Abstract
The web service DNATCO (dnatco.org) classifies local conformations of DNA molecules beyond their traditional sorting to A, B and Z DNA forms. DNATCO provides an interface to robust algorithms assigning conformation classes called ntC to dinucleotides extracted from DNA-containing structures uploaded in PDB format version 3.1 or above. The assigned dinucleotide ntC classes are further grouped into DNA structural alphabet ntA, to the best of our knowledge the first DNA structural alphabet. The results are presented at two levels: in the form of user friendly visualization and analysis of the assignment, and in the form of a downloadable, more detailed table for further analysis offline. The website is free and open to all users and there is no login requirement.
INTRODUCTION
The complexity and variability of DNA structures can no more be understood within the traditional ‘A–B–Z structural code’. DNA molecules are able to form sharp kinks in complexes with some transcription factors, spiral around the histone core proteins, accommodate sharp kinks in Holliday junctions, extend their backbone in intercalation complexes with aromatic drugs, or form stable quadruplex or hairpin structures. Surprisingly though, tools allowing to go beyond a simplified picture of DNA structure and reducing its complexity to a few qualitative descriptors are scarce. NDB (1) provides a comprehensive overview of the available structures and offers their limited structure classification, software tool 3DNA (2), concentrating on description of the geometry of base pairing.
Several years ago, some of us attempted to classify the geometry of the DNA backbone at the level of dinucleotides (3) and later on, we developed a robust automated pipeline to perform such an analysis (4). In this contribution, we present an improvement of this methodology implemented into a web-based tool that offers an objective analysis of the DNA local conformation based on a rigorous geometry-based algorithm. Conformations of dinucleotide steps are assigned to one of 57 conformers called ntC. To help interpret the results and the overall structural features of the analyzed structure, the assignment is also interpreted in terms of conceptually simpler structural alphabet ntA consisting of just 12 members, which were created by grouping structurally related ntC. The relatively high number of ntC classes, 57, resulted from the analysis of the available DNA structures and reflects the complexity of the DNA conformational space. In contrast, the particular way of grouping of ntC into letters of the ntA structural alphabet and the number of ntA letters are subjective.
MATERIALS AND METHODS
Conformations of dinucleotide steps (nomenclature defined in Figure 1) are analyzed by comparing their torsion angles to the torsions of 4439 dinucleotides in the so called ‘golden set’. The golden set is an ensemble of dinucleotides manually curated and classified into one of the 57 conformational classes called ntC. The geometry of the ntC classes is summarized in Supplementary Table S1 and is also available at the dnatco.org website. The assignment begins by uploading a PDB-formatted structure to the website and is performed in the torsional space by comparing values of nine DNA backbone torsion angles of the analyzed step and all 4439 steps in the golden set by the modified k-nearest neighbors algorithm according to the protocol by Čech et al. (4) with modifications. The currently used ntC definitions originate from the set reported by Svozil et al. (3), who described them in detail along with the methods how they were identified.
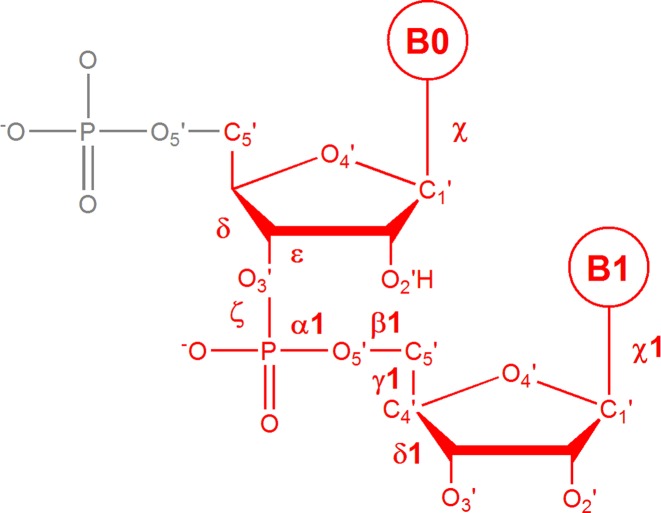
Figure 1.
The dinucleotide step as defined by ntC conformers is drawn in red. The step is described by seven backbone torsions from δ of the first nucleotide to δ1 of the second one, plus two torsion values around the glycosidic bonds of the first and second bases (χ and χ1).
THE DNATCO SERVER
Hardware and software
The previous version of the DNATCO server (available at dnatco.org/v1) was migrated from a home based hardware and is now hosted as a Linux based virtual machine in the environment provided by the ELIXIR CZ infrastructure. This ensures 24/7 availability and professional maintenance as well as easy scaling of the resources if necessary. The presented second version of the server ran about a year internally and had been tested over a year as a publicly available service accessible at the dnatco.org address.
The software part employs Apache web server and PHP5 for the server side scripting. The internal processing of uploaded PDB-formatted structures is performed using the VMD program (5) extracting only nucleic acid atoms for further analysis. The torsion angle measurement and the assignment of ntC conformers itself is performed by in house programs written in the Python programming language. The interactive display of analyzed 3D structures relies on JSmol (6), a JavaScript based molecular viewer running in a browser. The JavaScript allows straightforward transfer of the DNATCO web service to various platforms and devices including mobile devices without a need to install additional applets. The JSmol performance is known to depend on the browser version and the computer operation system used. The complete web service was successfully tested in the major web browser programs under Linux, OS X and Windows with Firefox having currently the best performance in the JSmol part.
The home page
The home page (snapshot in Supplementary Figure S1 and Table S1) briefly introduces the purpose of the web, defines the dinucleotide step, lists the geometries of the ntC conformers with their brief characterization and provides the tool to upload the structure to be analyzed. The top of the home page contains links to the tutorial section that describes the submission process step by step, explains the results, and also contains the link to a test run using the Dickerson-Drew dodecamer of PDB ID 1bna (7). The PDB formatted structure file can be uploaded either from user's disk or by typing a PDB four-letter code and pressing the respective SUBMIT button; the former way is useful for structures generated or modified by the user, the latter for analysis of the released PDB structures.
Names and brief annotations of the 57 ntC DNA conformers are tabulated on the home page together with values of the torsion angles defining their geometry. These are seven torsions defining the backbone conformation of the step from delta of the first nucleotide to delta of the second one, plus two torsion values around the glycosidic bonds of the first and second bases (Figure 1). ntC are identified by four-letter symbols. The first letter aims to characterize the main feature(s) of the first nucleotide, the second letter of the second one. A, B and Z letters imply stacked bases with the first/second nucleotide in the conformation bearing features typical of the A, B or Z DNA forms such as sugar pucker, torsion around the glycosidic bond and combination of the other torsions such as zeta and alpha as they have been described in various treaties, e.g. by Neidle (8). The first two letters ‘NS’ indicate that the bases are Not Stacked in the step. The third and fourth positions of the code are usually formed by numbers, which just guarantee the uniqueness of the ntC symbol; ‘S’ at either of these positions means that the first or second base is in the syn orientation. The nomenclature of ntC classes can best be understood from Supplemental Table S1 or a downloadable table at the dnatco.org home page, where the main structural features of each ntC are briefly annotated. A specific ‘conformational class’, NANT, was reserved for conformationally extreme steps that are not assigned to any of the above ntC classes; NANT formally represents the 58th conformer. Both torsional definitions of ntC and Cartesian coordinates of dinucleotides representing their structures can also be downloaded from dnatco.org.
Input and output
The input is a crystal, NMR or computer model structure containing DNA in the standard PDB format. DNA steps are identified based on atom names as defined by the PDB format, version 3.1 or above (sugar atoms as O4' not O4*, standard nucleotides DA, DG, DC, DT). If the PDB file contains multiple structures (NMR models or MD simulation snapshots) as MODELs, the currently available version of dnatco.org analyzes only the last MODEL. Conformer classes are also assigned for modified residues if they contain standard names for atoms defining the step torsions between δ of the first deoxyribose to δ+1 of the second one, and glycosidic torsions χ and χ+1; on the other hand, steps with non-standard or missing atoms that define these torsions cannot currently be considered in the assignment process.
The output of the assignment process is a comma separated summary of the assigned ntC and ntA classes. The standard CSV file contains the step ID, its assigned ntC and ntA, its nine torsion angles, and angular distances from the ntC averages. During the testing phase, we have analyzed over 1800 DNA structures, mostly experimental crystal and NMR structures from PDB.
When the structure is identified by its four letter PDB code, we compare the version stored on our server with the most recent version at the PDB website. If these two versions are identical, we use the pre-calculated results to speed up the analysis; otherwise, the full assignment process is performed. In either case, the results are displayed within a few minutes after the upload at the latest. An example of the result page can be obtained from the Tutorial section or simply by running the ‘test run’ on the website. The structures uploaded by users and their assignment results are protected by adding a hash value to the file names. The ntC assignment of structures deposited in the PDB database is accessible via PDB four-letter codes.
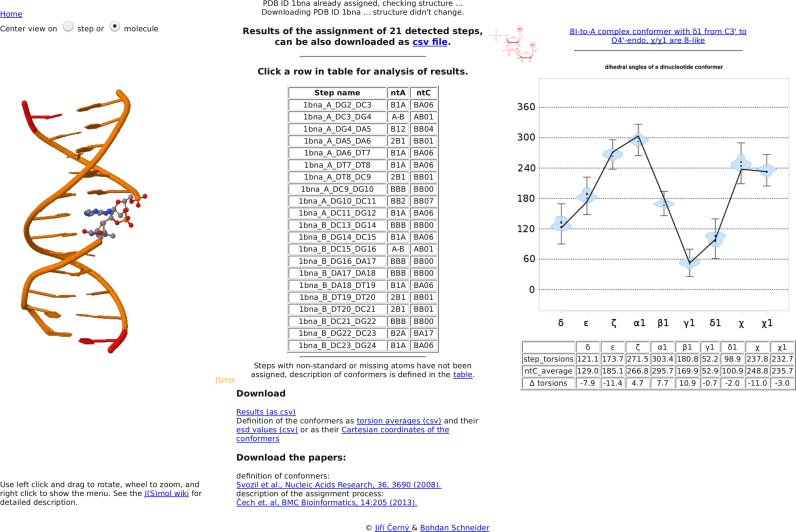
Results page
The results page (snapshot in Figure 2) is divided into three columns. The central column contains a table summarizing the results of the assignment of ntC classes. Each row of the table represents one complete step. The step name is displayed as PDBid_chain_base1_base2 and is followed by the corresponding ntA and ntC codes. The table is interactive; mouse over a row shows the detailed description of the assigned step with the ntC and ntA classes as well as values of the backbone torsion angles. Further analysis of a step can be obtained by clicking inside the table. The left panel contains an interactive 3D view of the analyzed DNA structure in the JSmol applet. The DNA structure is shown as a cartoon with the selected dinucleotide step highlighted in a ball and stick representation. At the same time, the right part of the page summarizes the results of the dinucleotide step assignment in graphical representation. The black line connects the torsion values of the selected step. For unassigned steps (ntC NANT), the chart contains only the line, while more information is shown for the assigned conformers: a violin plot summarizes the distribution of torsions for the assigned ntC in the golden set. Inside each violin plot, a thick black bar indicates the 1st and 3rd quartile of the golden set data with a white spot showing the mean value. The ‘error bars’ outside each violin plot depict the angular range still fulfilling the assignment algorithm criteria. The circularity of the torsion angles is taken into account, and for conformers with mean values approaching 0 or 360 degrees, the ‘error bar’ appears near 360 or 0 degrees, respectively. This diagram is a simple visual measure of the quality of the assignment, the table below the diagram is its numerical representation with the torsion values of the step, assigned ntC, and their differences. Further, a mouse over the thumbnail in the upper left corner of the chart will zoom in the figure with definitions of the backbone torsion angles. The results of the assignment in CVS format, representative Cartesian coordinates of the conformers, and the table of conformers defining the ntC and ntA classes can be downloaded using links at the bottom of the results page.
Figure 2.
Snapshot of the results page using the Dickerson–Drew dodecamer of PDB code 1bna as an example. The central column contains a table summarizing the assignment of ntC and ntA classes and links to downloadable files. Each row of the table represents one complete step listing the step name as PDBid_chain_base1_base2, ntA and ntC codes. The left panel contains an interactive 3D view of the analyzed DNA structure in the JSmol applet; the analyzed step is highlighted. The right part of the page summarizes the results of the dinucleotide step assignment in graphical representation and provides a table of torsion values for the analyzed step, the assigned ntC and their differences.
CONCLUSIONS
The website dnatco.org provides fine-grained classification of the DNA local structure based on the geometry of its backbone. Analysis of the nine dinucleotide torsions results in assignment of one of the 58 conformers called ntC, one of which is set aside for dinucleotides with exotic or as yet uncharacterized conformations. Based on the results of the ntC analysis, the web also provides a coarse-grained characterization of the structure by grouping ntC into a unique DNA structural alphabet ntA. The web service represents a valuable tool for DNA structure analysis and validation during the refinement of crystal and NMR structures, of structures deposited to PDB, as well as for analysis of results of molecular modeling and simulations by molecular dynamics.
The website is one of the services supported by the Czech ELIXIR node (elixir-czech.cz) with full financial coverage until the year 2019 guaranteed by the Czech national funding. Further sustainability and maintenance of the web service and the website for additional three years (until the end of year 2022) is likely to be obtained within the framework of European Infrastructural projects ESFRI ELIXIR from Czech national or pan-European funds. The Institute of Biotechnology CAS, employer of the authors of the website, provides additional long-term support. Further development of the service, namely its extension to RNA and more flexible treatment of modified nucleotides can therefore be realistically envisioned.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech National Infrastructure for Biological Data (ELIXIR CZ) [LM2015047]; ERDF [BIOCEV CZ.1.05/1.1.00/02.0109]; Institutional Research Project of the Institute of Biotechnology [RVO 86652036]. Funding for openaccess charge: Czech National Infrastructure for Biological Data (ELIXIR CZ) [LM2015047]; Institutional Research Project of the Institute of Biotechnology [RVO 86652036].
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman H.M., Westbrook J., Feng Z., Iype L., Schneider B., Zardecki C. The nucleic acid database. Acta Crystallogr. D Biol. Crystallogr. 2002;58:889–898. doi: 10.1107/s0907444902003487. [DOI] [PubMed] [Google Scholar]
- 2.Lu X.J., Olson W.K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svozil D., Kalina J., Omelka M., Schneider B. DNA conformations and their sequence preferences. Nucleic Acids Res. 2008;36:3690–3706. doi: 10.1093/nar/gkn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cech P., Kukal J., Cerny J., Schneider B., Svozil D. Automatic workflow for the classification of local DNA conformations. BMC Bioinformatics. 2013;14:205. doi: 10.1186/1471-2105-14-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 6.Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L. JSmol and the next-generation web-based representation of 3D molecular structure as applied to proteopedia. Isr. J. Chem. 2013;53:207–216. [Google Scholar]
- 7.Drew H.R., Wing R.M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R.E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neidle S. Principles of Nucleic Acid Structure. Cambridge: Academic Press, Elsevier; 2008. pp. 1–289. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.