the visualization of the enrichment results as clustergrams. This is achieved using Clustergrammer (https://github.com/MaayanLab/clustergrammer), an independent data visualization module we developed for multiple projects. Clustergrammer provides dynamic visualizations of Enrichr's enrichment analysis results. It enables a user to visualize the associations between their input genes and the overlapping genes of the top enriched terms. Clustergrammer visualizes these associations using a heat map in which the columns are the top enriched terms, and the rows are the input genes. The cells in the heat map indicate whether a gene from the input list overlaps with genes that belong to an enriched term. The enriched terms in the columns of the heat map are ranked based on their enrichment score. This score is indicated by the length of a transparent red bar that is displayed above the column labels. The input genes are hierarchically clustered based on their associations with the top enriched terms. Clustering is calculated using the Jaccard distance and average linkage. The heat map is interactive; a user can zoom and pan using scroll and drag functions. The rows and columns can be toggled between different orderings. The heat map can be re-ordered based on a single row or column by double-clicking on a label. The matrix is initialized to show the top 20 input genes that are associated with the top 10 enriched terms; however, these can be adjusted with sliders. This slider can be used to show more of the user's input genes. Users can search for an input gene using a search box to identify a gene of interest if the heat map contains many rows. Users can also save an image of the clustergram using the camera icon, or share the interactive visualization using the permanent link available by clicking the share icon.
Deployment with Docker, Mesos and Marathon
The Enrichr hosting and deployment process has changed drastically since its original publication. To account for the increased traffic through both Enrichr's web interface and API, the application and its dependencies are now packaged into a Docker container (34) running the Debian 8.0 operating system with Java 8 installed. Once packaged, the Docker container is deployed onto a 16-node cluster managed using Apache Mesos (35). To maximize uptime, Mesosphere's Marathon software is used on top of Apache Mesos as a cluster-wide initialization and control system (36). The Marathon software automatically controls restarting Enrichr and moving resources across cluster nodes.
Libraries management
One of the challenges related to enrichment analysis tools such as Enrichr is provenance: the ability to repeat enrichment results, even after libraries and computational methods for computing enrichment have been updated. To address this issue, we created a ‘Legacy’ category in which we place older libraries so that these can be accessed by users who wish to repeat their own results, or repeat a published result conducted by others. The Legacy category has gene set libraries with a year label. We plan to update libraries once a year to balance consistency of results with timely content. Initially, the gene set libraries of Enrichr were not made available for download. Since 2015, we have made all libraries accessible for direct download. This enables other computational biologists to explore the deep relationships between genes in annotated gene sets, and to develop new tools using these libraries.
Overview of Enrichr statistics
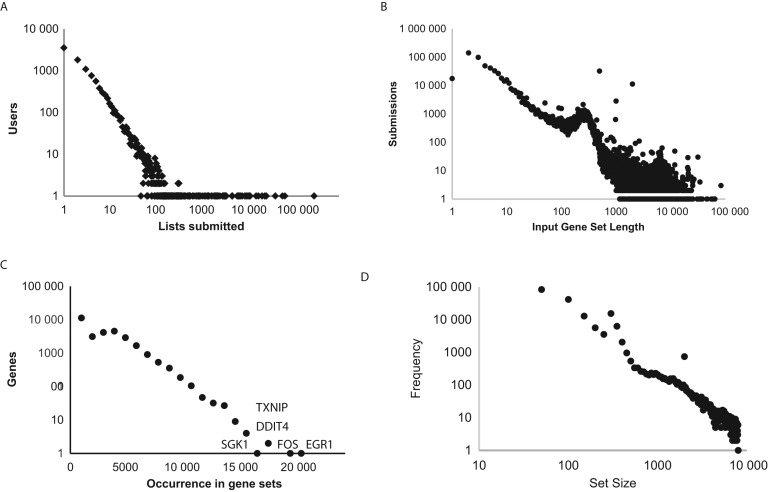
Enrichr currently contains 102 gene set libraries belonging to eight categories. In total, there are currently 180 184 annotated gene sets within Enrichr. So far, 1 050 236 gene sets have been uploaded for analysis with Enrichr. While most (∼65%) users submit only 1–3 lists to Enrichr, there are also many heavy users where the distribution of lists submitted per user fits a well-behaved power law (Figure 2A). The submitted lists' size also follows a power-law distribution, but contains a peak around ∼250 genes per list (Figure 2B). This peak is likely an artifact from submissions that arrive from the tool GEO2Enrichr, which has a default setting of posting the top 500 genes separated into up-regulated or down-regulated genes from signatures processed from GEO. Examining the occurrence of individual genes in a submitted gene sets, we observe a log-normal distribution (Figure 2C) with the most popular genes: EGR1, FOS, TXNIP, DDIT4 and SGK1. EGR1 and FOS are well-known immediate early genes (IEG), and their high presence likely confirms that these genes are most commonly found as differentially expressed. The appearance of TXNIP, DDIT4 and SGK1 as common genes is interesting since these genes have a lesser-known role to be most responsive. The identification of the common occurrence of genes in submitted lists and annotated gene sets can potentially be applied to correct for biases, and as a result improve knowledge extraction. More extensive analysis of gene occurrence and co-occurrence in submitted lists demonstrates that such collective knowledge can be used to discover gene functions and predict protein interactions (37). Finally, we plot the distribution of the lengths of the 180 184 annotated gene sets provided for search by Enrichr (Figure 2D). Overall, this distribution also fits a power law with few inflections that likely represent specific libraries with hard cut-offs for gene sets. It is still an open question what are the recommendations for optimal enrichment analysis when it comes to setting thresholds for gene set lengths. This is likely because the answer is context dependent, but more investigation can be done with appropriate benchmarks.
Figure 2.
Statistics of Enrichr. (A) Histogram of gene lists submitted per user. (B) Histogram of uploaded list lengths. (C) Histogram of appearance of genes in uploaded list. (D) Histogram of annotated gene set sizes within Enrichr.
COMPARISON TO OTHER SIMILAR TOOLS
Comparing libraries and resources in other tools
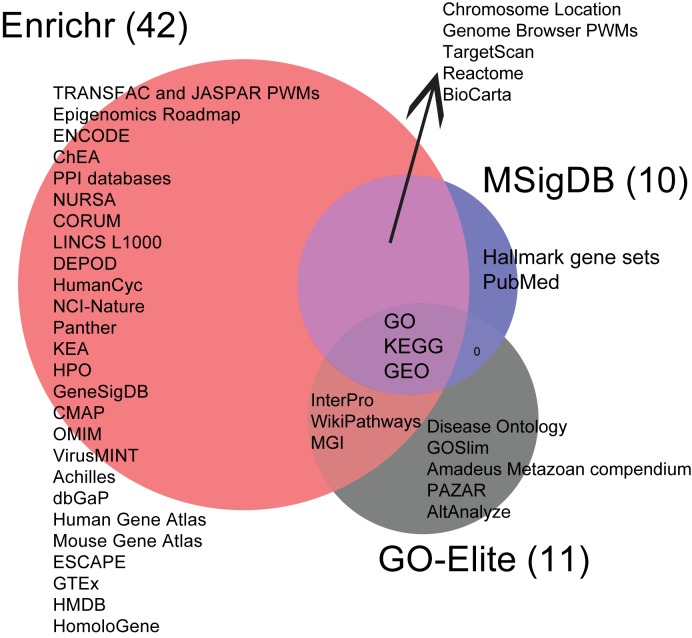
Next we aim to compare the resources and libraries offered for search by Enrichr with other similar tools. For this, we compared Enrichr with GO-Elite (38) and MSigDB (6), two leading resources that contain a comprehensive collection of gene set libraries. We summarized all the sources of gene set libraries for the three resources and plotted a Venn diagram to show the overlap among these resources (Figure 3A). Enrichr contains a large portion of MSigDB but is more comprehensive than both resources. MSigDB (6) contains eight collections of gene set libraries, two of which are also included in Enrichr (Computational and Oncogenic signatures). Many of the other collections of gene set libraries in MSigDB share the same sources with other gene set libraries currently present in Enrichr. These include, for example, the GO, pathway databases such as KEGG, Biocarta and Reactome, microRNAs/gene targets and gene sets created from position weight matrices. In addition, we note that MSigDB contains chemical and genetic perturbation gene sets manually curated from supporting materials of publications, whereas Enrichr contains differentially expressed genes after chemical and genetic perturbation curated from GEO. We compared the GEO data sets covered by Enrichr and MSigDB and found that there is some overlap while Enrichr has coverage of more data sets (Figure 3B).
Figure 3.
Comparing Enrichr resources with MSigDB and GO-Elite. (A) Venn diagram summarizing the various resources processed and served by Enrichr, MSigDB and GO-Elite. (B) Venn diagram to compare the number of processed gene sets of genetic and chemical perturbations curated from publications in Enrichr and MSigDB.
User interface pros and cons
There are many other gene set enrichment analysis tools that could be compared with Enrichr; for example, some leading tools are Fidea (39), DAVID (13), WebGestalt (12), g:Profiler (12) and GSEA (40). The advantages of Enrichr over some of these tools are its comprehensiveness, ease of use and interactive visualization of the results. Enrichr is lacking some of the flexibility available with those other tools. For example, Enrichr merges human, mouse and rat genes, which has advantages and disadvantages. Enrichr does not have an ID conversion tool, which is highly desired by many users. Enrichr also does not have the ability to upload a background list, and it does not have implementation of parametric tests such as Gene Set Enrichment Analysis (GSEA) (40), Parametric Analysis of Gene set Enrichment (PAGE) (9), and our own Principal Angle Enrichment Analysis (PAEA) (41). These features are planned.
FUTURE DIRECTIONS
As more genomics, transcriptomics and proteomics data accumulate, we plan to continue adding to Enrichr new gene set libraries. We also plan to continually improve the visualization of the enrichment results. It might be useful for users to view results across libraries, and to have a report of the most interesting enrichment results across all libraries. Enrichr currently supports only input from mammalian genes; in the future, we plan to add versions of Enrichr for yeast, worm and fly. The collection of terms for genes can be used to identify similarity between genes across resources, and this will improve the Find a Gene feature by suggesting similar genes. By examining the lists submitted to Enrichr, we noticed that approximately 10% of the submitted lists do not contain valid gene names. Users submit probe IDs, protein IDs, genes from other organisms, complete tables from spreadsheets with special characters, and other non-standard genes names. To accommodate these users, Enrichr needs to provide methods to convert these inputs into usable gene sets. The enrichment analysis concept can be expanded into new directions. For example, drug-set enrichment analysis (42) can be used to identify common functions for collections of drugs. In addition, enrichment analysis tools are increasingly becoming network-aware. The edge set enrichment analysis (43) method is one example of how network information can be incorporated into enrichment analysis. The collective analysis of the over one million gene sets submitted to Enrichr can be viewed as a potential resource for biological discovery. Each list can be classified into an attractor of similar lists and classified by methods of data acquisition but also biological regulatory layers, i.e. mRNA/proteins/SNPs, as well as biological roles. While we are committed to keeping user lists completely private, we also aim to explore the collective knowledge that is accumulating from all user submissions to Enrichr (37).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01GM098316, U54HL127624 and U54CA189201 to A.M.]. Funding for open access charge: Institutional funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Shahrour F., Díaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 3.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 4.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO:: TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung J.-H., Yang T.-H., Hu Z., Weng Z., DeLisi C. Gene set enrichment analysis: performance evaluation and usage guidelines. Brief. Bioinform. 2011;13:281–291. doi: 10.1093/bib/bbr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S.-Y., Volsky D.J. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y.A., Müller R., Meese E., Lenhof H.-P. GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 14.Chen E., Tan C., Kou Y., Duan Q., Wang Z., Meirelles G., Clark N., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero P. The Handbook of Metabolomics. NY: Humana Press; 2012. pp. 419–438. [Google Scholar]
- 16.Demir E., Cary M.P., Paley S., Fukuda K., Lemer C., Vastrik I., Wu G., D'Eustachio P., Schaefer C., Luciano J. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malovannaya A., Lanz R.B., Jung S.Y., Bulynko Y., Le N.T., Chan D.W., Ding C., Shi Y., Yucer N., Krenciute G. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan G., Li X., Kohn M. The human DEPhOsphorylation database DEPOD: a 2015 update. Nucleic Acids Res. 2015;43:D531–D535. doi: 10.1093/nar/gku1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J., et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R., et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43:D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T., et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44:D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunkin S.M., Ng L., Lau C., Dolbeare T., Gilbert T.L., Thompson C.L., Hawrylycz M., Dang C. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H., et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 27.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S., et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F., Pantel S.E., et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gundersen G.W., Jones M.R., Rouillard A.D., Kou Y., Monteiro C.D., Feldmann A.S., Hu K.S., Ma'ayan A. GEO2Enrichr: browser extension and server app to extract gene sets from GEO and analyze them for biological functions. Bioinformatics. 2015;31:3060–3062. doi: 10.1093/bioinformatics/btv297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark N., Hu K., Feldmann A., Kou Y., Chen E., Duan Q., Ma'ayan A. The characteristic direction: a geometrical approach to identify differentially expressed genes. BMC Bioinformatics. 2014;15:79. doi: 10.1186/1471-2105-15-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merkel D. Docker: lightweight linux containers for consistent development and deployment. Linux J. 2014;239:2. [Google Scholar]
- 35.Hindman B., Konwinski A., Zaharia M., Ghodsi A., Joseph A.D., Katz R.H., Shenker S., Stoica I. Mesos: A Platform for Fine-Grained Resource Sharing in the Data Center. NSDI. 2011;11:22–22. [Google Scholar]
- 36.Saha P., Govindaraju M., Marru S., Pierce M. Integrating Apache Airavata with Docker, Marathon, and Mesos. Concurrency and Computation: Practice and Experience. 2015;28:1952–1959. [Google Scholar]
- 37.Ma'ayan A., Clark N.R. Large collection of diverse gene set search queries recapitulate known protein-protein interactions and gene-gene functional associations. 2016. 1601.01653 arXiv:
- 38.Zambon A.C., Gaj S., Ho I., Hanspers K., Vranizan K., Evelo C.T., Conklin B.R., Pico A.R., Salomonis N. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Andrea D., Grassi L., Mazzapioda M., Tramontano A. FIDEA: a server for the functional interpretation of differential expression analysis. Nucleic Acids Res. 2013;41:W84–W88. doi: 10.1093/nar/gkt516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark N.R., Szymkiewicz M., Wang Z., Monteiro C.D., Jones M.R., Ma'ayan A. Principle Angle Enrichment Analysis (PAEA): Dimensionally reduced multivariate gene set enrichment analysis tool. 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE. 2015:256–262. doi: 10.1109/BIBM.2015.7359689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napolitano F., Sirci F., Carrella D., di Bernardo D. Drug-set enrichment analysis: a novel tool to investigate drug mode of action. Bioinformatics. 2016;32:235–241. doi: 10.1093/bioinformatics/btv536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han J., Shi X., Zhang Y., Xu Y., Jiang Y., Zhang C., Feng L., Yang H., Shang D., Sun Z. ESEA: discovering the dysregulated pathways based on edge set enrichment analysis. Sci. Rep. 2015;5:13044. doi: 10.1038/srep13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.